Cytotoxicity effects of three-dimensional graphene in NIH-3T3 fibroblasts
Bowei Zhanga,
Hongwei Ni*a,
Rongsheng Chenb,
Tongcun Zhangc,
Xi Lic,
Weiting Zhana,
Zhenyu Wangc and
Yao Xuc
aThe State Key Laboratory of Refractories and Metallurgy, Wuhan University of Science and Technology, Wuhan 430081, China. E-mail: nihongwei@wust.edu.cn; Fax: +86-27-68862811; Tel: +86-27-68862856
bSchool of Chemical Engineering and Technology, Wuhan University of Science and Technology, Wuhan 430081, China
cInstitute of Biology and Medicine, Wuhan University of Science and Technology, Wuhan 430065, China
First published on 19th April 2016
Abstract
The 3D configuration of graphene materials has been intensively investigated due to their novel properties in biomedical, electrical and optical applications. But few reports have been intentionally carried out to understand the biological influence of 3D graphene materials so far. Herein, we presented an evaluation of the in vitro cytotoxicity of 3D graphene sheets fabricated by carbonization of polydopamine (PDA) films on a template of aligned nanopore arrays (NPAs) on a stainless steel surface. The prepared 3D graphene sheets with a thickness of ∼20 nm displayed a nanoporous architecture that can be readily tuned by the NPA template to control the morphology of the 3D configuration. The in vitro toxicity of the nanoporous 3D graphene sheets with pore sizes of ∼50 nm and ∼240 nm was evaluated using NIH-3T3 fibroblasts as the representative mammal fibroblast cell type. The NPA structure exhibits enhanced properties in cell attachment, spreading, proliferation, and the assembly of focal adhesions and actin filament associated proteins. Morphology-dependent cytotoxicity aroused by the 3D graphene configuration should be attributed to the engulfment of the carbon nanospheres embedded in the 3D configuration and the lack of both focal adhesions and actin filament associated proteins assembling at the nanoscale.
Introduction
Graphene and graphene-related counterparts have aroused a wide range of potential applications as biomaterials, including biodevices, microbial detection, disease diagnosis, and drug delivery systems, due to their extraordinary mechanical, electronic, optical, geometric, and biological properties.1–3 But the biological applications and potential adverse influences of graphene and graphene-related materials remain unclear. Some in vitro and in vivo studies clearly indicated no particular risks.4,5 Other explorations, however, pointed out that graphene and graphene-related materials might render health hazards.6,7 Such graphene-based devices showed the requisite biocompatibility in situ yet inhalation toxicity aroused by multi-wall carbon nanotubes, graphene, and graphene nanoplatelets were also reported.8,9 Furthermore, even if the graphene and graphene-related materials are not devoid of possible immense risks to human health or the environment, the identification of different mechanisms that trigger toxicity is extremely useful for developing possible ways to control programmed cell death, to eliminate internalized graphene and graphene-related materials, and to activate weak immune responses.10–12 Consequently, before the full implementation of such nanomaterials in a large range of biological applications, concerns about potential toxicity and biocompatibility from both the scientific community and the public should be studied.13,14Graphene, a two-dimensional (2D) hexagonal honeycomb lattice of carbon atoms, is prone to aggregation or restacking due to intersheet van der Waals attractions, resulting in the loss of its unique properties.15,16 Recently, three dimensional (3D) graphene architectures have been intensively investigated to address these problems via various approaches, for example, integration of graphene onto 3D architectures, or producing wrinkles on graphene sheets.17–19 3D graphene configurations exhibit an amplified surface area and an interconnected graphene framework, thus offering novel opportunities in mechanical, electrical and optical properties, such as enhancing energy conversion efficiency and storage capacity, and opening tunable electrical bandgaps to expand application in the semiconductor industry.20–22 However, few reports have been intentionally carried out to understand the biological influences of 3D graphene materials so far.
Herein, we present an evaluation of the in vitro cytotoxicity of 3D graphene sheets, which were fabricated by carbonization of polydopamine (PDA) films on a template of aligned nanopore arrays (NPAs) on a stainless steel surface. As a biomolecule that contains catechol and amine functional groups, dopamine can spontaneously deposit PDA conformal films on virtually any surface in alkaline media.23 The PDA film can serve as a versatile carbon precursor to produce graphene-like films with a controllable thickness in nanoscale.24–26 The morphology of the NPA template on stainless steel foil can be readily tuned by an electrochemical process, resulting in controllable architecture of the 3D graphene materials. The in vitro toxicity of 3D graphene sheets prepared on the honeycomb NPAs with pore sizes of ∼50 nm and ∼240 nm was evaluated using NIH-3T3 fibroblasts as the representative mammal fibroblast cell type. Morphology-dependent cytotoxicity effects aroused by the 3D graphene configuration are observed. The cytotoxicity of the 3D graphene sheets should be attributed to the engulfment of the carbon nanospheres embedded in the 3D configuration and the lack of both focal adhesions and actin filament associated proteins assembling at the nanoscale.
Materials and methods
Formation of 3D graphene sheets
Medical 316L stainless steel foils (10 × 10 × 1 mm) were mechanically polished on abrasive papers (grade 1000 and 2000) with diamond pastes (from 3.5 to 0.25 μm). Anodization was then conducted using a two electrode configuration in an ethylene glycol (EG; 99.8% and anhydrous) solution containing 5 vol% perchloric acid (HClO4; 70%) for 10 min in an ice/water bath.27 The electrolytic solution was stirred by a rotating magnet. The applied voltages for anodization were 30 V and 60 V, respectively and designated as NPA30 and NPA60. After anodization, the foils were ultrasonically cleaned in acetone, ethanol, and deionized water for 10 min, respectively. The cleaned foils were immersed in a dopamine solution (2 mg mL−1 in 10 mM Tris–HCl buffer, pH 8.5) at room temperature for 2 h. After rinsing with deionized water and drying under a nitrogen flow, the foils were loaded onto a ceramic substrate placed at the center of a tube furnace. The furnace was purged with pure argon several times to remove residual air and moisture and was then heated to 650 °C for 3 h at a rate of 15 °C min−1 under argon flow. The carbonized PDA films on NPA30 and NPA60 were designated as NPA30G and NPA60G, respectively. Before cell culturing, the prepared foils were sterilized by ultraviolet irradiation for 30 min.Materials characterization
Field emission scanning electron microscopy (FE-SEM, FEI Nova 400 Nano) and transmission electron microscopy (TEM, JEM-2100UHR STEM/EDS, JEOL, Japan) were utilized to examine the surface topography and structure of the prepared materials. The higher-magnification images of the nanopore surfaces were obtained in “tapping mode” by atomic force microscope (AFM) Agilent 5500 (Agilent Technologies, Chandler, AZ) with a scanning range of 5 μm. The chemical composition of the carbonized PDA films was determined by X-ray photoelectron spectroscopy (XPS, ESCALAB MK-II). Raman spectra were recorded on a Lab Ram HR raman spectrometer (Horiba JobinYvon, France) with an excitation wavelength of 514 nm. Water contact angle measurements were carried out by the sessile-drop method at room temperature.Cell culture
NIH-3T3 fibroblasts were cultured in DMEM/High Glucose (Hyclone) with 10% fetal bovine serum (FBS, Gibico) and 0.1% penicillin and streptomycin, at 37 °C in an atmosphere of 5% CO2 with the medium changed every 3 d. The cells at passages 2 to 4 were used in the experiments.Protein absorption assay
A droplet (2 mL) of the medium (DMEM/High Glucose, Hyclone) containing 10% FBS (Gibico) was pipetted onto the sample and maintained at 37 °C for 2 h. The samples were transferred to new 24 well plates and washed with 1× PBS three times, and then the adsorbed proteins on the samples were detached into 400 μL of 1% sodium dodecyl sulfate (SDS) solution by shaking for 1 h. The amount of protein in the collected SDS solutions was measured via both a BCA kit (Pierce) and spectrophotometer.Initial cell adhesion
The cells were stained with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI, Beyotime), then seeded on the sample surfaces at a density of 2 × 104 cells per mL for 0.5, 1, and 2 h and the attached cells on the samples were fixed with 4% paraformaldehyde (Sigma) for 30 min at 4 °C. After washing with PBS, the samples were blocked with 90% glycerol/PBS, and counted from five random 10× fields on each sample using a fluorescence microscope (Olympus).Lactate dehydrogenase activity assay
The lactate dehydrogenase (LDH) activity in the culture media was used as an index of cytotoxicity. The cells were seeded at a density of 2 × 104 cells per well. After 3 days of culturing, the culture media was collected and centrifuged, and the supernatant was used for the LDH activity assay. The samples were placed in 24 well plates and tissue culture plates served as the control (C). NIH-3T3 fibroblasts were seeded on the surface of culture plate wells with the same cell density as other groups. The LDH activity was determined by a spectrophotometer according to the manufacturer’s instructions.Cell morphology
The fibroblasts were seeded at a density of 2 × 104 cells per well. After culturing for 2 d, the cells on the samples were washed in PBS, fixed with 4% paraformaldehyde, dehydrated in a graded ethanol series, freeze dried, coated with gold, and examined by FE-SEM.Cell proliferation
The cell proliferation and cytotoxicity assay kit (WST-1) was used to measure cell viability according to the manufacturer’s instructions. In brief, a 1 mL droplet of cell suspension was seeded onto each specimen with a density of 2 × 104 cells per mL and then cultured in DMEM/High Glucose containing 10% FBS. After 1 and 4 days, cell proliferation was assessed using the WST-1 cell proliferation reagent. At the prescribed time points, the prepared foils were rinsed three times with PBS gently and transferred to a new 24-well plate. The WST-1 solution was added and the prepared foils were incubated for 3 h at 37 °C to allow the formation of formazen. The optical density (OD) was measured at 450 nm using a spectrophotometer.Immunostaining
Fibroblasts were seeded at a density of 1 × 104 cells per well for 24 h of incubation and non-adherent cells were removed with two gentle washes with PBS. The adhered cells were fixed with 4% paraformaldehyde for 20 min followed by three PBS washes. The cell membranes were permeated by incubating in 0.1% Triton X-100 (Sigma) for 20 min, followed by three PBS washes, and were then blocked by 10% normal goat serum in PBS for 1 h, followed by an additional three PBS washes. Fixed cells were immunostained for vinculin and AFAP-1L1 (Santa Cruz Biotechnology), followed by incubation with Alexa Fluor 488 goat anti-mouse and goat anti-rabbit antibody for 1 h and three PBS washes. All pictures were collected using the Zeiss NOL-LSM 710 confocal microscope at 600× fields.Statistics
The data were analyzed using the SPSS 14.0 software (SPSS, USA). The means and standard deviations were calculated for the recorded data. Student’s t-test was employed to determine data sets that differed significantly from one another. The value of p < 0.05 was considered to be significant and p < 0.01 was considered to be highly significant.Results
Materials characterization
The morphology of the prepared foils was observed by FE-SEM; the mean pore sizes of NPA30 and NPA60 are about 50 nm (Fig. 1a) and 240 nm (Fig. 1b), respectively. The wall and surface of NPAs appeared smooth without defects and exhibited a perfect quasi-periodic arrangement of pores. Mechanically polished stainless steel substrates were used as the control (flat). Morphology of the porous surfaces remains almost unchanged after carbonization of the PDA coating (Fig. 1c). Raman spectra of the carbonized PDA film (Fig. 1e) showed two strong peaks of the D and G band at 1345 and 1590 cm−1,28 respectively. The dominant C 1s component at 284.6 eV (Fig. 1f) should be corresponding to sp2-hybridized carbon atoms of the graphene sheet,29 suggesting the formation of 3D graphene-like films on the nanopore arrays. As shown in Fig. 2, these dark domains consist of a graphene-like nanostructure which contain several tens (∼50) of stacking layers with a whole thickness of ∼20 nm. The size of the dark domain in Fig. 2a is consistent with the dimension of the carbon nanospheres shown in the higher-magnification inset in Fig. 1c. The HR-TEM image (Fig. 2c) illustrates that lattice spacing between these adjacent layers in these dark domains is about 0.34 nm, corresponding to the (002) planes of graphene, which is the typical distance between graphene layers.30,31 The carbon nanostructure except the dark graphene-like domains (Fig. 2d) is similar to amorphous carbon, with some short-range stacking layers.32The water surface contact angles are shown in Fig. 3. The flat 316L stainless steel surface (control) has a water contact angle of 41°. NPA30 and NPA60 show a water contact angle of 21.3° and 25.7°, respectively. Meanwhile, NPA30G and NPA60G display enhanced hydrophilicity relative to the mechanically polished (flat) and anodized stainless steel foils (NPAs) with a water contact angle of 17.8° and 17.2°, respectively.
 | ||
| Fig. 3 Water contact angles on samples. *p < 0.05 and **p < 0.01 when compared with flat control surfaces. The top panel shows the optical pictures. | ||
Protein absorption
The protein adsorption ability in the cell culture of 10% FBS in DMEM/High Glucose is shown in Fig. 4 to explain the subsequent cellular response. The 3D graphene films absorb more proteins than the flat control, especially NPA60G. NPA30 adsorbs a similar amount of protein compared to the flat control but the absorb protein amount of NPA60 is smaller than that of carbonized PDA films on NPAs (NPAsG) and more than that of NPA30. | ||
| Fig. 4 Protein adsorption onto the prepared foils after 2 h of incubation in DMEM/High Glucose containing 10% FBS. **p < 0.01 when compared with flat control surfaces. | ||
Initial cell adhesion
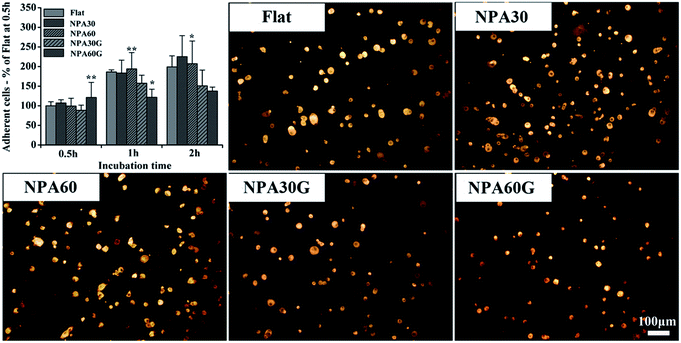
The initial cell adhesion assayed during the first 2 h of the cell/biomaterial interaction is illustrated in Fig. 5. The adherent cell numbers on the flat and NPA surfaces increase with incubation time. More cells appear to adhere on NPAs compared to the flat control although not all of them are statistically significant. But NPAsG show less adherent cell numbers at 1 h and 2 h. As the fluorescence images show, after incubation for 2 h, the average sizes of attached cells on NPAsG were smaller than that of both flat and NPA surfaces.Cytotoxicity assay
The cytotoxicity indicated by LDH activity in the culture media after 3 d of incubation is assayed and the results are displayed in Fig. 6. The flat and NPAs show no obvious cytotoxicity compared to the culture plate control but the NPAsG show high cytotoxicity. | ||
| Fig. 6 Amount of LDH released by cells to the culture medium in the first 3 d of culturing. *p < 0.05 and **p < 0.01 when compared with flat control surfaces. | ||
Cell morphology
The SEM pictures (Fig. 7) show the cell morphology on the samples after 2 days of incubation. The cells attach and spread well on the flat, NPA30, and NPA60 surfaces. There are many dense lamellipodia and filopodia stretching out to anchor to the surface forming good intercellular connection. However, on the surfaces of NPAsG, the cells cannot grow well and seem to have an apoptotic appearance. In contrast to the flat, cells on NPA30G spread poorly with a spindle shape and form thin lamellipodia-like structures that appeared to attach to the surface. On the surface of NPA60G, poorly spreading round cells can be seen. Therefore, while cells can attach to NPAsG using lamellipodia-like structures, they are not able to spread on the surfaces.Cell proliferation
The cell proliferation during 4 days of incubation is shown in Fig. 8. The cells proliferated well on the flat and NPAs; the cell proliferation was clearly higher on the nanopore surfaces than on the flat, especially on the NPA60 surface. It is obvious that NPA30G and NPA60G induce a dramatically smaller cell number at days 1 and 4. Cell numbers on the NPA30G and NPA60G were significantly reduced (71.8% and 68.6%, separately) compared to the cells on the flat after 1 day of culturing; the cell numbers on NPAsG decreased with incubation time suggesting apoptosis. After 4 days of culturing, cell numbers on NPA30G and NPA60G reduced to about 38% and 8% of the cell number on the flat at day 1, respectively. Consequently, the cell numbers show an obvious tendency to decline on NPAsG and decreased rapidly on NPA60G compared to NPA30G. | ||
| Fig. 8 Cell viability after 1 and 4 days of incubation measured by the WST-1 assay. *p < 0.05 and **p < 0.01 when compared with flat control surfaces. | ||
Immunostaining
To evaluate the role of adhesion molecules and physical forces in cell attachment, proliferation and survival, immunostaining specific to vinculin and AFAP-1L1 were performed (Fig. 9). We next investigated the influence of 3D carbonized PDA films on cell spreading after 24 h. Vinculin and AFAP-1L1 staining were widely distributed within cells grown on the flat and NPAs surfaces; the amount of vinculin increased in cells grown on the NPA surfaces but decreased seriously in cells grown on the NPAG surfaces. Overall, the nanopore surfaces of NPA30 and NPA60 enhanced cell attachment. These graphene-like coatings on NPAsG reduced the formation of focal adhesions in fibroblasts significantly.Discussion
In this paper, we found that cell spreading, proliferation, focal adhesions and AFAP-1L1 assembly are significantly enhanced on nanopore surfaces in contrast to the flat control while adhesion and viability of NIH-3T3 fibroblasts are greatly altered on the 3D graphene-like films. Cells adhered less and spread less on NPAsG than the corresponding flat and NPA substrates. The experimental results suggest that the 3D graphene-like films dramatically reduce cell adhesion, spreading, survival as well as late cell functions such as the formation of focal adhesions and AFAP-1L1.As mentioned previously, the surface aggregation state of nanomaterials significantly affects their cytotoxicity.33 As the stacking layers with a whole thickness of ∼20 nm emerge on NPAG surfaces in the form of carbon, nanosphere and nanomaterial properties differ from those of bulk materials, a possible side effect of these capabilities is interactions with biological systems, with the potential to generate toxicity.34,35 These raise the possibility that cells may adhere initially on the graphene platelets but undergo apoptosis due to engulfment of carbon nanoparticles at longer times. Results of initial adhesion during 0.5–2 h and the morphologies of cells cultured on samples for 2 days can clarify this problem to some degree. After NIH-3T3 fibroblasts were seeded on samples for 0.5 h, there is no appreciable difference of the adhered cell numbers on NPAs compared to the flat but the NPA60G sample shows larger adherent cell numbers. At 1 h and 2 h, however, there is almost no difference in adhered cell numbers of NPA30G and NPA60G but it is obviously less than on the flat. In addition, the DiI staining images at 2 h show that the average size of cells on NPAsG is smaller compared to the flat. The 3D carbonized PDA layers hinder the survival of cells as shown in SEM pictures. Actually, the SEM micrographs indicate that poorly spreading cells can be seen clearly and seem to have an apoptotic appearance on the surfaces of NPAsG compared to the cells spread completely on the surfaces of the 316L flat and NPAs. These results show that the carbonized PDA film did not support cell survival and provide evidence that carbon nanosphere engulfment may contribute to decreased survival of cells.1
In order to further understand the problem of toxicity aroused by 3D carbonized PDA graphene-like films, a LDH assay of the culture medium after 3 days of incubation and a WST-1 assay after 4 days were performed. They show that the NPAsG are cytotoxic compared to the culture plate control. Actually, 316L stainless steel is generally considered to be a kind of biomaterial that can provide an anti-corrosion and non-cytotoxic surface to induce the adhesion of cells.36 Our data suggest that the dramatically reduced cell numbers and poor stretching of cells are certainly due to cytotoxicity.37,38
Surface wettability is believed to be an important factor in cell/biomaterial interactions. We can find that NPA30G and NPA60G display enhanced hydrophilicity relative to flat and NPAs with a water contact angle of 17.8° and 17.2°. The surface wettability of biomaterials is a complex issue and is affected by many factors.39,40 In this study, the wettability differences between flat, NPA30 and NPA60 convey the effect of topography to the biological performance of biomaterials.41,42 The reasons why the coatings on NPAsG display enhanced hydrophilicity can be mainly attributed to the hydrophilic nature of amorphous carbon and nanoscale spheres, which formed on NPAG surfaces and aggregated to minimize their surface-energy.43 Nonetheless, our study is believed to indicate that the poor attachment of cells on graphene platelets has no obvious relationship with surface wettability.
Protein adsorption on biomaterials also plays a vital role in cell/biomaterial interactions.44 In contrast to 316L flat, high protein absorption on NPA30 and NPA60 can be ascribed to the nanoscale pore walls that provide effective nucleation sites for protein absorption. Besides the topographical factors, the highest protein adsorption on NPAsG can be attributed to the aromatic nature of graphene; it is rather common that interactions are rationalised in terms of π–π stacking with aromatic amino acids, such as histidine and tryptophan.45,46 As aforementioned, there is the expectation that protein adsorption is an important precursor to cell attachment.47 Thus, there is a relationship between the cells attached and stretched completely on NPAs and protein adsorption but the poor attachment of cells on the carbon films is not relevant to protein adsorption. However, it is of special interest to modulate protein adsorption and delivery kinetics via possible means such as surface nanostructure alteration.48 More protein loading and controlled release may thus be implemented, and at the same time for better performance in drug delivery.
The cell attachment and spreading on biomaterials is regulated by focal adhesions.49,50 Physical forces, derived from or applied to cells, are important signals in determining cellular structure, proliferation and survival. The actin filament associated proteins interact with dynamin to promote endocytosis and also transduce mechanical stretch into c-src protein tyrosine kinase activation.51,52 Cell spreading, proliferation and subsequent focal adhesion formation are closely related to the geometry of the surface.53 The detailed mechanics of various cellular nano-sensing between cells and how the geometry of pore sizes enhance cell attachment and proliferation are still not fully understood, which are still the subjects of intense research.54,55 The nanoporous surfaces of NPAs significantly enhanced cell spreading, proliferation, focal adhesions and actin filament associated protein assembly in contrast to the flat. This may be ascribed to the mechanism that nanopores affect cell adhesion by fostering an interaction between integrin anchoring sites and the edges of pores for cell attachment.56 The phenomena that cell proliferation on NPA60 was slightly higher than that on NPA30 and that cell viability decreased faster on NPA60G compared to NPA30G were shown. These results indicate that the cellular responses are modulated by nanopore features in a size-dependent manner.
Focal adhesions are not allowed to assemble in cells that depend on anchorage for survival, leading to weak attachment to the surface, lack of cell spreading and subsequent apoptosis. This suggests that the lack of initial spreading may cause cell death.57 As initial adhesion is required to polymerize actin filaments,58 the lack of lamellipodia is probably due to an inability of cells to establish a strong initial adhesion to the substrate, thereby altering the dynamics of cell spreading. Cavalcanti-Adam et al. observed altered cell spreading dynamics on RGD (arginine–glycine–aspartic acid) nanopatterned substrates.59 Our results can therefore be explained by a mechanism in which abnormal assembly of focal adhesions and actin filament associated proteins contributes to restrained cell spreading on the 3D multilayer graphene platelet films. Because a lack of cell spreading can cause cell death, decreased spreading may explain the observed decrease in cell survival on the 3D graphene sheets.
Conclusions
In the present study, we fabricated honeycomb nanopore arrays (NPAs) on 316L stainless steel substrates with different pore dimensions of 50 nm and 240 nm, respectively. Graphene-like carbonized polydopamine (PDA) films are coated on nanopore arrays (NPAsG). Our results indicate that in the 3D graphene-like coatings there are many graphene-like nanoaggregates with thicknesses of about 20 nm, which are composed of about 50 stacking layers. Fibroblast attachment, spreading and proliferation are highly promoted by NPAs, and NPA60 enhances cell proliferation significantly. The obtained results suggest that the cytotoxicity aroused by 3D graphene-like coatings is dimensional configuration-dependent according to the evaluations. The mechanism is likely due to carbon nanosphere engulfment and contributed to the lack of both focal adhesions and actin filament associated proteins assembling at the nanoscale.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 51171133, No. 21105077 and No. 51471122).References
- A. Magrez, S. Kasas, V. Salicio, N. Pasquier, J. Won Seo, M. Celio, S. Catsicas, B. Schwaller and L. Forró, Nano Lett., 2006, 6, 1121 CrossRef CAS PubMed.
- N. Mohanty and V. Berry, Nano Lett., 2008, 8, 4469 CrossRef CAS PubMed.
- X. Sun, Z. Liu, K. Welsher, J. T. Robinson, A. Goodwin, S. Zaric and H. Dai, Nano Res., 2008, 1, 203 CrossRef CAS PubMed.
- H. Shen, L. Zhang, M. Liu and Z. Zhang, Theranostics, 2012, 2, 283 CrossRef CAS PubMed.
- J. Liu, L. Cui and D. Losic, Acta Biomater., 2013, 9, 9243 CrossRef CAS PubMed.
- E. L. K. Chng, Z. Sofer and M. Pumera, Chem.–Eur. J., 2014, 20, 6366 CrossRef CAS PubMed.
- Y. Chong, C. Ge, Z. Yang, J. Antonio Garate, Z. Gu, J. K. Weber, J. Liu and R. Zhou, ACS Nano, 2015, 9, 5713 CrossRef CAS PubMed.
- K.-H. Liao, Yu-S. Lin, C. W. Macosko and C. L. Haynes, ACS Appl. Mater. Interfaces, 2011, 3, 2607 CAS.
- M. S. Artiles, C. S. Rout and T. S. Fisher, Adv. Drug Delivery Rev., 2011, 63, 1352 CrossRef CAS PubMed.
- A. Bianco, Angew. Chem., Int. Ed., 2013, 52, 4986 CrossRef CAS PubMed.
- Q. Mu, G. Su, L. Li, B. O. Gilbertson, L. H. Yu, Q. Zhang, Y. Sun and B. Yan, ACS Appl. Mater. Interfaces, 2012, 4, 2259 CAS.
- G. Chen, H. Yang, C. Lu, Y. Chao, S. Hwang, C. Chen, K. Lo, L. Sung, W. Luo, H. Tuan and Y. Hu, Biomaterials, 2012, 33, 6559 CrossRef CAS PubMed.
- Y. Zhang, S. F. Ali, E. Dervishi, Y. Xu, Z. Li, D. Casciano and A. S. Biris, ACS Nano, 2010, 4, 3181 CrossRef CAS PubMed.
- C. Ge, J. Du, L. Zhao, L. Wang, Y. Liu, D. Li, Y. Yang, R. Zhou, Y. Zhao, Z. Chai and C. Chen, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 16968 CrossRef CAS PubMed.
- Z. Fan, J. Yan, L. Zhi, Q. Zhang, F. Wei and J. Feng, Adv. Mater., 2010, 22, 3723 CrossRef CAS PubMed.
- Y. Liu, J. Zhou, J. Tang and W. Tang, Chem. Mater., 2015, 27, 7034 CrossRef CAS.
- X. H. Xia, D. L. Chao, Y. Q. Zhang, Z. X. Shen and H. J. Fan, Nano Today, 2014, 9, 785 CrossRef CAS.
- Y. Xu, G. Shi and X. Duan, Acc. Chem. Res., 2015, 48, 1666 CrossRef CAS PubMed.
- J. Leem, M. C. Wang, P. Kang and S. W. Nam, Mechanically Self-Assembled, Nano Lett., 2015, 15, 7684 CrossRef CAS PubMed.
- J. Li, T. F. Chung, Y. P. Chen and G. J. Cheng, Nano Lett., 2012, 12, 4577 CrossRef CAS PubMed.
- X. Yao, G. Guo, X. Ma, Y. Zhao, C. Ang, Z. Luo, K. T. Nguyen, P. Li, Q. Yan and Y. Zhao, ACS Appl. Mater. Interfaces, 2015, 7, 26085 CAS.
- F. Symalla, S. Shallcross, I. Beljakov, K. Fink, W. Wenzel and V. Meded, Phys. Rev. B: Condens. Matter Mater. Phys., 2015, 91, 205412 CrossRef.
- H. Lee, S. H. Dellatore, W. M. Miller and P. H. Messersmith, Science, 2007, 318, 426 CrossRef CAS PubMed.
- J. Kong, W. A. Yee and L. Yang, Chem. Commun., 2012, 48, 10316 RSC.
- H. Jiang, L. Yang and C. Li, Energy Environ. Sci., 2011, 4, 1813 CAS.
- R. Liu, S. M. Mahurin and C. Li, Angew. Chem., Int. Ed., 2011, 50, 6799 CrossRef CAS PubMed.
- B. Zhang, H. Ni, R. Chen, W. Zhan, C. Zhang, R. Lei and Y. Zha, Appl. Surf. Sci., 2015, 351, 1161 CrossRef CAS.
- Z. Wang, H. Ni, R. Chen, W. Zhan, C. Zhang, R. Lei and B. Zhang, Electrochem. Commun., 2014, 47, 75 CrossRef CAS.
- M. Pumera, T. Sasaki and H. Iwai, Chem.–Asian J., 2008, 3, 2046 CrossRef CAS PubMed.
- N. G. Shang, P. Papakonstantinou and M. McMullan, Adv. Funct. Mater., 2008, 18, 3506 CrossRef CAS.
- R. Chen, L. Hu and K. Huo, Chem.–Eur. J., 2011, 17, 14552 CrossRef CAS PubMed.
- X. Yu, H. Fan, Y. Liu, Z. Shi and Z. Jin, Langmuir, 2014, 30, 5497 CrossRef CAS PubMed.
- S. Liu, L. Wei, L. Hao, N. Fang, M. W. Chang, R. Xu, Y. Yang and Y. Chen, ACS Nano, 2009, 3, 3891 CrossRef CAS PubMed.
- T. Xia, M. Kovochich and A. E. Nel, Nano Lett., 2006, 6, 1794 CrossRef CAS PubMed.
- N. Lewinski, V. Colvin and R. Drezek, Small, 2008, 4, 26 CrossRef CAS PubMed.
- R. D. K. Misra, W. W. Thein-Han, T. C. Pesacreta, K. H. Hasenstein, M. C. Somani and L. P. Karjalainen, Adv. Mater., 2009, 21, 1280 CrossRef CAS.
- X. Han, R. Gelein, N. Corson, P. Wade-Mercer, J. Jiang, P. Biswas, J. N. Finkelstein, A. Elder and G. Oberdörster, Toxicology, 2011, 287, 99 CrossRef CAS PubMed.
- P. Ngamwongsatit, P. P. Banada, W. Panbangred and A. K. Bhunia, J. Microbiol. Methods, 2008, 73, 211 CrossRef CAS PubMed.
- M. Lundgren, N. L. Allan and T. Cosgrove, Langmuir, 2007, 23, 1187 CrossRef CAS PubMed.
- G. Palasantzas and J. T. M. De Hosson, Acta Mater., 2001, 49, 3533 CrossRef CAS.
- M. L. Careman, T. G. Estes and A. W. Feinberg, Biofouling, 2006, 22, 1 CrossRef PubMed.
- S. Lavenus, M. Berreur and V. Trichet, Eur. Cells Mater., 2011, 22, 84 CAS.
- Y. Zhou, B. Wang and X. Song, Appl. Surf. Sci., 2006, 253, 2690 CrossRef CAS.
- M. S. Lord, M. Foss and F. Besenbacher, Nano Today, 2010, 5, 66 CrossRef CAS.
- M. Prato and S. Marchesan, Chem. Commun., 2015, 51, 4347 RSC.
- S. Wang, E. S. Humphreys, S. Y. Chung, D. F. Delduco, S. R. Lustig, H. Wang, K. N. Parker, N. W. Rizzo, S. Subramoney, Y. Chiang and A. Jagota, Nat. Mater., 2003, 2, 196 CrossRef CAS PubMed.
- M. A. Cole, N. H. Voelcker, H. Thissen and H. J. Griesser, Biomaterials, 2009, 30, 1827 CrossRef CAS PubMed.
- C. Galli, M. C. Coen, R. Hauert, V. L. Katanaev, P. Gröning and L. Schlapbach, Colloids Surf., B, 2002, 26, 255 CrossRef CAS.
- X. Zhang, G. Jiang and Y. Cai, Nat. Cell Biol., 2008, 10, 1062 CrossRef CAS PubMed.
- C. A. Franzen, E. Amargo and V. Todorović, Cancer Prev. Res., 2009, 2, 830 CrossRef CAS PubMed.
- L. M. Saba, B. Bennett and B. Tabakoff, Neuropharmacology, 2011, 60, 1269 CrossRef CAS PubMed.
- B. Han, X. H. Bai, M. Lodyga, J. Xu, B. B. Yang, S. Keshavjee, M. Post and M. Liu, J. Biol. Chem., 2004, 279, 54793 CrossRef CAS PubMed.
- X. Zhu, J. Chen, L. Scheideler, R. Reichl and J. Geis-Gerstorfer, Biomaterials, 2004, 25, 4087 CrossRef CAS PubMed.
- H. Pan, Y. Hung, Y. Sui and G. S. Huang, Biomaterials, 2012, 33, 20 CrossRef CAS PubMed.
- M. Biggs, R. G. Richards and M. Dalby, Nanomedicine, 2010, 6, 619 CAS.
- H.-A. Pan, J.-Y. Liang and G. Steven Huang, Biomaterials, 2013, 34, 841 CrossRef CAS PubMed.
- J. Lee, B. H. Chu, K. H. Chen, F. Ren and T. P. Lele, Biomaterials, 2009, 30, 4488 CrossRef CAS PubMed.
- N. Xia, C. K. Thodeti, T. P. Hunt, Q. Xu, M. Ho, G. M. Whitesides, R. Westervelt and D. E. Ingber, FASEB J., 2008, 22, 1649 CrossRef CAS PubMed.
- E. A. Cavalcanti-Adam, T. Volberg, A. Micoulet, H. Kessler, B. Geiger and J. P. Spatz, Biophys. J., 2007, 92, 2964 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2016 |