Efficient antibacterial activity via protein degradation of a 3D layered double hydroxide–reduced graphene oxide nanohybrid†
EswaraVara Prasadarao Komaralaa,
Sejal Doshia,
Aslam Mohammedb and
Dhirendra Bahadur*a
aDepartment of Metallurgical Engineering and Materials Science, Indian Institute of Technology Bombay, Mumbai 400076, India. E-mail: dhirenb@iitb.ac.in; Fax: +91 22 2572 3480; Tel: +91 22 2576 7632
bDepartment of Physics, Indian Institute of Technology Bombay, Mumbai 400076, India
First published on 15th April 2016
Abstract
Graphene and its derivatives have typical physicochemical properties that show strong cytotoxic effects on bacterial cell lines. In this study, we have investigated the antibacterial activity of a nanohybrid of three-dimensional Mg–Al layered double hydroxide–reduced graphene oxide (LDH–rGO) on the Gram negative coccobacilli bacterium Escherichia coli (E. coli). A biocompatibility test by a sulphorhodamine-B (SRB) assay inferred that LDH–rGO is biocompatible up to a concentration of 2 mg mL−1. The concentration and time-dependent antibacterial studies confirmed that 125 μg mL−1 of LDH–rGO is sufficient to inhibit 95.4 ± 1.1% E. coli cells in 60 min. A systematic study shows that the nanohybrid is capable of developing oxidative stress by producing reactive oxygen species (ROS), while LDH and rGO does not. However, both rGO and the nanohybrid are capable of developing oxidative stress by oxidizing glutathione. Further, the effect of oxidative stress on one of the biological entities (protein) of E. coli was also studied. We thereby propose the antibacterial mechanism of the nanohybrid on the basis of size, morphology, glutathione loss, and protein degradation.
1. Introduction
Graphene, a two-dimensional carbon nanomaterial has attracted worldwide attention due to its exceptional properties, such as larger specific surface area, high thermal and electrical conductivity, and mechanical strength.1,2 It has generated excessive interest in many fields of research like nanoelectronics,3 energy storage,4 composite materials,5 and biomedicine.6 Graphene oxide (GO) produced by a chemical approach can develop lots of oxygen functional groups.7,8 These functional groups help to form a stable colloidal suspension of GO8 and also provide a variety of bonding interactions to synthesize nanohybrids with other materials, especially with inorganic nanoparticles through different approaches.9,10 In recent times, several studies on graphene and its derivatives discuss their strong toxicity to a variety of bacterial cell lines.11–15 Liu et al. have compared the antibacterial activity of different graphene derivatives towards E. coli cells through membrane and oxidative stress.16 They have found that GO dispersion showed highest antibacterial activity of 89.7% with a concentration of 40 μg mL−1. Pandey et al. have reported that gentamicin drug-loaded methanol-derived graphene has shown 82.2% antibacterial activity on E. coli at a concentration of 40 μg mL−1 by controlled release of the drug.17On the other hand, among all the inorganic nanomaterials, layered double hydroxides (LDHs) have shown a strong potential in various applications such as catalysis,18 magnetic precursors,19 energy storage,20 and biomedical field21 due to its high anion exchange capability and biocompatibility. The general formula of LDHs is [M1−x2+Mx3+(OH)2]·[Ax/nn−·mH2O], where M2+ and M3+ are divalent and trivalent metal cations respectively and An− are the intercalated anions. The exchangeable intercalated anions of LDHs by a variety of biochemical anions and diverse drug molecules facilitate their extensive usage in biomedical applications.22,23 The positive charge on LDHs24 and the negative charge on graphene derivatives25 aid the ease of formation of the nanohybrids by electrostatic interactions between these two materials. These LDH–graphene based nanohybrid materials have been used as electrode materials for super capacitor,26,27 heavy metal removal,28 and catalysis29 applications. However, these nanohybrids were scarcely studied for antibacterial activity. Wang et al. reported the antibacterial activity of graphene oxide–Mg–Al LDH intercalated with benzyl penicillin hybrid films.30 About 95% of M. lysodeikticus were inhibited with a GO/LDH mass ratio of 4.
Although the graphene components have shown promise in a vast spectrum of applications, their cytotoxicity (chemically derived graphene components) and tendency of agglomeration limits their use in the field of nanobiomedicine. These problems can be addressed by introducing a biocompatible component and the subsequent formation of the nanohybrids. We have used LDH, which is a known biocompatible material and this in conjugation with rGO to form a nanohybrid. These nanohybrids are more biocompatible and gives an improved colloidal dispersion.31 Moreover, LDH surface is electrostatically positive which aids in a better adhesion of nanohybrids to the negatively charged bacterial cell wall. This electrostatic charge-based adhesion enhances the activity of the nanohybrid over its individual components. In the present study, we have synthesized LDH–rGO nanohybrid by one pot hydrothermal crystallization method. We studied the antibacterial activity of the LDH–rGO toward Gram negative coccobacilli Escherichia coli (E. coli) bacteria cell lines. Further, we have also studied the time and concentration-dependent antibacterial activity of LDH–rGO. The interaction of LDH and the nanohybrid towards the bacterial cell has been studied using SEM. The oxidative stress caused by the nanohybrid was measured by reactive oxygen species (ROS) release and glutathione (GSH) loss studies. To the best of our knowledge, this is the first report on the study of antibacterial activity of LDH–rGO nanohybrid without using any drugs and also where we have explored the effect of nanohybrid on protein degradation of the bacterial cell line. On the basis of these results, we have proposed a possible mechanism for the antibacterial activity of LDH–rGO nanohybrid.
2. Materials and methods
2.1 Synthesis of Mg–Al LDH nanoflowers
Mg–Al LDH nanoflowers, were synthesized using a method described in the literature, with some modifications.32 In a typical synthesis, 2 mmol of Mg(NO3)2·6H2O, 1 mmol of Al(NO3)3·9H2O and 15 mmol of urea were dissolved in a solution of 45 mL of ethylene glycol and 5 mL of MilliQ water, and stirred vigorously for 30 min at room temperature. The resultant solution was transferred to a stainless steel autoclave and aged at 160 °C for 6 h. The precipitate was collected by centrifugation and washed with MilliQ water and ethanol copiously. Mg–Al LDH nanoflowers powder (∼105 mg, white in colour) was obtained after the precipitate was dried in oven at 100 °C for 24 h.2.2 Synthesis of graphene oxide
GO was prepared by a modified Hummer's method.33 In brief, 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of H2SO4
1 ratio of H2SO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H3PO4 (360
H3PO4 (360![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 mL) acid mixture was added to 3 g of graphite and 18 g of KMnO4. The reaction was performed in an ice bath to control the temperature of the above exothermic reaction. Then, the reaction was cooled to room temperature, and then was heated at 55 °C for 12 h under continuous stirring. Later, the reaction was put in an ice bath with 30% hydrogen peroxide and cooled to room temperature. It was then filtered using nylon filter paper with a pore size of 0.2 μm, using a vacuum filtration assembly. The filtered solid sample was washed multiple times with water, HCl, and ethanol. The material remained after washing was coagulated with 35 mL of diethyl ether. After that, the suspension was filtered again using vacuum filtration assembly. The sample obtained after the filtration was vacuum-dried at 60 °C to get graphite oxide powder. Furthermore, the aqueous suspension of graphite oxide (0.1 mg mL−1) was sonicated for 1 h to obtain GO. The aqueous solution was dried in oven to get GO powder.
40 mL) acid mixture was added to 3 g of graphite and 18 g of KMnO4. The reaction was performed in an ice bath to control the temperature of the above exothermic reaction. Then, the reaction was cooled to room temperature, and then was heated at 55 °C for 12 h under continuous stirring. Later, the reaction was put in an ice bath with 30% hydrogen peroxide and cooled to room temperature. It was then filtered using nylon filter paper with a pore size of 0.2 μm, using a vacuum filtration assembly. The filtered solid sample was washed multiple times with water, HCl, and ethanol. The material remained after washing was coagulated with 35 mL of diethyl ether. After that, the suspension was filtered again using vacuum filtration assembly. The sample obtained after the filtration was vacuum-dried at 60 °C to get graphite oxide powder. Furthermore, the aqueous suspension of graphite oxide (0.1 mg mL−1) was sonicated for 1 h to obtain GO. The aqueous solution was dried in oven to get GO powder.
2.3 Synthesis of LDH–rGO nanohybrid
A LDH–rGO nanohybrid was synthesized by adding 5 mL of synthesized GO aqueous solution (5 mg mL−1) with the precursors of LDH (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 mole ratio of Mg(NO3)2·6H2O, Al(NO3)3·9H2O and urea) in a mixture of 45 mL ethylene glycol and 5 mL MilliQ water, followed by continuous stirring at room temperature for 30 min. The obtained mixture was moved to 100 mL stainless steel autoclave and heated at 160 °C for 6 h. During the hydrothermal treatment, the GO nanosheets were reduced along with the formation of LDH. The sample was washed three times with MilliQ water and ethanol, respectively. The LDH–rGO nanohybrid (∼126 mg, ash gray in colour) was collected and dried in oven at 100 °C for 24 h. Compared to the weight of LDH (∼105 mg), the nanohybrid shows ∼20% more weight which is contributing from the graphene component. All precautions were taken to avoid the loss of samples after each synthesis process.
15 mole ratio of Mg(NO3)2·6H2O, Al(NO3)3·9H2O and urea) in a mixture of 45 mL ethylene glycol and 5 mL MilliQ water, followed by continuous stirring at room temperature for 30 min. The obtained mixture was moved to 100 mL stainless steel autoclave and heated at 160 °C for 6 h. During the hydrothermal treatment, the GO nanosheets were reduced along with the formation of LDH. The sample was washed three times with MilliQ water and ethanol, respectively. The LDH–rGO nanohybrid (∼126 mg, ash gray in colour) was collected and dried in oven at 100 °C for 24 h. Compared to the weight of LDH (∼105 mg), the nanohybrid shows ∼20% more weight which is contributing from the graphene component. All precautions were taken to avoid the loss of samples after each synthesis process.
2.4 Biocompatibility test
Though it is well known that LDH is biocompatible,34 however, the change in biocompatibility that results from the addition of rGO needs to be checked. Sulforhodamine-B (SRB) assay was performed to check biocompatibility of LDH–rGO on L929 cells.35 Briefly, 1 × 104 L929 cells were seeded per well in 96-well plates and incubated for 24 h in a 5% CO2 environment at 37 °C. Later, different concentrations of LDH, rGO and LDH–rGO (2.0–0.03125 mg mL−1) were dispersed in Dulbecco's Modification of Eagle's Medium (DMEM) growth medium and again incubated for 24 h more at 37 °C and a 5% CO2 environment. Then the cells were gently washed with phosphate saline buffer (PBS; pH 7.3) and further treated with a 10% trichloroacetic acid solution by incubating at 4 °C for 1 h followed by staining with 0.4% SRB dissolved in 1% acetic acid and incubate in the dark. Then the optical density was measured at 560 nm using a microplate reader.2.5 Antibacterial activity
2.6 Cell morphology observation
SEM was used for morphological observations. Before preparing the sample for SEM imaging, the treated cells were centrifuged twice with ethanol at 2000 rpm. This process was repeated twice with sterile MilliQ water. The obtained cells were mounted on a cover-slip and dried at room temperature. The dried cells were sputter-coated to make them conducting for SEM imaging.2.7 Oxidative stress studies
2.8 Total protein assay (by SDS-PAGE method)
The total protein was extracted by incubating 1 mL of E. coli cells that were grown overnight, with 125 μg mL−1 of samples for 2 h. The control was E. coli cells without any treatment. Further, 5 mL of a phosphate buffer was added, followed by centrifugation at 8000 rpm for 20 min. Now, the supernatant was collected and washed four times with PBS, and the final volume was raised to 50 mL. Then, 1 mL of 20% trichloroacetic acid was added to 1 mL of the obtained extract. After 30 min, the extract was centrifuged again and then it was washed with acetone. The pellet obtained was dissolved in 5 mL of 0.1 N NaOH and used for sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). After gel preparation, the protein sample to be tested was denatured and added in 2 mL gel sample buffer and the final volume was made to 10 mL with MilliQ water. 25 μL of this sample was added to electrophoretic gel wells. HimediaHTP001 XPert™ protein marker that has a range of 10–250 kDa molecular weight was used as a standard protein marker. The current was maintained at 10–15 mA for 10–15 min, and then increased to 30 mA. The gel was collected and stained with Coomassie blue dye until bands were observed, after which the excess stain was detained.The biochemical assay for total cellular protein degradation detection was carried out by the Folin–Lowry method, in which E. coli cells were mixed with a freshly prepared alkaline copper sulfate reagent, followed by the addition of Folin's reagent and then the absorbance was measured at 660 nm.
3. Results and discussion
3.1 Characterization of LDH–rGO nanohybrid
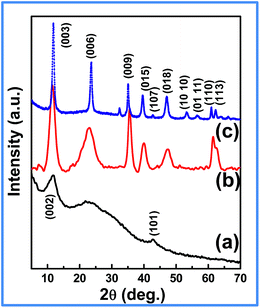
Fig. 1 shows the XRD patterns of GO, LDH and LDH–rGO nanohybrid. A strong reflection peak centered at 2θ = 10.8° (002) (Fig. 1a) with the lattice parameter of 0.78 nm (which is higher than graphite, 0.34 nm) confirms the oxidation of graphite to GO.36 The diffraction pattern of LDH (Fig. 1b) exhibits the characteristic reflections of (003), (006), (009), (015), (018), (110) and (113) of hydrotalcite-like compound (ICDD: 01-089-5434). The d spacing of (003) diffraction peak of LDH is about 0.76 nm, which indicates the intercalation of CO32− anions into the brucite layers of LDH.32 It has been noted that, the nanohybrid does not contain the characteristic peak of GO (Fig. 1c), which could be ascribed to the formation of rGO in the nanohybrid and is further confirmed by XPS and Raman analyses. However, the nanohybrid consists of all the diffraction peaks of LDH, suggesting the successful formation of LDH phase in the nanohybrid.29 Further, the diffraction pattern of the nanohybrid exhibits sharp and intense peaks as compared to that of LDH phase. After the synthesis of nanohybrid, the crystallite size (calculated by Scherrer formula: L = 0.94 × λ/β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ. For LDH, L = 5.2 nm and for LDH–rGO, L = 8.5 nm) of the LDH component has significantly increased as is obvious from FWHM. Hence the identification of rGO peak in the XRD spectrum of LDH–rGO is difficult as this phase gives a weak and broad peak, which in the present case is overshadowed by strong LDH peaks. From literature also, it has been observed that the XRD patterns of LDH–rGO composites does not show any reflections of rGO.9,20
θ. For LDH, L = 5.2 nm and for LDH–rGO, L = 8.5 nm) of the LDH component has significantly increased as is obvious from FWHM. Hence the identification of rGO peak in the XRD spectrum of LDH–rGO is difficult as this phase gives a weak and broad peak, which in the present case is overshadowed by strong LDH peaks. From literature also, it has been observed that the XRD patterns of LDH–rGO composites does not show any reflections of rGO.9,20
The morphology of GO, LDH and LDH–rGO nanohybrid is investigated by TEM as shown in Fig. 2. The TEM image of GO (Fig. 2a) shows transparent ultrathin sheets, which indicates an effective exfoliation of graphite oxide to graphene oxide. Pure LDH (Fig. 2b) shows three dimensional flower-type morphology consists of curved and agglomerated nanoflakes with an average size of 0.5–1 μm. In case of LDH–rGO (Fig. 2c), the flower-type structure of LDH essentially disappeared, and the size changed significantly. The selected area electron diffraction (SAED) patterns (Fig. 2e) marked as 1 and 2 give the individual features of rGO and LDH phase, whereas region 3 shows the mixed pattern of both LDH and rGO. This indicates that, during the in situ crystallization process both LDH and rGO phases interact as shown in region 3, Fig. 2c. This interaction avoids the restacking of rGO nanosheets in the nanohybrid. Although the contrast difference between LDH nanoflakes and rGO nanosheets is little, the rGO sheets can be found in between the LDH nanoflakes which are evident with their definite feature as shown in their corresponding SAED patterns. The EDS spectrum of the nanohybrid shown in Fig. 2d consists of Mg, Al, C and O elements, indicates that both LDH and rGO phases may co-exist. The atomic percentage of Mg and Al is 12.61% and 5.99% respectively. The Mg/Al atomic ratio is nearly 2, which is in agreement with their respective precursor's mole ratio. The zeta potential values of LDH, GO and LDH–rGO were found to be 24.3, −34.1 and 20.33 mV respectively. The electrostatic interaction between LDH and GO might play a role in the formation of nanohybrid. The positive nature of both LDH and nanohybrid helps in better interaction with the bacterial cell lines.
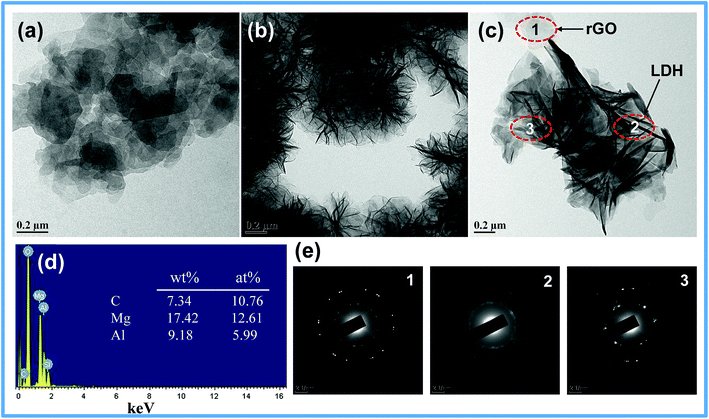 | ||
| Fig. 2 TEM images of (a) GO, (b) LDH. (c) LDH–rGO, (d) EDS spectrum and (e) selected area electron diffraction at different regions of LDH–rGO nanohybrid. | ||
The chemical and structural changes of GO and LDH–rGO nanohybrid were analyzed by XPS and Raman spectroscopy. Fig. 3 and 4 shows the survey, C1s XPS spectra, and Raman spectra of GO and LDH–rGO nanohybrid. Compared to that of GO, the XPS spectrum of LDH–rGO (Fig. 3a) not only exhibits a relatively low C1s and O1s peaks, but also exhibits additional peaks at 24, 48.8, 73.6, 88 and 118.4 eV corresponding to O2s, Mg2p, Al2p, Mg2s and Al2s respectively (Fig. 3b), which confirms the presence of Mg–Al LDH in the nanohybrid. The deconvoluted C1s XPS spectrum of GO (Fig. 3c) shows three different carbon bonds: non-oxygenated C(C–C) ring at 284.5 eV, oxygenated C corresponds to C–O and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O at 286.6 and 285.5 eV respectively indicating a significant degree of oxidation.9,37 The core-level C1s components show about 36% and 67% of non-oxygenated carbon in GO and nanohybrid respectively. On the other hand, the intensity of carbonyl carbon (C–O) peak decreases sharply in the nanohybrid (Fig. 3d). The XPS results thus suggest that the graphene oxide has been reduced during the formation of LDH–rGO nanohybrid. In addition, the Mg2p and Al2p XPS spectra of LDH–rGO shows a meagre shift in the binding energies as compared to that of LDH (Fig. S1†). The shift in the peaks (Mg–O–C and Al–C) indicates the interaction of Mg and Al atoms with the graphene oxide sheets during the synthesis of LDH–rGO nanohybrid. This interaction further leads to growth of LDH phase on the GO sheets and avoids the agglomeration (in case of LDH, the flower structure is due to the agglomeration of LDH nanoflakes32) and results in increase in the size of the nanohybrid (Fig. 2c).
O at 286.6 and 285.5 eV respectively indicating a significant degree of oxidation.9,37 The core-level C1s components show about 36% and 67% of non-oxygenated carbon in GO and nanohybrid respectively. On the other hand, the intensity of carbonyl carbon (C–O) peak decreases sharply in the nanohybrid (Fig. 3d). The XPS results thus suggest that the graphene oxide has been reduced during the formation of LDH–rGO nanohybrid. In addition, the Mg2p and Al2p XPS spectra of LDH–rGO shows a meagre shift in the binding energies as compared to that of LDH (Fig. S1†). The shift in the peaks (Mg–O–C and Al–C) indicates the interaction of Mg and Al atoms with the graphene oxide sheets during the synthesis of LDH–rGO nanohybrid. This interaction further leads to growth of LDH phase on the GO sheets and avoids the agglomeration (in case of LDH, the flower structure is due to the agglomeration of LDH nanoflakes32) and results in increase in the size of the nanohybrid (Fig. 2c).
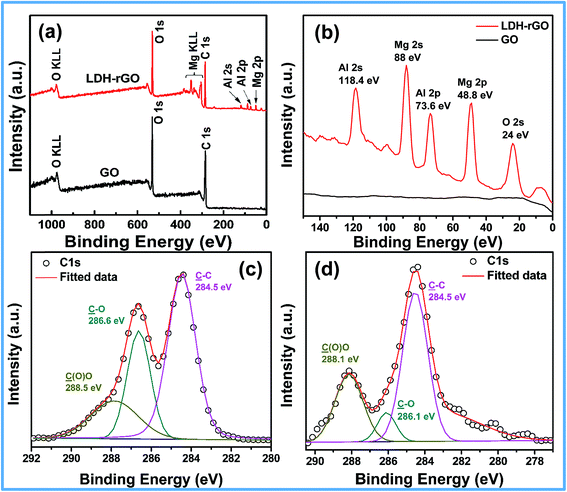 | ||
| Fig. 3 XPS survey spectra (a) full spectra and (b) expanded in lower binding energy of GO and LDH–rGO. C1s XPS spectra of (c) GO and (d) LDH–rGO nanohybrid. | ||
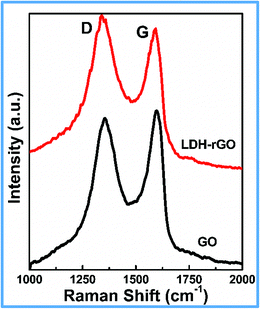
Fig. 4 shows the Raman spectra of GO and LDH–rGO nanohybrid. The spectra show two prominent peaks at around 1350 and 1600 cm−1 respectively corresponding to the D and G bands of carbon materials.38 The quantitative measurement of intensity ratio of the D and G bands, that is, ID/IG, gives the degree of disorder and is inversely proportional to sp2 domain size of carbon in graphite materials.39,40 The ID/IG ratios of GO and LDH–rGO nanohybrid were found to be 0.94 and 1.12 respectively. This indicates that the interaction of LDH flakes with the rGO could lead to more disordered structure. The increase in the ratio could also be attributed to the decrease in the average size of the sp2 domain size, which causes some unpaired defects caused by the removal of oxygen groups of GO (formation of rGO) by LDH during their growth.41,42 The reduction of GO and formation of LDH phase in the nanohybrid is further confirmed by FTIR analysis (Fig. S2†). The O–H stretching of GO appears at 3440 cm−1, the C–H stretching mode at 2926 and 2851 cm−1, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (carboxylic acid) at 1715 cm−1, C
O (carboxylic acid) at 1715 cm−1, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C at 1618 cm−1 originating from aromatic graphitic domains, COO− stretching at 1368 cm−1, C–O (epoxy/alkoxy) at 1050 cm−1, in agreement with GO spectra (Fig. S2a†).43,44 For LDH (Fig. S2b†), the O–H stretching of water molecules and the hydrogen-bonded –OH group peaks are centered at around 3440 cm−1 and 1635 cm−1 respectively. The absorption peak at 1365 cm−1 is related to intercalated CO32− anions. The metal–oxygen (M–O) absorption bands of LDH layers are observed below 800 cm−1.45 In the case of LDH–rGO, (Fig. S2c†) the O–H stretching at 3440 cm−1 and the presence of CO32− ions at around 1365 cm−1 remain the same. The observed band at 1560 cm−1 corresponds to the vibrations of backbone carbon. However, in comparison to the GO spectra, the peak at 1050 cm−1 corresponding to the alkoxy/epoxy groups disappeared completely. It means that the growth of LDH causes the removal of oxygen functional groups from GO. An intense peak obtained at 800 cm−1 in the nanohybrid belongs to bending and stretching of Mg–O and Al–O bonds. The stretching is much higher in the nanohybrid compared to LDH phase, which is due to the interaction of LDH and rGO during the in situ synthesis process. The results of FTIR confirms the reduction of GO and the formation of LDH phase with some interaction with rGO in the LDH–rGO nanohybrid.
C at 1618 cm−1 originating from aromatic graphitic domains, COO− stretching at 1368 cm−1, C–O (epoxy/alkoxy) at 1050 cm−1, in agreement with GO spectra (Fig. S2a†).43,44 For LDH (Fig. S2b†), the O–H stretching of water molecules and the hydrogen-bonded –OH group peaks are centered at around 3440 cm−1 and 1635 cm−1 respectively. The absorption peak at 1365 cm−1 is related to intercalated CO32− anions. The metal–oxygen (M–O) absorption bands of LDH layers are observed below 800 cm−1.45 In the case of LDH–rGO, (Fig. S2c†) the O–H stretching at 3440 cm−1 and the presence of CO32− ions at around 1365 cm−1 remain the same. The observed band at 1560 cm−1 corresponds to the vibrations of backbone carbon. However, in comparison to the GO spectra, the peak at 1050 cm−1 corresponding to the alkoxy/epoxy groups disappeared completely. It means that the growth of LDH causes the removal of oxygen functional groups from GO. An intense peak obtained at 800 cm−1 in the nanohybrid belongs to bending and stretching of Mg–O and Al–O bonds. The stretching is much higher in the nanohybrid compared to LDH phase, which is due to the interaction of LDH and rGO during the in situ synthesis process. The results of FTIR confirms the reduction of GO and the formation of LDH phase with some interaction with rGO in the LDH–rGO nanohybrid.
3.2 Antibacterial activity
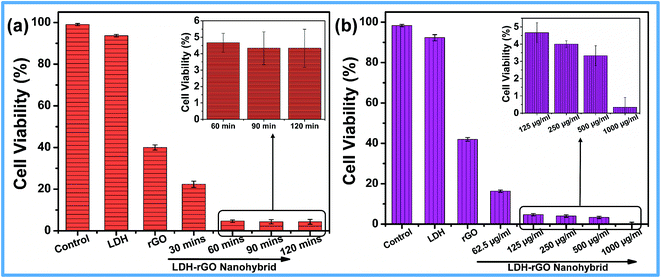
Reduced graphene oxide (rGO), was prepared by the same method as nanohybrid prepared without Mg and Al precursors. All the characterization data of rGO are given in ESI (Fig. S3–S6†). The as-prepared rGO was used as control for all antibacterial activity studies. Before being used for antibacterial activity, these materials were tested for biocompatibility with L929 cells. Fig. S7† shows that around 94.9 ± 0.5% of cells are viable with 2 mg mL−1 of LDH–rGO, indicating that it is a biocompatible material. For antibacterial activity, a suspension of (125 μg mL−1) LDH, rGO and LDH–rGO was incubated with E. coli cells (106 to 107 CFU mL−1). An isotonic saline solution without any material was used as a control. As shown in Fig. 5, the LDH dispersion shows negligible antibacterial activity with a decrease of cell viability by 7.4 ± 1.0%, whereas, for rGO this value is 51.7 ± 0.9% for E. coli cells. However, when treated with LDH–rGO, the loss of E. coli cells increased to 95.6 ± 1.1%, which is nearly double the loss of E. coli caused by rGO. Hence, it suggests that in situ reduction of GO in the nanohybrid plays a crucial role in the antibacterial activity as compared to the individual rGO. The antibacterial activity of the nanohybrid was compared with other graphene and LDH based materials12,16,17,30,46 and are given in Table 1. The results clearly show that, LDH–rGO nanohybrid exhibits better antibacterial activity than any of the graphene components or drug loaded LDH–graphene hybrids.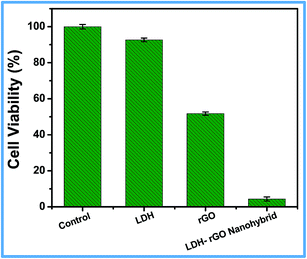 | ||
| Fig. 5 Cell viability of E. coli cells treated with LDH, rGO and LDH–rGO nanohybrid for 1 h with 125 μg mL−1 concentration. | ||
| Sample | Antibacterial activity (approx.%) | Quantity used (μg mL−1) | Reference |
|---|---|---|---|
| Reduced graphene oxide nanosheets | 90 | 85 | 12 |
| Reduced graphene oxide | 46 | 40 | 16 |
| Gentamicin-loaded methanol derived graphene | 82 | 40 | 17 |
| Graphene oxide–benzyl penicillin intercalated Mg–Al LDH hybrid films | 95 | GO/LDH = 4 | 30 |
| Reduced graphene nanowalls | 84 | — | 46 |
| LDH–rGO nanohybrid | 96 | ∼25 (wt of rGO in the nanohybrid) | This work |
Further, for the time-dependent antibacterial studies, dispersions of all samples (125 μg mL−1) were incubated for 2 h at the mid-log phase of E. coli. The cell viability was counted at every 30 min. Fig. 6a shows that with LDH–rGO the cell viability decreases to 22.4 ± 1.5% after 30 min and to 4.6 ± 1.1% after 60 min, and then reaches to saturation. It is interesting to note that the maximum cell death occurs with the nanohybrid in the first 30 min. Furthermore, different concentrations of LDH–rGO (0–1000 μg mL−1) were treated with E. coli cells for 60 min to evaluate cell viability (Fig. 6b). It was observed that for 125 μg mL−1 LDH–rGO, the cell viability decreases to 4.6 ± 0.6%. Beyond this concentration, there is no change in viability. It was now essential to study the morphology of bacterial cells after treatment with the LDH–rGO nanohybrid in order to understand the bacteriostatic or bactericidal effect.
3.3 Interaction of LDH and LDH–rGO nanohybrid with bacterial membrane
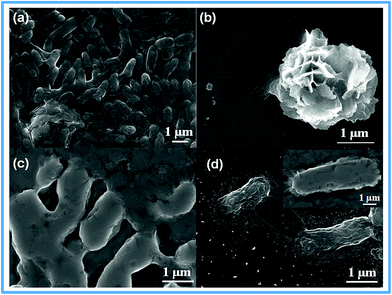
SEM was used to understand the interaction of LDH and LDH–rGO nanohybrid with bacteria that causes the cell death. Fig. 7a depicts E. coli cells that were grown in nutrient media. The bacterial cell wall (Fig. 7b) on which LDH has been capped, is intact, showing bacteriostatic effect whereas E. coli cells have the capacity to grow back in sometime. It was also observed that there was a mild destruction of the cell wall and it started losing cellular integrity. Fig. 7c shows that the E. coli cells started disintegrating in the initial stage of interaction with the nanohybrid. Fig. 7d shows the presence of completely ruptured cells with a few spikes (inset) as a result of the treatment of E. coli cells with the nanohybrid. This is the bactericidal action of LDH–rGO on the bacterial cells. This also suggests that the mode of action for bacterial death in case of LDH–rGO takes place through the cell wall route. Further, ROS-dependent and -independent oxidative stress and protein estimation studies have been performed in order to specify the mode of action.3.4 Oxidative stress due to effect of LDH–rGO nanohybrid
Oxidative stress is one of the many modes of actions of nanomaterials when they interact directly with bacterial cells. Out of the several paths, ROS-dependent and ROS-independent oxidative stress have been considered in this study.The XTT assay47 was performed to assess the ROS-dependent oxidative stress through superoxide radical production. TiO2 under UV radiation was used as a positive control for XTT tests. Fig. 8a shows the release of reactive oxygen species O2˙ of E. coli cells treated with different samples. It has been shown that, LDH–rGO exhibits high ROS release as compared to those treated with LDH and rGO. Even morphological studies have shown complete destruction of the cell wall with the hybrid, which is consistent with this analysis.
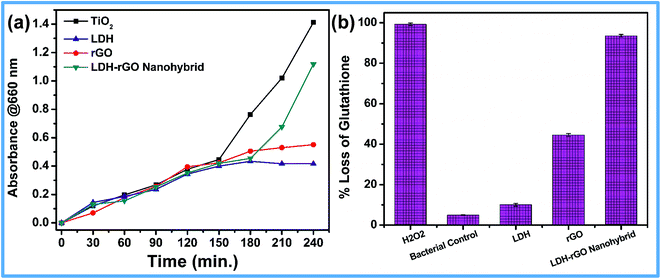 | ||
| Fig. 8 Oxidative stress through (a) super oxide (O2˙) release and (b) oxidation of glutathione by E. coli cells due to the effect of LDH, rGO and LDH–rGO nanohybrid. | ||
Further, Ellman's assay was used to estimate the oxidation of glutathione that was caused due to ROS-independent oxidative stress by LDH–rGO. Glutathione (GSH) solution without any material was used as a negative control, and H2O2 was used as a positive control. This assay also quantifies the concentration of thiol groups in GSH.48,49 In general, GSH consists of tripeptide with thiol groups that contain an antioxidant property in bacteria. GSH concentrations that range between 0.1 and 10 mM protects the cellular organelles from oxidative stress. In this case, thiol groups (–SH) oxidizes to disulfide bond (S–S), forming glutathione disulfide. In this test, oxidation of GSH was evaluated when it was incubated with LDH–rGO dispersions (125 μg mL−1) for 60 min. The control data shows that a negligible amount of GSH has been oxidized. A gradual increase in GSH oxidation occurred when 0.4 mM GSH was incubated with 125 μg mL−1 of LDH–rGO. Fig. 8b shows a substantial increase in GSH oxidation by LDH–rGO (93.5 ± 0.6%), as compared to that by LDH (10.0 ± 0.7%) and rGO (44.5 ± 0.7%). The oxidation of GSH indirectly suggests that LDH–rGO nanohybrid is capable of intervening ROS-independent oxidative stress in bacterial cells. The XTT and Ellman's assay results show that LDH–rGO is capable of mediating both ROS-dependent and ROS-independent oxidative stress in E. coli cells.
3.5 Protein degradation as an effect of LDH–rGO nanohybrid
We have further examined whether the oxidative stress can cause any damage to the cellular components. The surplus ROS generation damages the respiratory chains in order to activate the apoptotic process.50 This causes the weakening of cellular components such as DNA, proteins or any other targets, causing the loss of cell viability. We have studied the protein degradation due to the effect of LDH–rGO on E. coli cells that was performed using the Folin–Lowry test.51Fig. 9a shows the protein degradation study that indicates that the LDH in track 2 has no degraded protein, since it shows no antibacterial activity. However, track 4 shows protein degradation, with only two bands left at a higher molecular weight between 80 and 100 kDa, as compared with the control. Consequently, rGO also exhibits fade marks of proteins, from which it can be interpreted that most of the proteins are denatured. Fig. 9b shows that the total protein content value is decreased to 67.2 μg mL−1 as compared to that of control (236 μg mL−1) when E. coli cells were treated with LDH–rGO. This is in agreement with the antibacterial activity (Fig. 5) on E. coli conducted earlier, in which LDH–rGO has shown the least viable cells. This is probably the reason that only 4.6% E. coli could survive (that is, 95.4% mortality) when exposed to 125 μg mL−1 LDH–rGO, confirming the cell inhibition due to degradation of total cellular protein.
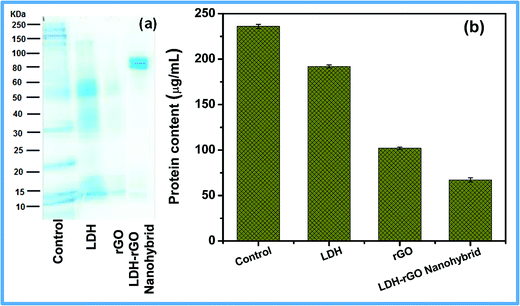 | ||
| Fig. 9 (a) SDS-PAGE shows protein degradation on the cell wall membrane and (b) quantitative analysis of total protein degradation of E. coli cells. | ||
3.6 Mechanism of antibacterial activity of LDH–rGO nanohybrid
The mechanism of antibacterial activity can be explored by correlating three different aspects presented in Table 2. Among all, the LDH–rGO nanohybrid shows a higher efficiency in killing E. coli cells. The higher antibacterial activity can be explained on the basis of size/agglomeration, GSH loss, development of oxidative stress and protein degradation.| Sample | Cell viabilitya (%) | GSH lossb (%) | Protein remained after degradationc (μg mL−1) |
|---|---|---|---|
| a Extracted from Fig. 5.b Extracted from Fig. 8b.c Extracted from Fig. 9b.d Control is E. coli cells without any material. | |||
| Controld | 99.9 ± 0.7 | 4.9 ± 0.1 | 236.0 ± 2.3 |
| LDH | 92.7 ± 1.0 | 10.0 ± 0.7 | 192.0 ± 1.7 |
| rGO | 51.7 ± 0.9 | 44.5 ± 0.7 | 102.0 ± 1.3 |
| LDH–rGO nanohybrid | 4.3 ± 1.1 | 93.6 ± 0.6 | 67.2 ± 2.2 |
The activity for graphene components mostly depend on aggregation of graphene components.52 Higher size and more agglomerated structures show lesser antibacterial activity and vice versa.16 If we compare both structures, rGO is in an agglomerated form (Fig. S6†) and rGO in the nanohybrid (Fig. 2c) is in a dispersed form and hence more antibacterial activity is seen with the nanohybrid. In addition, LDH shows better adherence to E. coli cells (Fig. 7b) because of its positive nature, which assists in a better interaction of the nanohybrid.
The antibacterial mechanism of the nanohybrid seems to be due to the synergistic effect of both ROS-dependent and ROS-independent oxidative stresses, which disrupts any cellular structure or component. Here, we have examined protein degradation study to understand the effect of the nanohybrid on the cellular component. The nanohybrid was able to degrade 75% of bacterial proteins as compared to that of control. After cell interaction with the nanohybrid, the sharp edges of both LDH and rGO act as cutters and cause oxidative stress. Subsequently, the nanohybrid oxidizes the thiol groups and degrades the proteins which causes the cell death.
4. Conclusions
The LDH–rGO nanohybrid is successfully synthesized by a hydrothermal method. All the characterizations confirm the de-oxygenation of GO and formation of LDH in the nanohybrid. The nanohybrid shows biocompatibility with L929 cells up to 2 mg mL−1. Antibacterial activity on E. coli cells was carried out extensively. The results show that the LDH–rGO nanohybrid exhibits the highest antibacterial activity and is capable of causing ∼96% cell death for E. coli cells. The antibacterial activity of the nanohybrid is both time and concentration-dependent. Oxidative stress studies suggest that the nanohybrid is capable of developing both ROS-dependent and ROS-independent stresses. In addition, it was found that all proteins were degraded with only two bands left at a higher molecular weight. The mechanism for the antibacterial activity was explained on the basis of size of graphene component, GSH loss, and protein degradation. All antibacterial studies of the nanohybrid suggest the oxidative stress causes GSH loss, protein degradation, and finally cell death. The biocompatible nature and higher antibacterial activity of the LDH–rGO nanohybrid compared to other graphene based materials render it potentially useful as a water purifying agent.Acknowledgements
The authors are thankful to Nanomission, Department of Science and Technology (DST), India for their financial support, and SAIF, IIT Bombay for providing instrumental facilities.References
- A. K. Geim and K. S. Novoselov, Nat. Mater., 2007, 6, 183–191 CrossRef CAS PubMed.
- A. K. Geim, Science, 2009, 324, 1530–1534 CrossRef CAS PubMed.
- F. Schwierz, Nature Nanotechnology, 2010, 5, 487–496 CrossRef CAS PubMed.
- L. Dai, Acc. Chem. Res., 2013, 46, 31–42 CrossRef CAS PubMed.
- X. Sun, H. Sun, H. Li and H. Peng, Adv. Mater., 2013, 25, 5153–5176 CrossRef CAS PubMed.
- D. Bitounis, H. Ali-Boucetta, B. H. Hong, D.-H. Min and K. Kostarelos, Adv. Mater., 2013, 25, 2258–2268 CrossRef CAS PubMed.
- S. Park and R. S. Ruoff, Nature Nanotechnology, 2009, 4, 217–224 CrossRef CAS PubMed.
- D. R. Dreyer, S. Park, C. W. Bielawski and R. S. Ruoff, Chem. Soc. Rev., 2010, 39, 228–240 RSC.
- Z. Gao, J. Wang, Z. Li, W. Yang, B. Wang, M. Hou, Y. He, Q. Liu, T. Mann, P. Yang, M. Zhang and L. Liu, Chem. Mater., 2011, 23, 3509–3516 CrossRef CAS.
- H. Li, G. Zhu, Z.-H. Liu, Z. Yang and Z. Wang, Carbon, 2010, 48, 4391–4396 CrossRef CAS.
- J. Chen, H. Peng, X. Wang, F. Shao, Z. Yuan and H. Han, Nanoscale, 2014, 6, 1879–1889 RSC.
- W. Hu, C. Peng, W. Luo, M. Lv, X. Li, D. Li, Q. Huang and C. Fan, ACS Nano, 2010, 4, 4317–4323 CrossRef CAS PubMed.
- J. Li, G. Wang, H. Zhu, M. Zhang, X. Zheng, Z. Di, X. Liu and X. Wang, Sci. Rep., 2014, 4, 4359 Search PubMed.
- F. Perreault, A. F. de Faria, S. Nejati and M. Elimelech, ACS Nano, 2015, 9, 7226–7236 CrossRef CAS PubMed.
- Y. Tu, M. Lv, P. Xiu, T. Huynh, M. Zhang, M. Castelli, Z. Liu, Q. Huang, C. Fan, H. Fang and R. Zhou, Nature Nanotechnology, 2013, 8, 594–601 CrossRef CAS PubMed.
- S. Liu, T. H. Zeng, M. Hofmann, E. Burcombe, J. Wei, R. Jiang, J. Kong and Y. Chen, ACS Nano, 2011, 5, 6971–6980 CrossRef CAS PubMed.
- H. Pandey, V. Parashar, R. Parashar, R. Prakash, P. W. Ramteke and A. C. Pandey, Nanoscale, 2011, 3, 4104–4108 RSC.
- C. G. Silva, Y. Bouizi, V. Fornés and H. García, J. Am. Chem. Soc., 2009, 131, 13833–13839 CrossRef PubMed.
- H. S. Panda, R. Srivastava and D. Bahadur, J. Phys. Chem. C, 2009, 113, 9560–9567 CAS.
- J. Xu, S. Gai, F. He, N. Niu, P. Gao, Y. Chen and P. Yang, J. Mater. Chem. A, 2014, 2, 1022–1031 CAS.
- Y. Kuthati, R. K. Kankala and C.-H. Lee, Appl. Clay Sci., 2015, 112–113, 100–116 CrossRef CAS.
- E. Ruiz-Hitzky, P. Aranda, M. Darder and G. Rytwo, J. Mater. Chem., 2010, 20, 9306–9321 RSC.
- V. Rives, M. del Arco and C. Martín, Appl. Clay Sci., 2014, 88–89, 239–269 CrossRef CAS.
- D. G. Evans and R. C. T. Slade, in Layered Double Hydroxides, ed. X. Duan and D. G. Evans, Springer Berlin Heidelberg, Berlin, Heidelberg, 2006, pp. 1–87 Search PubMed.
- S. Mao, H. Pu and J. Chen, RSC Adv., 2012, 2, 2643–2662 RSC.
- L. Wang, D. Wang, X. Y. Dong, Z. J. Zhang, X. F. Pei, X. J. Chen, B. Chen and J. Jin, Chem. Commun., 2011, 47, 3556–3558 RSC.
- Y. Wimalasiri, R. Fan, X. S. Zhao and L. Zou, Electrochim. Acta, 2014, 134, 127–135 CrossRef CAS.
- T. Wen, X. Wu, X. Tan, X. Wang and A. Xu, ACS Appl. Mater. Interfaces, 2013, 5, 3304–3311 CAS.
- M. Y. Miao, J. T. Feng, Q. Jin, Y. F. He, Y. N. Liu, Y. Y. Du, N. Zhang and D. Q. Li, RSC Adv., 2015, 5, 36066–36074 RSC.
- Y. Wang, D. Zhang, Q. Bao, J. Wu and Y. Wan, J. Mater. Chem., 2012, 22, 23106–23113 RSC.
- M.-Q. Zhao, Q. Zhang, J.-Q. Huang and F. Wei, Adv. Funct. Mater., 2012, 22, 675–694 CrossRef CAS.
- X.-Y. Yu, T. Luo, Y. Jia, R.-X. Xu, C. Gao, Y.-X. Zhang, J.-H. Liu and X.-J. Huang, Nanoscale, 2012, 4, 3466–3474 RSC.
- D. C. Marcano, D. V. Kosynkin, J. M. Berlin, A. Sinitskii, Z. Sun, A. Slesarev, L. B. Alemany, W. Lu and J. M. Tour, ACS Nano, 2010, 4, 4806–4814 CrossRef CAS PubMed.
- U. Costantino, V. Ambrogi, M. Nocchetti and L. Perioli, Microporous Mesoporous Mater., 2008, 107, 149–160 CrossRef CAS.
- J. Gupta, P. Bhargava and D. Bahadur, J. Mater. Chem. B, 2015, 3, 1968–1978 RSC.
- J. Xu, S. Gai, F. He, N. Niu, P. Gao, Y. Chen and P. Yang, Dalton Trans., 2014, 43, 11667–11675 RSC.
- X. Yan, J. Chen, J. Yang, Q. Xue and P. Miele, ACS Appl. Mater. Interfaces, 2010, 2, 2521–2529 CAS.
- L. Zhang, J. Wang, J. Zhu, X. Zhang, K. San Hui and K. N. Hui, J. Mater. Chem. A, 2013, 1, 9046–9053 CAS.
- O. Akhavan and E. Ghaderi, Carbon, 2012, 50, 1853–1860 CrossRef CAS.
- Y. Guo, X. Sun, Y. Liu, W. Wang, H. Qiu and J. Gao, Carbon, 2012, 50, 2513–2523 CrossRef CAS.
- M. Li, J. E. Zhu, L. Zhang, X. Chen, H. Zhang, F. Zhang, S. Xu and D. G. Evans, Nanoscale, 2011, 3, 4240–4246 RSC.
- X. Wang, S. Zhou, W. Xing, B. Yu, X. Feng, L. Song and Y. Hu, J. Mater. Chem. A, 2013, 1, 4383–4390 CAS.
- V. H. Pham, T. V. Cuong, S. H. Hur, E. Oh, E. J. Kim, E. W. Shin and J. S. Chung, J. Mater. Chem., 2011, 21, 3371–3377 RSC.
- Z. Wang, X. Zhang, J. Wang, L. Zou, Z. Liu and Z. Hao, J. Colloid Interface Sci., 2013, 396, 251–257 CrossRef CAS PubMed.
- C. Chen, P. Gunawan and R. Xu, J. Mater. Chem., 2011, 21, 1218–1225 RSC.
- O. Akhavan and E. Ghaderi, ACS Nano, 2010, 4, 5731–5736 CrossRef CAS PubMed.
- H. Ukeda, S. Maeda, T. Ishii and M. Sawamura, Anal. Biochem., 1997, 251, 206–209 CrossRef CAS PubMed.
- G. L. Ellman, Arch. Biochem. Biophys., 1959, 82, 70–77 CrossRef CAS PubMed.
- C. C. Winterbourn and D. Metodiewa, Free Radicals Biol. Med., 1999, 27, 322–328 CrossRef CAS PubMed.
- M. Valko, C. J. Rhodes, J. Moncol, M. Izakovic and M. Mazur, Chem.-Biol. Interact., 2006, 160, 1–40 CrossRef CAS PubMed.
- S. K. Sawhney and R. Singh, Introductory Practical Biochemistry, Narosa publishing house, New Delhi, India, 2002 Search PubMed.
- S. Gurunathan, J. W. Han, A. A. Dayem, V. Eppakayala, M.-R. Park, D.-N. Kwon and J.-H. Kim, J. Ind. Eng. Chem., 2013, 19, 1280–1288 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra04511a |
| This journal is © The Royal Society of Chemistry 2016 |