Stable isotope labeling to study the nitrogen metabolism in microcystin biosynthesis†
Abstract
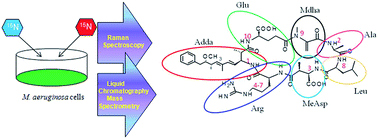
Cyanobacterial blooms have occurred frequently in the surface waters of the world, posing a potential threat to humans and ecosystems due to their toxic secondary metabolite, microcystin. Genetic tools cannot easily explain the biosynthesis of MC in specific strains due to the iterative nature of MC production and environmental factors. In this study, the fully 15N-labeled MC-LR was biosynthesized successfully in Microcystis aeruginosa by in vivo stable isotopic enrichment. The biosynthesis and metabolic flux of MC-LR were explored through Raman spectroscopy and liquid chromatography-mass spectrometry (LC-MS). The results showed that the essential amino acids that were incorporated into MC-LR were all labeled by 15N. The Raman spectra revealed that most of the bands in 6th cell transfer MC-LR were shifted to lower wavenumbers due to the substitution by the heavier isotope 15N. MS indicated that fully 15N-labeled Arg and MC-LR were the dominant species from the 2nd and 6th cell transfer M. aeruginosa, respectively. Through MS–MS fragmentation analysis, Adda was the first unit to be synthesized, followed by Ala, MeAsp, Arg, Leu, Mdha and Glu. These results provide new evidence and an ideal tool to explore the biosynthesis of MC-LR in the specific strain of M. aeruginosa.

 Please wait while we load your content...
Please wait while we load your content...