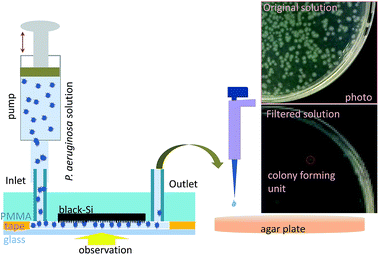
A bactericidal microfluidic device constructed using nano-textured black silicon
Xuewen
Wang†
ab,
Chris M.
Bhadra†
a,
Thi Hoang
Yen Dang
a,
Ričardas
Buividas
a,
James
Wang
a,
Russell J.
Crawford
a,
Elena P.
Ivanova
*a and
Saulius
Juodkazis
*ab
aFaculty of Science, Engineering and Technology, Swinburne University of Technology, John St., Hawthorn, Vic. 3122, Australia. E-mail: sjuodkazis@swin.edu.au; eivanova@swin.edu.au; Fax: +61 3 9214 5435; Tel: +61 3 9214 8718
bMelbourne Centre for Nanofabrication (MCN), Australian National Fabrication Facility (ANFF), Clayton, VIC 3168, Australia
First published on 1st March 2016
Abstract
Nano-structured black silicon (bSi) was used as a substratum for the construction of a microfluidic device to test the bactericidal action of this nano-textured surface against Pseudomonas aeruginosa bacteria. A narrow 15 µm high and 1 cm wide flat flow channel was constructed that allowed the bacteria to come into contact with the bactericidal nano-spikes present on the surface of the bSi. The narrow channel within the device was designed such that a single layer of bacterial cells could reside at any given time above the bSi substratum during flow. The large 1 × 2 cm2 surface area of the bSi was shown to be efficient in being able to kill the bacterial cells, achieving an approximate 99% killing efficiency. The flow rate required to fill the bSi chamber was found to be 0.1 µL s−1, with a 10 min equilibration time being allowed for the bacterial cells to interact with the bSi surface. Complete rupturing of E. coli cells was achieved after 15 cycles, allowing the effective release of cellular proteins from within the bacterial cells (65.2 µg mL−1 from 3 × 108 cells per mL). The channel was then able to be re-used after washing of the cell with 10 successive cycles of sterile MilliQ water. Larger volumes of bacterial suspensions have the potential to be treated using a similar flow channel configuration if the dimensions of the flow channel are scaled accordingly. This bactericidal microfluidic device provides a novel platform for studies carried out under both static and dynamic (flow) conditions.
Introduction
Antibacterial surfaces1–3 are becoming imperative in applications designed to curb the negative consequences associated with resistance to antibiotics present in food, water, and soil.4 Bacterial resistance arising from extensive exposure to antibiotics has the potential to compromise our immune system, particularly with regard to our ability to effectively resist bacterial infections. Many natural and synthetic surfaces achieve their self-cleaning, anti-fouling and/or bactericidal properties through various mechanisms; they can be highly oxidative, becoming bactericidal when activated by UV-light5 self-cleaning due to their surfaces being rendered hydrophobic via modification of their chemical or mechanical properties,6–8 or anti-fouling due to their surface structures sterically hindering the attachment of pathogens.4,9 Other applications of micro- and nano-structured surfaces in the biomedical industry include dermal patches, which possess painless needles that allow the controlled release of drugs, and bandages that possess bio-compatible microfibers that trigger increased levels of healing when exposed to ultraviolet UV light.10 Surfaces that display mechanical means for antifouling and antibacterial properties are a topic of significant research as they provide a substrate from which a fundamental understanding of the mechanism takes place.11–14 These surfaces have wide applications in the production of sanitary surfaces such as mobile telephones and other household items.15The search for inexpensive methods for the fabrication of large area nano-textured surfaces is currently underway. Silicon is a substratum that has been used extensively in the semiconductor and solar cell industries.16–20 Being a relatively inexpensive product, silicon represents one of the best substrata for the fabrication of large areas of nano-textured surfaces, where reproducible surfaces are currently able to be prepared over surfaces of several centimetres in diameter. Such substrata can also have electrical and photo-electrochemical device level functionalities incorporated into their surface, which is useful when producing micro-chips.21 Methods for preparing these nano-textured surfaces using a silicon substratum include plasma etching, where the deposition of electrical contacts is required for the fabrication of wafer sized bSi surfaces.22–26 The unique nano-topography of bSi forms due to the self-organized hard mask that results from the first few seconds of etching. These are specific to the chemistry and chamber materials being used for the production of the bSi.27 It is used in highly efficient solar cells, as it represents a low reflectance or broadband absorbing surface. More recently it has been used for the production of sensors that are based on surface enhanced Raman scattering (SERS).28 bSi substrata have recently been produced that possess a similar surface topography to that of dragonfly wings, and have been found to exhibit a similar bactericidal efficiency when coming in contact with pathogenic bacteria and spores.29
Given this demonstrated bactericidal functionality of the bSi surface, a microfluidic device was constructed incorporating a bSi substrate to investigate whether this bactericidal action would be effectively translated within a flow channel of a microfluidic device. Applications of such devices, if effective, would be of great benefit in many different fields, such as in the pharmaceutical industry for the detection and/or monitoring of bacterial contamination.
Experimental
The bSi was prepared using a plasma etching process.28,30 An Oxford PlasmaLab 100 ICP380 plasma etcher was used for patterning the surface of p-type boron-doped 4-inch diameter silicon wafers of specific resistivity 10–20 Ω cm−1, having a 〈100〉 oriented surface (Atecom Ltd, Taiwan). The resulting surface possessed pencil-like nano-spikes that were approximately 500 nm in height and 95 nm in pillar diameter (at half maximum). The lateral distribution was relatively random, with a distance between neighbouring spikes being approximately 450 ± 200 nm. The lateral distribution of the needles was determined from fast Fourier transform (FFT) processing of the SEM images (Fig. 1(a)). The static water contact angle on the bSi was measured to be approximately 101°, displaying a similar hydrophobicity to that previously reported for bSi prepared under the same conditions.29,31 Unmodified silicon wafers were used as control surfaces.The bSi and silicon wafers were precisely cut using a femtosecond laser (Pharos, Light Conversion Ltd.) at a wavelength of λ = 515 nm, pulse duration of 230 fs, pulse energy 7 µJ per pulse at repetition rate of 100 kHz and scan speed 1 mm s−1. The resulting wafer was mounted on 3-axis stage with 5 nm repetition accuracy (Areotech Ltd.). The beam was focused to a 0.9 µm spot by an objective lens with a numerical aperture NA = 0.7 (d = 1.22λ/NA). The line scribing process was repeated 7 times and took 40 min to scribe a single 4-inch wafer into 20 × 10 mm2 pieces, which would be used for construction of the micro-fluidic chip (Fig. 1). The scribing depth reached approximately 60 µm which was sufficient for clean cleavage of the wafer (Fig. 1). The lateral width of the laser cut was only 2–3 times wider than the focal spot diameter. An arbitrary substratum shape could be prepared using this procedure (see circular cuts in Fig. 1(a)).
The microfluidic chip was assembled via a simple method using an adhesive tape spacer, which allowed the shape and height of the microfluidic channel to be defined.30,32 In such a design, an adhesive double sided tape (ARclad IS-8026-15, Adhesives Research Inc.) was placed on the glass substratum with the channel layout being defined by a laser cutter (CO2 laser VLS 2.30, Versa Laser). The chip was completed by placing the top plate of the bSi in position and sealing the device with silicone. The tubing, obtained from syringe needles, was added and sealed in position, as required. Duplicate chips, fabricated using the control silicon wafer substrates, were used as negative controls. The channel height of both the bSi and control chips was 15 µm, determined by the thickness of the adhesive tape used in the construction of the device. This height was selected in order to accommodate the rod-shaped P. aeruginosa cells, which had dimensions of approximately 2 µm × 1 µm.33
P. aeruginosa ATCC 9027 and E. coli K 12 cells, obtained from the American Type Culture Collection (ATCC, USA), were used in this study. Bacterial stocks were prepared in 20% glycerol nutrient broth (Oxoid) and stored at −80 °C until needed. Prior to each experiment, the bacterial cultures were refreshed from the stock solution on nutrient agar (Oxoid), and a fresh bacterial suspension was prepared from bacterial cells, which were grown overnight in 100 mL of nutrient broth (in 0.5 L Erlenmeyer flasks at 37 °C with shaking at 120 rpm). Bacterial cells were collected at the logarithmic stage of growth (data not shown). The P. aeruginosa bacterial suspension was adjusted to OD600 = 0.1 and diluted to produce bacterial suspension with an infectious dose of 105 cells per mL in a 10 mM phosphate buffer solution (PBS), pH 7.4. A peristaltic pump (Minipuls evolution, Gilson Inc.) was used to introduce an infectious dose of P. aeruginosa cells into the micro-fluidic chip at a flow speed of 0.1 µL s−1. A 3.2 µL aliquot was taken from the output and incubated on agar plates for 12 hours at 37 °C (Memmert, Heraeus CO2 incubator) to allow the colony forming units to be determined. All experiments were performed at room temperature (ca. 25 °C), with at least three independent experiments being performed. Viability assays were performed using standard plate counts,34 where colonies were counted and the number of colony forming units (cfu) per millilitre was calculated. The calculated cfu numbers were assumed to be equivalent to the number of live cells present in suspension.34 The bactericidal efficiency was measured as the number of inactivated cells per cm2 of sample per minute, relative to the control surfaces. All experiments were completed within 3 h.
The bacterial solution was passed through the channel with a 10 min pause between repeated passes through the device (Fig. 2). Each experiment was repeated three times. The filling time required for the entire volume of the channel was found to be 45 s. Repeated cycles were timed in such a way that an equivalent volume of solution was used to fill the cell using a forward and reverse rotation of the pump.
The bactericidal effect of the microfluidic bSi channel was also evaluated after each cycle by staining the dead and live cells, which were then visualised. Non-viable bacterial cells are stained red with propidium iodide, whereas the living bacteria are stained green with SYTO 9 (Molecular Probes, Invitrogen, Grand Island, NY, USA). Imaging was carried out using a Fluorview FV10i Confocal System with a water immersion objective lens (UPLSAPO 60W) with a NA of 1.2 and working distance of 2 mm. This allowed a large field of view at a very high resolution 0.61λ/NA ≈ 0.5 µm for the optimised red-green spectral range of imaging; here λ is the wavelength of fluorescence. In addition, the cells of the surface of the flow channel were visualised using scanning electron microscopy (SEM). SEM images were obtained using a field-emission FESEM (ZEISS SUPRA 40VP) tool at 3 kV under magnification values of 1 k×, 5 k× and 20 k× respectively, as previously described.29
The flow channel was tested to evaluate its efficiency in achieving total cellular protein release from the ruptured E. coli K 12 cells (Fig. 2). Before each experiment, the bacterial suspension was adjusted to OD600 = 0.1. A peristaltic pump (Minipuls evolution, Gilson Inc.) was used to introduce the E. coli suspension (cell density of 3 × 108 cells per mL) into the micro-fluidic chip. The bacterial cell suspension was subjected to 20 repeated cycles through the microfluidic device at a flow speed of 0.1 μL s−1. A 50 µL portion (in triplicate) of the suspension was collected after each cycle and the resulting total protein concentration was quantified using a Bradford protein assay35–38 using a NanoDrop 2000 (ThermoFisher, Australia). The total concentration of proteins from E. coli cells lysed using enzyme treatment and sonication was determined as described elsewhere39–41 for comparative purposes.
To test whether this microfluidic device was able to be re-used, a washing procedure was adopted whereby the device was initially flushed using PBS buffer solution for 4 s, followed by washing with distilled water at speed of 5.7 µL s−1 for up to 20 cycles. A forward and backward flow switching procedure was carried out using the peristaltic pump for 2 s intervals in each flow direction, with a total washing time of 4 s being used. Several microfluidic channels were fabricated to allow a comparison of the consistency between tests.
Results
The flow cell dimensions were optimised to achieve efficient elimination of bacterial cells by restricting the instances of several bacterial cells being present within the cells on top of each other, maximising their exposure to the nano-textured surface of the black silicon. The fabricated microfluidic device contained a 2 × 1 cm2 section of bSi, with a 15 µm gap above the bactericidal surface of the bSi, which is almost a twofold reduction in available volume compared with previous cell designs.42 This reduction in volume was essential in order to ensure an efficient interaction occurred between the bSi surface and the bacterial suspension during flow. The wall effect causes a larger viscous drag near the substrate43 with a faster flow being present in the centre of the cell. This means that there was a greater probability that bacteria could be located at the centre, or mid-height, of the channel. When the flow was paused for the bacteria to come into contact with the bSi surface, the larger width and large surface area of bSi were also key features of the microfluidic chip. The peristaltic pump was pushing the P. aeruginosa cells through the channel, and a uniform advancing front of air–liquid interface was observed under the microscope, confirming the uniform height of the channel over the entire area of the bSi.The efficient bactericidal action of the bSi surface was confirmed using standard staining techniques using propidium iodide (red) for non-viable and SYTO 9 (green) for viable bacterial cells, respectively (Fig. 3). The SEM images of the bacterial cells on the bSi surface revealed changed cell morphology confirming that structural damage to the cells had occurred.
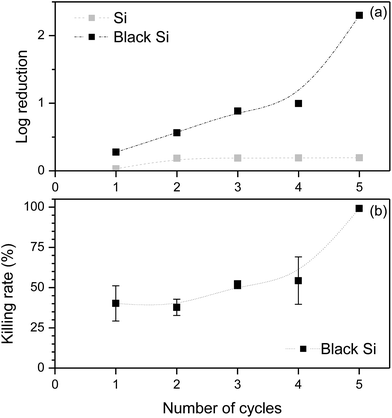
To quantify the bactericidal performance of the flow channel, a portion of bacterial solution that had passed through the microfluidic device was sampled after each cycle and plated onto agar plates. The results presented in Fig. 4(a) demonstrate that the elimination of bacterial cells from the initial suspension was dependent on the number of filtering cycles to which the initial suspension was subjected. A slight reduction in the concentration of bacterial cells was also observed for the control surface, this being likely due to adhesion of the bacterial cells onto the flow channel walls. There was, however, no evidence of damaged bacterial cells present on the microfluidic channel with the control silicon surface, as confirmed by confocal and SEM image analysis (Fig. 3).
The bactericidal efficiency of the bSi-containing microfluidic device was calculated by subtracting the extent of bacterial removal using the control microfluidic device under the same experimental conditions, the results of which are presented in Fig. 4(b). The bacterial killing rate was calculated as a log10 reduction value to analyse the bactericidal rate on a comparative scale, which revealed that up to 99% of the cells were killed after 5 consecutive cycles through the bSi-containing microfluidic device.
It is crucial to integrate cell lysis and fractionation steps to achieve a total micro analytical system for the analysis of cells and their constituent proteins on-chip, without adding extra steps.44 To determine the same functionality, the microfluidic channel was used to quantify the release of the total cellular proteins from ruptured E. coli cells from the cell suspension, which was being circulated through the channel (Fig. 5). The protein concentration was monitored after each cycle and over the entire 15 cycles. An additional 5 cycles were performed to ensure the complete extraction of proteins. Approximately 65.2 µg mL−1 of cellular protein was extracted after 15 cycles, as confirmed using Bradford's assay (Table 1). These results are in agreement with an estimated amount of 60–66 µg mL−1 of total cellular proteins, which can be obtained from 3 × 108 cells per mL, taking into account that a single E. coli cell contains 0.2 pg protein.45 Notably, it appeared that the combined enzyme and sonication treatment was less efficient at extracting the cellular proteins than the mechanical rupture method that occurred within the microfluidic device, which resulted in a total cellular protein yield of 52.7 µg mL−1.
To assess whether the bSi-containing microfluidic device was able to be cleaned and re-used, a single flush of the device using PBS buffer solution was carried out at a 5.7 µL s−1 flow rate for 4 s followed by 10 successive cycles of MilliQ water. Each flush was carried out at a rate of 5.7 µL s−1 for a period of 4 s, followed by a forward and backward flow for 2 s each. After the washing cycle, there was no evidence of any viable bacteria being present within the device, as confirmed by the direct colony counting technique. The total time required for cleaning the microfluidic device was 1 minute.
Discussion
The 10 minute flow stoppage that occurred during the bacterial flow through the microfluidic device was undertaken to allow the bacterial cells sufficient time to come into contact with the bSi surface within the cell. The average thermal velocity that occurs through this process was estimated by assuming equality between the kinetic and thermal energies taking place. This was calculated using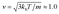 mm s−1, where kb is the Boltzmann constant, T = 293 K is the absolute temperature at normal conditions, and m ∼10.0 pg is the mass of a single P. aeruginosa cell. The mean displacement of bacteria over time (t) occurs due to Brownian motion, and is calculated according to
mm s−1, where kb is the Boltzmann constant, T = 293 K is the absolute temperature at normal conditions, and m ∼10.0 pg is the mass of a single P. aeruginosa cell. The mean displacement of bacteria over time (t) occurs due to Brownian motion, and is calculated according to 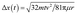 is, where µ = 8.9 × 10−4 Pa s is the dynamic viscosity of water. It takes only t ∼ 2 min for bacteria to move over Δx(t) = 15 µm which is comparable with the height of the microfluidic channel. During the total exposure period, it is therefore almost certain that the bacteria would have come into contact with the bSi surface within the microfluidic device.
is, where µ = 8.9 × 10−4 Pa s is the dynamic viscosity of water. It takes only t ∼ 2 min for bacteria to move over Δx(t) = 15 µm which is comparable with the height of the microfluidic channel. During the total exposure period, it is therefore almost certain that the bacteria would have come into contact with the bSi surface within the microfluidic device.
Further studies are now required to systematically investigate the bactericidal action of the bSi-containing microfluidic device against other bacterial types, and to determine the maximum flow rate that can be used that will still achieve an efficient level of bactericidal action.
In our previous study,42 where a very high flow speed of more than 1 m s−1 were used when pumping polystyrene bead suspensions through a microfluidic device with channels containing sharp micron-sized features, the beads were seen to be able to avoid contact with the sharp features by following the laminar flow within the channel. Since the bSi components of the microfluidic devices used in the current study were prepared from 4-inch wafers, it is possible to construct a microfluidic device that contains wider and longer bSi sections, and perhaps to also contain multiple bSi channels for sequentially treating bacterial solutions. Studies involving such components would reveal whether it is possible to design a device for testing the bacterial contamination of grey water. The bactericidal action of the bSi component of the microfluidic devices could be further improved by incorporating ultraviolet light-emitting diodes, making such devices applicable to a wide range of water disinfection and sterilisation applications.46–50 The microfluidic device reported here has demonstrated that it has a high sensitivity for the refractive index of the solution being used; Δλ/Δn = 390 nm per RIU (refractive index units)20 and could be further assessed for its ability to recognise bacterial (or other) contamination by incorporating a section within the device that contains Fabry–Pérot mirrors on the upper and lower walls of the channel. Such a microfluidic device would allow the in situ monitoring of refractive index changes that could be related to the removal of bacteria from solution.
An additive pressure (ΔP), scales linearly with the flow velocity (v) in the microfluidic device, for a liquid of viscosity (µ), inside a channel with the transverse dimension height (t), width (w) and length (l) according to: ΔP ∝ µvl/(tw).51 Hence for a longer and narrower channel, ΔP would increase with the second power of the decreasing geometrical length and height dimensions. To maintain a high throughput flow at as large as practical, the width of the channel should be increased.
Micro and nano-fabricated devices have been designed to expedite applied and basic research into cell biology and morphology in dynamic flow through these devices.52–55 Protein analysis and quantification in clinical samples, such as blood serum or whole-cell lysates presents certain challenges56 such as in the pre-treatment or fractionation of complex samples for integration into the micro-analysis systems. Several groups have developed microchip- or capillary-based two-dimensional separation systems, which are an integration of micellar electrokinetic chromatography (MEKC) or isoelectric focusing (IEF) with capillary electrophoresis.57–61 These reports are, however limited by the fact that they are quite complex systems, and a number of additional steps become involved in the separation and identification of the desired protein analyte. A study has also been published where E. coli cells have been lysed using a simple channel mechanism.62 This study, however, uses detergent addition as an additional step to lyse the cells in flow, with the height and depth of the cell being 1000 µm and 100 µm, respectively. Such width and height could be detrimental to the attachment pattern of bacteria, which are not more than 5 µm in their average dimension. Another study incorporated a surface chemistry technique to detect E. coli cells in clinical samples, where E. coli cells have been made to interact with specific antibodies for detection.63 The device reported in this current study avoids these extra steps, as the bactericidal bSi surfaces have already been incorporated inside the channel to lyse the cells. Moreover, the height of the channel is such that it ensures the bacteria coming into a contact with the bSi. Moreover, the width of channel can be controlled by mechanically applied pressure.
Conclusions
In this study, a simple method was used to fabricate a microfluidic device containing a channel that was 15 µm in height over a relatively large 2 cm2 area. Incorporation of bSi into the device design resulted in a device that was bactericidal when flow into the cell was paused for 10 min after filling the cell, which took 45 s. Approximately 99% of P. aeruginosa cells from the infectious dose were eliminated after 5 successive fill-stop cycles through the device. The bacterial killing rate was found to be 2.3 × 103 cfu min−1 cm−2 for one cycle. The newly designed microfluidic device was also used for the effective extraction of cellular protein from the ruptured E. coli bacterial cells. The device was shown to be able to be re-used after washing using a simple high speed flushing process.The proposed simple microfluidic device containing the nanotextured bSi could be used for surface enhanced Raman spectroscopy applications under the required flow conditions. The device also has the capacity of incorporating electrodes to carry out electrochemical surface cleaning (oxidation of adventitious carbon) or/and removal of oxide.52 Such flow devices that have the potential for controlling the electrochemical potential are expected to be useful for investigations into detailed surface chemical reactions and catalysis, e.g., in the generation of hydrogen.64,65 The evolution of oxygen through such electrodes could be achieved if an oxidative stress could be applied to the bacterial cells. The incorporation of UV-LED treatment sections into the device would also be possible.
Acknowledgements
Partial support was obtained for this research through an Australian Research Council DP130101205 grant. In addition, the collaborative research project being undertaken with Altechna Ltd., of which this work is a part, is acknowledged. The technical assistance of Dr SHT Nguyen is gratefully acknowledged. X.W.W. and C.M.B. are recipients of Swinburne University Postgraduate Awards. SJ acknowledges the start-up funding of the Nanotechnology facility, available through a strategic infrastructure grant provided from Swinburne University of Technology.References
- W. Zhuang, D. Yuan, J. R. Li, Z. Luo, H. C. Zhou, S. Bashir and J. Liu, Adv. Healthcare Mater., 2012, 1, 225–238 CrossRef CAS PubMed.
- E. P. Ivanova, J. Hasan, H. K. Webb, V. K. Truong, G. S. Watson, J. A. Watson, V. A. Baulin, S. Pogodin, J. Y. Wang and M. J. Tobin, Small, 2012, 8, 2489–2494 CrossRef CAS PubMed.
- A. H. Broderick, A. S. Breitbach, R. Frei, H. E. Blackwell and D. M. Lynn, Adv. Healthcare Mater., 2013, 2, 993–1000 CrossRef CAS PubMed.
- A. Jain, L. S. Duvvuri, S. Farah, N. Beyth, A. J. Domb and W. Khan, Adv. Healthcare Mater., 2014, 3, 1969–1985 CrossRef CAS PubMed.
- M. Sakai, M. Nishimura, Y. Morii, T. Furuta, T. Isobe, A. Fujishima and A. Nakajima, Journal of Materials Science, 2012, 47, 8167–8173 CrossRef CAS.
- T. S. Santra and F. G. Tseng, Micromachines, 2013, 4, 333–356 CrossRef.
- Y. Kikuchi, K. Sunada, T. Iyoda, K. Hashimoto and A. Fujishima, J. Photochem. Photobiol., A, 1997, 106, 51–56 CrossRef CAS.
- K. Kairyte, A. Kadys and Z. Luksiene, J. Photochem. Photobiol., B, 2013, 128, 78–84 CrossRef CAS PubMed.
- M. V. Graham and N. C. Cady, Coatings, 2014, 4, 37–59 CrossRef CAS.
- C. Guimarães, J. An, M. Humar, W. Goth and A. Yun, Proc. SPIE BiOS, 2015, 93410R Search PubMed.
- J. Hasan, R. J. Crawford and E. P. Ivanova, Trends Biotechnol., 2013, 31, 295–304 CrossRef CAS PubMed.
- H. K. Webb, R. J. Crawford and E. P. Ivanova, in Antibacterial Surfaces, Springer, 2015, pp. 1–8 Search PubMed.
- G. D. Bixler and B. Bhushan, Crit. Rev. Solid State Mater. Sci., 2015, 40, 1–37 CrossRef CAS.
- M. V. Graham, A. P. Mosier, T. R. Kiehl, A. E. Kaloyeros and N. C. Cady, Soft Matter, 2013, 9, 6235–6244 RSC.
- K.-C. Park, H. J. Choi, C.-H. Chang, R. E. Cohen, G. H. McKinley and G. Barbastathis, ACS Nano, 2012, 6, 3789–3799 CrossRef CAS PubMed.
- A. Mavrokefalos, S. E. Han, S. Yerci, M. S. Branham and G. Chen, Nano Lett., 2012, 12, 2792–2796 CrossRef CAS PubMed.
- Y. Liu, T. Lai, H. Li, Y. Wang, Z. Mei, H. Liang, Z. Li, F. Zhang, W. Wang and A. Y. Kuznetsov, Small, 2012, 8, 1392–1397 CrossRef CAS PubMed.
- H. Hauser, B. Michl, S. Schwarzkopf, V. Kubler, C. Muller, M. Hermle and B. Blasi, IEEE Journal of Photovoltaics, 2012, 2, 114–122 CrossRef.
- S. Y. Reece, J. A. Hamel, K. Sung, T. D. Jarvi, A. J. Esswein, J. J. Pijpers and D. G. Nocera, Science, 2011, 334, 645–648 CrossRef CAS PubMed.
- B.-R. Huang, Y.-K. Yang, T.-C. Lin and W.-L. Yang, Sol. Energy Mater. Sol. Cells, 2012, 98, 357–362 CrossRef CAS.
- H. Savin, P. Repo, G. von Gastrow, P. Ortega, E. Calle, M. Garín and R. Alcubilla, Nat. Nanotechnol., 2015, 10, 624–628 CrossRef CAS PubMed.
- L. Sainiemi, V. Jokinen, A. Shah, M. Shpak, S. Aura, P. Suvanto and S. Franssila, Adv. Mater., 2011, 23, 122–126 CrossRef CAS PubMed.
- M. Otto, M. Kroll, T. Käsebier, R. Salzer, A. Tünnermann and R. B. Wehrspohn, Appl. Phys. Lett., 2012, 100, 191603 CrossRef.
- M. Gaudig, J. Hirsch, T. Schneider, A. N. Sprafke, J. Ziegler, N. Bernhard and R. B. Wehrspohn, J. Vac. Sci. Technol., A, 2015, 33, 05E132 Search PubMed.
- K. Nguyen, D. Abi-Saab, P. Basset, E. Richalot, M. Malak, N. Pavy, F. Flourens, F. Marty, D. Angelescu and Y. Leprince-Wang, Microsyst. Technol., 2012, 18, 1807–1814 CrossRef CAS.
- K. Nguyen, P. Basset, F. Marty, Y. Leprince-Wang and T. Bourouina, J. Appl. Phys., 2013, 113, 194903 CrossRef.
- H. Jansen, M. De Boer, S. Unnikrishnan, M. Louwerse and M. Elwenspoek, J. Micromech. Microeng., 2009, 19, 033001 CrossRef.
- G. Seniutinas, G. Gervinskas, R. Verma, B. D. Gupta, F. Lapierre, P. R. Stoddart, F. Clark, S. L. McArthur and S. Juodkazis, Opt. Express, 2015, 23, 6763–6772 Search PubMed.
- E. P. Ivanova, J. Hasan, H. K. Webb, G. Gervinskas, S. Juodkazis, V. K. Truong, A. H. Wu, R. N. Lamb, V. A. Baulin and G. S. Watson, Nat. Commun., 2013, 4, 2838 Search PubMed.
- G. Gervinskas, D. Day and S. Juodkazis, Sens. Actuators, B, 2011, 159, 39–43 CrossRef CAS.
- S. Pogodin, J. Hasan, V. A. Baulin, H. K. Webb, V. K. Truong, T. H. P. Nguyen, V. Boshkovikj, C. J. Fluke, G. S. Watson and J. A. Watson, Biophys. J., 2013, 104, 835–840 CrossRef CAS PubMed.
- E. P. Ivanova, V. K. Truong, G. Gervinskas, N. Mitik-Dineva, D. Day, R. T. Jones, R. J. Crawford and S. Juodkazis, Biosens. Bioelectron., 2012, 35, 369–375 CrossRef CAS PubMed.
- I. Vallet-Gely and F. Boccard, PLoS Genet., 2013, 9, e1003492 CAS.
- J. Postgate, Methods Microbiol., 1969, 1, 611–628 Search PubMed.
- H.-K. Ku, H.-M. Lim, K.-H. Oh, H.-J. Yang, J.-S. Jeong and S.-K. Kim, Anal. Biochem., 2013, 434, 178–180 CrossRef CAS PubMed.
- S. C. Silvério, S. Moreira, A. M. Milagres, E. A. Macedo, J. A. Teixeira and S. I. Mussatto, Anal. Biochem., 2012, 421, 719–724 CrossRef PubMed.
- N. Carlsson, A. Borde, S. Wölfel, B. Åkerman and A. Larsson, Anal. Biochem., 2011, 411, 116–121 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- P. Shrestha, T. M. Holland and B. C. Bundy, BioTechniques, 2012, 53, 163–174 CAS.
- L. Benov and J. Al-Ibraheem, J. Biochem. Mol. Biol., 2002, 35, 428–431 CrossRef CAS PubMed.
- R. Mehigh, Sigma Origins 3, 2001, pp. 15–16 Search PubMed.
- G. Gervinskas, D. J. Day and S. Juodkazis, Opt. Mater. Express, 2012, 2, 279–286 CrossRef.
- M. Miwa, S. Juodkazis and H. Misawa, Jpn. J. Appl. Phys., 2000, 39, 1930 CrossRef CAS.
- K. Aran, A. Fok, L. A. Sasso, N. Kamdar, Y. Guan, Q. Sun, A. Ündar and J. D. Zahn, Lab Chip, 2011, 11, 2858–2868 RSC.
- G. Carta and A. Jungbauer, Protein Chromatography: Process Development and Scale-Up, Wiley, 2010 Search PubMed.
- K. Y. Nelson, D. W. McMartin, C. K. Yost, K. J. Runtz and T. Ono, Environ. Sci. Pollut. Res., 2013, 20, 5441–5448 CrossRef CAS PubMed.
- M. Shatalov, A. Lunev, X. Hu, O. Bilenko, I. Gaska, W. Sun, J. Yang, A. Dobrinsky, Y. Bilenko and R. Gaska, Int. J. High Speed Electron. Syst., 2012, 21, 1250011 CrossRef.
- A. Venancio-Marques, F. Barbaud and D. Baigl, J. Am. Chem. Soc., 2013, 135, 3218–3223 CrossRef CAS PubMed.
- M. Würtele, T. Kolbe, M. Lipsz, A. Külberg, M. Weyers, M. Kneissl and M. Jekel, Water Res., 2011, 45, 1481–1489 CrossRef PubMed.
- M. Mori, A. Hamamoto, A. Takahashi, M. Nakano, N. Wakikawa, S. Tachibana, T. Ikehara, Y. Nakaya, M. Akutagawa and Y. Kinouchi, Med. Biol. Eng. Comput., 2007, 45, 1237–1241 Search PubMed.
- P. Tabeling, Introduction to microfluidics, Oxford University Press, 2010 Search PubMed.
- R. Buividas, N. Fahim, J. Juodkazytė and S. Juodkazis, Appl. Phys. A: Mater. Sci. Process., 2014, 114, 169–175 CrossRef CAS.
- F. Wu and C. Dekker, Chem. Soc. Rev., 2016, 45, 268–280 RSC.
- D. Gao, H. Liu, Y. Jiang and J.-M. Lin, TrAC, Trends Anal. Chem., 2012, 35, 150–164 CrossRef CAS.
- E. K. Sackmann, A. L. Fulton and D. J. Beebe, Nature, 2014, 507, 181–189 CrossRef CAS PubMed.
- J. El-Ali, P. K. Sorger and K. F. Jensen, Nature, 2006, 442, 403–411 CrossRef CAS PubMed.
- Y.-C. Wang, M. H. Choi and J. Han, Anal. Chem., 2004, 76, 4426–4431 CrossRef CAS PubMed.
- R. D. Rocklin, R. S. Ramsey and J. M. Ramsey, Anal. Chem., 2000, 72, 5244–5249 CrossRef CAS PubMed.
- J. Mok, M. N. Mindrinos, R. W. Davis and M. Javanmard, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 2110–2115 CrossRef CAS PubMed.
- I. M. Lazar and J. L. Kabulski, Lab Chip, 2013, 13, 2055–2065 RSC.
- R. Hu, X. Feng, P. Chen, M. Fu, H. Chen, L. Guo and B.-F. Liu, J. Chromatogr. A, 2011, 1218, 171–177 CrossRef CAS PubMed.
- E. A. Schilling, A. E. Kamholz and P. Yager, Anal. Chem., 2002, 74, 1798–1804 CrossRef CAS PubMed.
- S. Wang, F. Inci, T. L. Chaunzwa, A. Ramanujam, A. Vasudevan, S. Subramanian, A. C. F. Ip, B. Sridharan, U. A. Gurkan and U. Demirci, Int. J. Nanomed., 2012, 7, 2591 CAS.
- J. Juodkazytė, B. Šebeka and S. Juodkazis, Appl. Surf. Sci., 2014, 290, 13–17 CrossRef.
- K. Juodkazis, J. Juodkazytė, E. Jelmakas, P. Kalinauskas, I. Valsiūnas, P. Miečinskas and S. Juodkazis, Opt. Express, 2010, 18, A147–A160 CrossRef CAS PubMed.
Footnote |
| † X. W. W. and C. M. B. have contributed equally. |
| This journal is © The Royal Society of Chemistry 2016 |