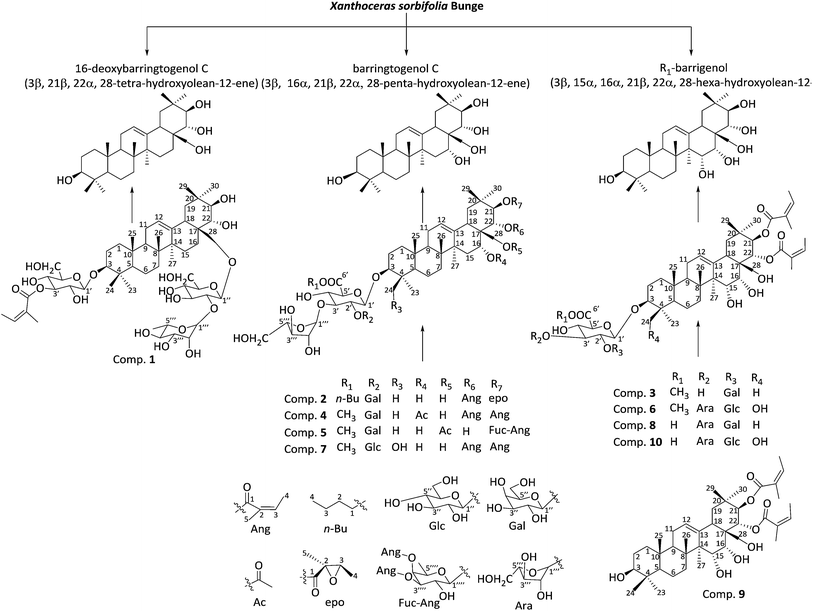
Barrigenol triterpenes from the husks of Xanthoceras sorbifolia Bunge and their antitumor activities†
Da Wanga,
Dan Sua,
Xian-Zhe Lia,
Dan Liua,
Rong-Gang Xib,
Hui-Yuan Gao*a and
Xiao-Bo Wang*b
aKey Laboratory of Structure-Based Drug Design & Discovery of Ministry of Education, Shenyang Pharmaceutical University, Shenyang 110016, People's Republic of China. E-mail: sypugaohy@163.com; Fax: +86 2423986460; Tel: +86 2423986481
bDepartment of Pharmacy, 210th Hospital of People's Liberation Army, Dalian, 116021, People's Republic of China. E-mail: wxbbenson0653@sina.com; Fax: +86 41139675460; Tel: +86 41185841066
First published on 11th March 2016
Abstract
Barrigenol-like triterpenoids containing angeloyl residues in their structures are unusual natural products. To discover the chemical constituents responsible for the antitumor activity of Xanthoceras sorbifolia Bunge, the present study was carried out using the husk of this crop. Ten angeloyl barrigenol triterpenoids, including seven new (1–7) and three known compounds (8–10), were isolated from the active fraction via the bioassay-guided method. The structures of the compounds were established on the basis of spectral analysis, especially according to the data afforded by two digital-NMR and high-resolution mass spectra experiments. Compounds 1–10 exhibited varying degrees of cytotoxicity toward the human hepatoma cell line (HepG2), the human colorectal cancer cell line (HCT116) and the human glioma cell line (U87-MG). New compounds 3, 6, and 7 and known compounds 8, and 10 showed inhibitory activities similar to those of the positive control (doxorubicin hydrochloride). Cell cycle and apoptosis analysis of compound 8 revealed that it could suppress U87-MG cell proliferation by inducing apoptosis in the early period of exposure and then promote arrest at the G0/G1 phase.
Introduction
Barrigenol-type triterpenoids, which mainly come from the Barringtoniaceae, Hippocastanaceae and Sapindaceae family plants, exhibit antitumor activity, neuroprotective function, and anti-inflammatory activity.1–3 Barrigenol triterpenoids may be mainly classified into five types according to differences in substituent groups linked at the C-15, C-16, C-21, and C-22 positions:3,4 barrigenol R1, barrigenol A1, barrigenol A2, barringtogenol C, and 16-deoxybarringtogenol C (Fig. 1). Barrigenol derivatives with angeloyl or tiggeloyl groups at C-21 and/or C-22 position show more interesting bioactivities than the corresponding derivatives without these residues. Sodium aescinate, a common example of this type of triterpenoid, is made up of a mixture of barrigenol type triterpenoids and the major active components of Aesculus hippocastanum seeds, this drug is widely applied in the clinical treatment of brain swelling and venous reflux disease.4,5Xanthoceras sorbifolia Bunge, a woody oil crop from the Sapindaceae family, is also known as the yellow horn or Chinese flowering chestnut in China. This plant is also notable for its seed oil (rich in unsaturated fatty acids, 85–93%) and considerable value in the food industry;6 however, the husks of its fruit is generally discarded as agriculture residues. The wood and fruit of the plant may also be used as an ethno-medicine to help cure rheumatism, gout, and enuresis in children.7 Husk extracts of this plant have been shown to exhibit anti-inflammatory, anti-HIV, anti-tumor, and anti-Alzheimer's activities.3 Previous chemical studies showed that triterpenoids, especially barrigenol type triterpenoids, are the main active components of the plant responsible for these activities.8–11 Thus, determining more active barrigenol triterpenoids and exploiting the medicinal and economic values of this plant is a worthwhile endeavor. In this study, alcohol extracts of the husk of X. sorbifolia were found to show potent inhibitory activity toward human carcinoma cell lines through the bioassay-guided method, further chemical study led to seven new barrigenol type triterpenoids (1–7) and three known compounds, namely, xanifolia Y (8),11 xanifolia ACH-Y (9),10 xanifolia Y2 (10)8 (Fig. 1). The inhibitory activities of these compounds against three tumor cell lines were further evaluated in vitro.
Results and discussion
The ethanol extract of X. sorbifolia husks was dispersed in distilled water and extracted successively with different solvents. The n-BuOH fraction showed marked inhibitory effects against the proliferation of human colorectal cancer (HCT-116), human hepatoma (HepG2), and human glioma (U87-MG) cell lines with IC50 values of 15.32, 6.74, and 16.31 μg ml−1, respectively. This fraction was subsequently subjected to silica gel column chromatography, Sephadex LH-20 column chromatography, preparative RP-18 HPLC, and flash ODS column chromatography to yield a total of 10 barrigenol triterpenoids.Characterization of compounds 1–10
Compound 1 was obtained as white amorphous powder (CH3OH), which gave a molecular formula of C53H86O19 with 11 degrees of unsaturation, as deduced by a pseudomolecular ion peak of positive HR-ESI-MS spectrum (m/z 1049.5656 [M + Na]+, calcd for C53H86O19Na, 1049.5661). The IR spectrum of this compound showed the absorption bands of carbonyl (1719 cm−1) and hydroxyl (3423 cm−1) groups. 1H-NMR (600 MHz, pyridine-d5) displayed seven diagnostic methyl proton signals at δH, 0.92, 1.01, 1.13, 1.23, 1.26, 1.30, 1.33 (3H, s); one olefinic proton at δH 5.49 (1H, brs); three anomeric proton signals of sugar moieties at δH 4.79 (1H, d, J = 7.9 Hz), δH 4.97 (1H, d, J = 7.7 Hz), and δH 6.68 (1H, brs); and a group of signals for an angeloyl unit at δH 1.90 (3H, s), δH 2.00 (3H, d, J = 7.1 Hz), δH 5.87 (1H, q, J = 7.1 Hz) (Table 1). The 13C-NMR spectrum (Table 1) of 1 showed seven methyl carbon signals at δC 15.7, 16.8, 16.9, 19.6, 26.3, 28.1, 30.4; a pair of olefinic carbon signals at δC 123.9, 143.2; three anomeric carbon signals at δC 100.5, 103.6, and 106.7; and a group of angeloyl signals at δC 15.9, 20.8, 129.0, 136.8, 168.2. These data confirmed that compound 1 was a barringtogenol triterpenoid with an aglycon of 16-deoxybarringtogenol C, three sugar moieties, and an angeloyl group in its structure.12 The presence of two D-glucose residues and one L-rhamnose residue was confirmed using a combination of HSQC, HMBC spectral analysis, and chemical reactions. Acid hydrolysis of compound 1 with aqueous 2 M HCl yielded D-glucose and L-rhamnose, which were identified through HPLC analysis using an optical rotation detector.13 Two anomeric proton signals at δH 4.97 (d, J = 7.7 Hz) and 4.79 (d, J = 7.9 Hz) were assigned to two D-glucose units, and the anomeric proton signal at δH 6.68 (1H, brs) was assigned to L-rhamnose. In the HMBC spectrum (Fig. 2) of 1, long-range correlations between the anomeric proton signal of Glc-H-1′ (δH 4.97) and C-3 (δC 89.0), as well as between Glc-H-1′′ (δH 4.79) and C-28 (δC 74.3), suggested that two glucopyranosyl groups were linked at the C-3 and C-28 positions, respectively. Moreover, the presence of a cross-peak between the proton signal of Glc-H-3′ (δH 6.03) and the carbon signal at δC 168.2 (Ang-C-1) suggested that compound 1 was an unusual barringtogenol triterpenoid containing the angeloyl residue in its sugar part. 13C-NMR spectrum showed that the carbon signal of Glc-C-2′′ moved to 80.3, thereby implying that the α-L-rhamnopyranosyl was attached to this glucose unit at the C-2′′ position. The stereochemistry of 1 was confirmed by a NOESY experiment (Fig. 3). The presence of NOE correlations between H-3 (δH 3.3) and CH3-23 (δH 1.34), between H-5 (δH 0.79) and H-3, between H-5 and H-9 (δH 1.68), as well as between H-18 (δH 2.81) and CH3-30 (δH 1.23) were in good agreement with the ring conjunction reported for barringtogenol derivatives.2 NOE correlations between H-22 (δH 4.35) and CH3-30 (δH 1.23), as well as between H-21 (δH 3.78) and CH3-29 (δH 1.26), indicted that C-21-OH presented an β form whereas C-22-OH presented an α form. Based on the above analysis, compound 1 was identified as 3-O-(3′-O-angeloyl)-β-D-glucopyranosyl-28-O-[α-L-rhamnopyranosyl(1→2)]-β-D-glucopyranosyl-3β,21β,22α,28-tetrahydroxy-olean-12-ene, which is a 16-deoxybarringtogenol C-type barringtogenol triterpenoid.| No. | 1b | No. | 5b | 6b | 7c | ||||
|---|---|---|---|---|---|---|---|---|---|
| δH J (Hz) | δC | δH J (Hz) | δC | δH J (Hz) | δC | δH J (Hz) | δC | ||
| a “o” overlapped.b 600 MHz for 1H-NMR, 150 MHz for 13C-NMR.c 300 MHz for 1H-NMR, 150 MHz for 13C-NMR. | |||||||||
| 1 | 0.80 m, 1.36 m | 39.5 | 1 | 0.82 m, 1.35 m | 38.7 | 0.83 m, 1.40 m | 38.6 | 0.79 m, 1.38 o | 38.4 |
| 2 | 1.81 m, 2.01 m | 25.8 | 2 | 1.80 m, 1.99 m | 26.5 | 1.83 m, 2.18 m | 26.6 | 1.88 o, 2.19 m | 26.5 |
| 3 | 3.37 (dd, 11.7, 4.4) | 89.0 | 3 | 3.18 (dd, 11.6, 4.3) | 90.0 | 3.41 (dd, 11.5, 4.0) | 91.5 | 3.42 o | 91.6 |
| 4 | — | 38.7 | 4 | — | 40.4 | — | 43.6 | — | 43.6 |
| 5 | 0.79 (d, 12.1) | 55.6 | 5 | 0.69 (d, 11.7) | 55.7 | 0.91 (d, 10.6) | 55.9 | 0.85 o | 56.0 |
| 6 | 1.46 o, 1.61 m | 18.4 | 6 | 1.21 m, 1.43 m | 18.4 | 0.91 m, 1.66 m | 18.8 | 1.19 m, 1.52 m | 18.4 |
| 7 | 1.11 o, 1.22 m | 32.8 | 7 | 1.19 m, 1.50 m | 33.0 | 2.02 m, 2.18 m | 36.5 | 1.22 m, 1.54 m | 33.1 |
| 8 | — | 40.3 | 8 | — | 39.6 | — | 41.0 | — | 39.9 |
| 9 | 1.68 o | 47.9 | 9 | 1.65 m | 46.6 | 1.67 m | 46.9 | 1.66 m | 46.7 |
| 10 | 36.8 | 10 | — | 36.7 | — | 36.8 | — | 36.4 | |
| 11 | 1.83 o, 1.76 m | 24.0 | 11 | 1.81 m, 1.86 m | 23.8 | 1.84 m, 1.89 m | 24.2 | 1.80 m, 1.88 m | 24.0 |
| 12 | 5.49 brs | 123.9 | 12 | 5.40 brs | 123.5 | 5.52 brs | 125.2 | 5.97 brs | 124.1 |
| 13 | — | 143.2 | 13 | — | 142.9 | — | 143.7 | — | 142.7 |
| 14 | — | 41.9 | 14 | — | 41.8 | — | 47.7 | — | 41.6 |
| 15 | 1.68 o, 1.85 o | 26.5 | 15 | 1.57 m, 1.86 o | 34.8 | 4.26 o | 67.5 | 1.60 m, 1.88 o | 34.8 |
| 16 | 5.56 brs | 18.4 | 16 | 4.80 o | 67.9 | 4.47 (d, 4.6) | 73.5 | 4.50 o | 68.6 |
| 17 | — | 43.2 | 17 | — | 46.9 | — | 48.3 | — | 48.0 |
| 18 | 2.81 (dd, 13.9, 4.6) | 41.3 | 18 | 2.78 (dd, 4.0, 13.9) | 40.0 | 3.13 o | 41.3 | 3.12 o | 40.0 |
| 19 | 1.47 o, 3.12 o | 46.4 | 19 | 3.08 (t, 13.6), 1.35 o | 47.6 | 1.44 m, 3.13 o | 46.9 | 1.41 m, 3.12 o | 47.2 |
| 20 | — | 36.4 | 20 | — | 37.1 | — | 36.3 | — | 36.3 |
| 21 | 3.78 (d, 9.9) | 76.8 | 21 | 4.80 (d, 9.3) | 92.0 | 6.76 (d, 10.3) | 78.4 | 6.70 (d, 10.2) | 78.7 |
| 22 | 4.35 o | 72.7 | 22 | 4.37 (d, 9.5) | 71.4 | 6.39 (d, 10.3) | 73.3 | 6.30 (d, 10.2) | 73.5 |
| 23 | 1.33 s | 28.1 | 23 | 1.23 s | 27.9 | 1.37 s | 22.2 | 1.37 s | 22.2 |
| 24 | 1.01 s | 16.9 | 24 | 1.11 s | 16.6 | 3.33 m, 4.25 m | 63.1 | 3.33 m, 4.24 m | 63.2 |
| 25 | 0.92 s | 15.7 | 25 | 0.78 s | 15.6 | 0.69 s | 15.7 | 0.75 s | 15.5 |
| 26 | 1.13 s | 16.8 | 26 | 0.93 s | 16.9 | 0.97 s | 17.3 | 0.89 s | 16.6 |
| 27 | 1.30 s | 26.3 | 27 | 1.79 s | 27.5 | 1.87 s | 21.0 | 1.84 s | 27.5 |
| 28 | 4.12 (d, 9.4), 4.39 (d, 9.4) | 74.3 | 28 | 4.32 (d, 10.9), 4.21 (d, 10.9) | 66.4 | 3.53 (d, 10.1), 3.79 (d, 10.1) | 63.3 | 3.44 (d, 10.4), 3.68 (d, 10.4) | 63.5 |
| 29 | 1.26 s | 30.4 | 29 | 1.45 s | 29.8 | 1.14 s | 29.5 | 1.14 s | 29.5 |
| 30 | 1.23 s | 19.6 | 30 | 1.32 s | 20.5 | 1.39 s | 20.2 | 1.38 s | 20.3 |
| C-3 | Glc | C-3 | GlcA | GlcA | GlcA | ||||
| 1′ | 4.97 (d, 7.7) | 106.7 | 1′ | 4.83 (d, 7.6) | 105.1 | 4.90 (d, 7.7) | 104.7 | 4.87 (d, 7.5) | 104.8 |
| 2′ | 4.10 m | 73.6 | 2′ | 4.41 o | 78.6 | 4.26 o | 78.2 | 4.27 o | 78.7 |
| 3′ | 6.03 (t, 9.5) | 79.4 | 3′ | 4.13 o | 85.4 | 4.23 o | 86.3 | 4.21 o | 86.3 |
| 4′ | 4.62 brs | 69.5 | 4′ | 4.22 o | 71.2 | 4.36 o | 71.5 | 4.33 o | 71.5 |
| 5′ | 4.00 m | 76.8 | 5′ | 3.91 o | 76.7 | 4.51 (d, 9.6) | 76.4 | 4.47 o | 76.4 |
| 6′ | 4.58 m, 4.49 m | 62.3 | 6′ | — | 170.0 | — | 169.8 | — | 169.8 |
| C-28 | Glc | C-6′ | O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3 H3 |
O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3 H3 |
O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3 H3 |
||||
| 1′′ | 4.79 (d, 7.9) | 103.6 | 1 | 3.71 s | 52.2 | 3.81 s | 52.3 | 3.82 s | 52.3 |
| 2′′ | 4.51 m | 80.3 | C-2′ | Gal | Glc | Glc | |||
| 3′′ | 3.85 m | 77.9 | 1′′ | 5.28 (d, 7.7) | 104.8 | 5.51 (d, 7.8) | 103.9 | 5.47 (d, 7.8) | 103.9 |
| 4′′ | 4.36 brs | 71.7 | 2′′ | 4.43 o | 73.4 | 4.07 (t, 8.5) | 75.5 | 4.05 (t, 8.5) | 75.5 |
| 5′′ | 4.38 m | 78.1 | 3′′ | 4.05 (dd, 11.3, 3.3) | 75.1 | 4.84 m | 77.8 | 4.79 o | 77.8 |
| 6′′ | 4.54 m, 4.44 m | 62.6 | 4′′ | 4.40 o | 70.0 | 4.59 m | 69.5 | 4.61 m | 69.5 |
| C-2′′ | Rha | 5′′ | 3.90 o | 76.7 | 4.28 o | 78.2 | 4.27 o | 78.2 | |
| 1′′′ | 6.68 brs | 100.5 | 6′′ | 4.41 o, 4.49 m | 61.8 | 4.34 o, 4.47 o | 61.3 | 4.33 o, 4.47 o | 61.3 |
| 2′′′ | 4.70 o | 72.6 | C-3′ | Ara | Ara | Ara | |||
| 3′′′ | 4.65 o | 72.3 | 1′′′ | 5.97 (d, 2.2) | 111.0 | 6.06 (d, 2.2) | 111.1 | 5.98 (d, 2.1) | 111.2 |
| 4′′′ | 4.32 o | 74.9 | 2′′′ | 4.91 o | 83.5 | 5.01 m | 83.6 | 4.95 m | 83.6 |
| 5′′′ | 4.85 o | 69.1 | 3′′′ | 4.76 o | 77.6 | 4.83 m | 77.5 | 4.78 o | 77.8 |
| 6′′′ | 1.89 o | 18.9 | 4′′′ | 4.12 o | 85.9 | 4.85 m | 85.4 | 4.77 o | 85.4 |
| C-3′ | Ang | 5′′′ | 4.10 m, 4.25 o | 62.3 | 4.21 m, 4.36 o | 62.3 | 4.15 o, 4.28 o | 62.3 | |
| 1 | — | 168.2 | C-21 | Fuc | Ang | Ang | |||
| 2 | — | 129.0 | 1′′′′ | 4.92 (d, 7.9) | 106.2 | — | 167.7 | — | 167.7 |
| 3 | 5.87 (q, 7.1) | 136.8 | 2′′′′ | 4.54 (d, 2.9) | 69.7 | — | 128.9 | — | 128.9 |
| 4 | 2.00 (d, 7.1) | 15.9 | 3′′′′ | 5.63 (dd, 10.0, 3.5) | 74.3 | 6.01 (q, 7.1) | 136.5 | 5.98 (q, 7.2) | 137.1 |
| 5 | 1.90 s | 20.8 | 4′′′′ | 5.68 (d, 3.7) | 70.9 | 2.13 (d, 7.1) | 15.7 | 2.13 (d, 7.2) | 15.8 |
| 5′′′′ | 3.90 o | 69.4 | 2.05 s | 21.2 | 2.04 s | 20.8 | |||
| 6′′′′ | 1.11 (d, 6.3) | 16.3 | |||||||
| C-22/4′′′′ | C-4′′′′-Ang | C-22-Ang | C-22-Ang | ||||||
| 1 | — | 167.3 | — | 168.1 | — | 168.1 | |||
| 2 | — | 128.0 | — | 129.1 | — | 129.0 | |||
| 3 | 5.92 (q, 7.2) | 138.8 | 5.81 (q, 7.1) | 137.4 | 5.45 o | 137.2 | |||
| 4 | 1.94 m | 15.9 | 2.00 (d, 7.1) | 15.7 | 2.03 (d, 7.3) | 15.9 | |||
| 5 | 1.83 s | 20.7 | 1.77 s | 20.6 | 1.94 s | 21.0 | |||
| C-3′′′′ | Ang | ||||||||
| 1 | — | 167.3 | |||||||
| 2 | — | 127.8 | |||||||
| 3 | 5.87 (q, 7.2) | 138.4 | |||||||
| 4 | 1.93 m | 15.9 | |||||||
| 5 | 1.82 s | 20.2 | |||||||
| C-28 | Ac | ||||||||
| 1, 2 | 1.92 s | 170.1, 20.7 | |||||||
The molecular formula of compound 2 was C61H96O23 with 14 degrees of unsaturation, as deduced by a pseudomolecular ion peak [M + Na]+ at 1219.6238 (calcd for C61H96O23Na, 1219.6240) in the positive HR-ESI-MS. Absorptions at 1738 and 3422 cm−1 in the IR spectrum were ascribed to carbonyl and hydroxyl groups, respectively. 1H-NMR (300 MHz, pyridine-d5) (Table 2) revealed seven characteristic proton signals for the seven methyls of oleane [δH 0.84 (3H, s, CH3-25), 0.94 (3H, s, CH3-26), 1.15 (3H, s, CH3-29), 1.19 (3H, s, CH3-24), 1.34 (3H, s, CH3-23), 1.36 (3H, s, CH3-30), and 1.85 (3H, s, CH3-27)]; an olefinic group [δH 5.49 (1H, brs)]; an angeloyl [δH 1.97 (3H, s), 2.18 (3H, d, J = 7.2 Hz), 6.09 (1H, q, J = 7.2 Hz)]; an oxygenated methine [δH 3.1 (1H, overlap, epoxyangeloyl-H-3)]; an n-butyl group [δH 0.84 (3H, t, J = 7.3 Hz, n-butyl-H-4), 1.33 (2H, m, n-butyl-H-3), 1.65 (2H, m, n-butyl-H-2), 4.29 (2H, t, J = 6.5 Hz, n-butyl-H-1)]; three sugar units [δH 4.92 (1H, d, J = 7.4 Hz, D-glucuronic acid-H-1′), δH 5.33 (1H, d, J = 7.7 Hz, D-galactose-H-1′′), and δH 5.99 (1H, d, J = 2.2 Hz, L-arabinose-H-1′′′)]; and two methyls at δH 1.44 (3H, d, J = 5.3 Hz) and δH 1.69 (3H, s) were assigned to an epoxyangeloyl group. 13C-NMR (150 MHz, pyridine-d5) spectrum displayed 61 carbon resonances (Table 2), including carbons for 7 methyls (δC 16.1, 16.6, 16.8, 20.9, 27.5, 27.9, 29.5), 2 carbons for an olefinic bond [δC 124.4 (C-12), 142.6 (C-13)]; 17 carbons for three sugar units [δC 71.3, 76.4, 78.7, 86.0, 105.2, 169.6 (D-glucuronic acid); δC 61.8, 69.7, 73.4, 75.1, 76.7, 104.8 (D-galactose); and δC 62.2, 77.5, 83.6, 85.4, 111.1 (L-arabinose)]; five carbons for an angeloyl unit [δC 15.6, 20.2, 128.5, 139.4, 167.5], five carbons for an epoxyangeloyl group [δC 13.9, 19.9, 59.9, 60.2, 169.8]; four carbons for an n-butoxy group (δC 13.7, 19.2, 30.8, 65.1); and other carbons for alkyl groups. All spectral data indicated that 2 was also a barringtogenol triterpenoid with a barringtogenol C aglycon in its structure.1 Acid hydrolysis of this compound with aqueous HCl (2 M) yielded D-galactose and L-arabinose, which were also identified via HPLC analysis using an optical rotation detector.13 The location and linkage sequence of the sugar unites were established by HMBC correlations between the anomeric proton signal of glucuronic acid (δH 4.92) and C-3 carbon signal at δC 89.9, between δH 5.33 (Gal-H-1′′) and δC 78.7 (C-2′), and between δH 5.99 (Ara-H-1′′′) and δC 86.0 (C-3′) (Fig. 2). The configurations of the anomeric carbons of the sugar units of 2 could be determined by their anomeric protons coupling constants. Similarly, the positions of other moieties were speculated by HMBC correlations between δH 6.30 (H-22) and δC 167.5 (Ang-C-1), as well as between δH 6.65 (H-21) and δC 169.8 (epo-C-1); a long-range correlation between the proton signal of the n-butoxy group at δH 4.29 (n-Bu-1) and that of the carboxyl carbon δC 169.6 (C-6′) of D-glucuronic acid was also observed. These correlations indicted that the n-butoxy group was linked to the C-6′ position of D-glucuronic acid (Fig. 2).
| No. | 2c | 3b | 4b | |||
|---|---|---|---|---|---|---|
| δH J (Hz) | δC | δH J (Hz) | δC | δH J (Hz) | δC | |
| a “o” overlapped.b 600 MHz for 1H-NMR, 150 MHz for 13C-NMR.c 300 MHz for 1H-NMR, 150 MHz for 13C-NMR. | ||||||
| 1 | 0.87 m, 1.44 m | 38.7 | 0.84 m, 1.43 m | 38.6 | 0.80 m, 1.36 m | 38.6 |
| 2 | 1.81 m, 2.01 m | 26.5 | 1.86 m, 2.10 m | 26.4 | 1.81 m, 2.01 m | 26.5 |
| 3 | 3.33 (dd, 11.8, 4.3) | 89.9 | 3.28 (dd, 12.4, 4.1) | 88.8 | 3.16 (dd, 11.8, 4.4) | 89.9 |
| 4 | — | 39.7 | — | 39.2 | — | 39.6 |
| 5 | 0.79 m | 55.7 | 0.78 (1H, d, 11.6) | 55.2 | 0.69 (d, 12.1) | 55.6 |
| 6 | 1.49 m, 1.53 m | 18.4 | 1.30 m, 1.52 m | 18.4 | 1.46 m, 1.50 m | 18.2 |
| 7 | 1.26 m, 1.55 m | 33.0 | 2.09 m, 2.00 m | 36.6 | 1.11 o, 1.22 m | 32.9 |
| 8 | — | 40.0 | — | 41.1 | — | 39.9 |
| 9 | 1.73 m | 46.9 | 1.68 m | 46.8 | 1.68 o | 46.8 |
| 10 | 36.8 | 36.4 | 36.7 | |||
| 11 | 1.86 m, 1.78 m | 23.8 | 1.87 m, 1.79 m | 23.7 | 1.85 m, 1.76 m | 23.7 |
| 12 | 5.49 brs | 124.4 | 5.50 brs | 125.1 | 5.42 brs | 125.1 |
| 13 | — | 142.6 | — | 143.4 | — | 140.9 |
| 14 | — | 41.5 | — | 47.4 | — | 41.1 |
| 15 | 1.60 o, 1.86 o | 34.8 | 4.21 (d, 3.6) | 67.2 | 1.68 m, 1.85 m | 30.7 |
| 16 | 4.49 m | 68.6 | 4.42 (d, 3.6) | 73.0 | 5.56 brs | 72.1 |
| 17 | — | 48.2 | — | 48.1 | — | 47.1 |
| 18 | 3.08 o | 39.7 | 3.09 m | 40.7 | 3.09 (dd, 14.4, 4.5) | 39.5 |
| 19 | 3.07 o, 1.34 o | 47.0 | 3.09 m, 1.39 m | 46.6 | 1.47 o, 3.12 o | 46.9 |
| 20 | — | 36.5 | — | 36.0 | — | 36.1 |
| 21 | 6.65 (d, 10.1) | 80.6 | 6.70 (d, 10.3) | 78.3 | 6.01 (d, 10.4) | 78.0 |
| 22 | 6.30 (d, 10.2) | 73.0 | 6.31 (d, 10.3) | 73.3 | 6.31 (d, 10.4) | 72.4 |
| 23 | 1.34 s | 27.9 | 1.27 s | 27.7 | 1.29 s | 27.8 |
| 24 | 1.19 s | 16.6 | 1.10 s | 16.6 | 1.16 s | 16.6 |
| 25 | 0.84 s | 16.1 | 0.85 s | 15.6 | 0.78 s | 15.5 |
| 26 | 0.94 s | 16.8 | 1.01 s | 17.2 | 0.74 s | 16.7 |
| 27 | 1.85 s | 27.5 | 1.84 s | 21.0 | 1.47 s | 26.9 |
| 28 | 3.40 (d, 10.9), 3.67 (d, 10.9) | 63.3 | 3.49 (d, 10.7), 3.75 (d, 10.7) | 62.8 | 3.48 (d, 10.6), 3.63 (d, 10.6) | 63.3 |
| 29 | 1.15 s | 29.5 | 1.10 s | 29.2 | 1.11 s | 29.4 |
| 30 | 1.36 s | 20.9 | 1.32 s | 20.3 | 1.32 s | 19.7 |
| C-3-GlcA-1′ | 4.92 (d, 7.4) | 105.2 | 4.98 (d, 7.5) | 105.0 | 4.88 (d, 7.6) | 105.2 |
| 2′ | 4.44 o | 78.7 | 4.95 o | 83.4 | 4.37 brs | 78.6 |
| 3′ | 4.19 o | 86.0 | 4.31 o | 77.1 | 4.22 m | 85.9 |
| 4′ | 4.39 o | 71.3 | 4.54 o | 72.5 | 4.31 m | 71.4 |
| 5′ | 4.37 m | 76.4 | 4.04 m | 76.6 | 4.45 (d, 9.5) | 76.3 |
| 6′ | — | 169.6 | — | 170.2 | — | 170.0 |
C-6′-O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3 H3 |
— | — | 3.69 s | 51.7 | 3.77 s | 52.3 |
| C-6′-n-Butyl-1 | 4.29 (t, 6.5) | 65.1 | — | — | — | — |
| 2 | 1.65 m | 30.8 | — | — | — | — |
| 3 | 1.33 m | 19.2 | — | — | — | — |
| 4 | 0.84 (t, 7.3) | 13.7 | — | — | — | — |
| C-2′-Gal-1′′ | 5.33 (d, 7.7) | 104.8 | 5.21 (d, 7.5) | 106.8 | 5.35 (d, 7.8) | 104.8 |
| 2′′ | 4.51 o | 73.4 | 4.58 o | 74.4 | 4.51 o | 73.4 |
| 3′′ | 4.14 m | 75.1 | 4.15 (dd, 9.5, 2.8) | 74.6 | 4.14 (dd, 9.5, 3.3) | 75.2 |
| 4′′ | 4.62 brs | 69.7 | 4.68 (d, 2.5) | 69.2 | 4.62 (d, 3.0) | 69.7 |
| 5′′ | 4.00 m | 76.7 | 4.50 m | 76.4 | 4.00 (d, 6.3) | 76.7 |
| 6′′ | 4.58 m, 4.49 o | 61.8 | 4.6 o, 4.38 o | 61.0 | 4.48 o, 4.58 o | 61.8 |
| C-3′-Ara-1′′′ | 5.99 (d, 2.2) | 111.1 | — | — | 6.06 (d, 2.2) | 111.1 |
| 2′′′ | 4.99 brs | 83.6 | — | — | 4.99 brs | 83.6 |
| 3′′′ | 4.83 brs | 77.5 | — | — | 4.83 m | 77.6 |
| 4′′′ | 4.84 brs | 85.4 | — | — | 4.84 brs | 85.5 |
| 5′′′ | 4.18 o, 4.32 o | 62.2 | — | — | 4.18 m, 4.32 m | 62.3 |
| C-21 | Epo | Ang | — | Ang | ||
| 1 | — | 169.8 | — | 167.8 | — | 167.7 |
| 2 | — | 60.2 | — | 128.6 | — | 128.5 |
| 3 | 3.10 o | 59.9 | 5.76 (q, 7.3) | 137.1 | 6.00 (q, 7.3) | 138.5 |
| 4 | 1.44 (d, 5.3) | 13.9 | 2.11 (d, 7.3) | 15.3 | 2.08 (d, 7.3) | 15.9 |
| 5 | 1.69 s | 19.9 | 1.73 s | 19.9 | 2.05 s | 20.9 |
| C-22-Angyl-1 | — | 167.5 | — | 167.4 | — | 167.2 |
| 2 | — | 128.5 | — | 128.8 | — | 128.3 |
| 3 | 6.09 (q, 7.2) | 139.4 | 5.96 (q, 7.3) | 136.1 | 5.94 (q, 7.3) | 138.2 |
| 4 | 2.18 (d, 7.2) | 15.6 | 2.00 (d, 7.3) | 15.5 | 2.01 o | 15.7 |
| 5 | 1.97 s | 20.2 | 2.00 s | 20.7 | 1.99 s | 20.9 |
| C-16-OCOCH3 | — | — | — | — | 2.53 s | 170.1, 22.0 |
Cross-peaks in NOESY spectrum of compound 2 between H-3 (δH 3.33), H-5 (δH 0.79), and H-9 (δH 1.73), as well as between H-18 (δH 3.08) and H-30 (δH 1.36), indicated that the stereochemistry of compound 2 was identical to that of compound 1 (Fig. 3). The NOE correlation between H-21 (δH 6.65) and CH3-29 (δH 1.15) also suggested that H-21 presented an α orientation. The NOE correlations between H-16 (δH 4.49) and H-22 (δH 6.30), as well as between H-22 (δH 6.30) and CH3-30 (δH 1.36), indicated that H-16 and H-22 presented an β orientations. The NOE correlation cross-peak between δH 3.10 (epoxyangeloyl-3-H) and δH 1.69 (epoxyangeloyl-5-H) confirmed the relative configuration of epoxyangeloyl as cis. Based on the above data, compound 2 was determined to be 3-O-[α-L-arabinofuranosyl(1→3)]-β-D-galactopyranosyl(1→2)-β-D-6′-n-butyl-glucuronic acid-21-O-epoxyangeloyl-22-O-angeloyl-3β,16α,21β,22α,28-pentahydroxy-olean-12-ene.
Compound 3 was obtained as white amorphous powder (CH3OH); its positive-ion HR-ESI-MS spectrum exhibited a pseudomolecular ion peak at m/z 1045.5294 [M + Na]+ (calcd for C53H82O19Na, 1045.5348), in agreement with a molecular formula of C53H82O19 with 13 degrees of unsaturation. Absorptions at 1679 and 3426 cm−1 in the IR spectrum were ascribed to carbonyl and hydroxyl groups, respectively. 1H-NMR (600 MHz, pyridine-d5) and 13C-NMR (150 MHz, pyridine-d5) spectral data suggested that compound 3 featured the same aglycone (21,22-di-O-angeloyl-R1-barrigenol) present in compound 8 (Table 2).11 Compared with compound 8, compound 3 included a methoxy group [δH 3.69 (3H, s), δC 51.7] and two sugar units in its structure; these sugar units were identified as D-galactose (δC 61.0, 69.2, 74.4, 74.6, 76.4, 106.8) and D-glucuronic acid (δC 72.5, 76.6, 77.1, 83.4, 105.0, 170.2) using the same method described for compound 1. Direct connections between the protons and carbons of compound 3 were identified by its HSQC spectrum. In the HMBC spectrum (Fig. 2), the methoxy group was confirmed to be located at the C-6 position of glucuronic acid by the presence of a long-range correlation between δH 3.69 and the carbonyl carbon δC 170.2. Moreover, the anomeric proton signal (δH 5.21, 1H, J = 7.5 Hz) of galactose showed a long-range correlation with the C-2 (δC 83.4) position of glucuronic acid. HMBC correlations between H-21 and H-22 (δH 6.70, 6.31) and two carbonyl carbons assigned to two angeloyls at δC 167.4 and 167.8, respectively, were also observed (Fig. 2). Compound 3 was thus elucidated to be 3-O-[β-D-galactopyranosyl(1→2)]-β-D-6′-methyl-glucuronic acid-21,22-O-diangeloyl-3β,15α,16α,21β,22α,28-hexahydroxyl-olean-12-ene and subsequently named 6′-methylether-O-xanifolia-Y5.
Compound 4 was isolated as a white powder (CH3OH), and its molecular formula C60H92O23 (15 degrees of unsaturation) was obtained via HR-ESI-MS (m/z 1203.5925 [M + Na]+, calcd 1203.5927 for C60H92O23Na). Characteristic absorption bands at 1720, 1743, and 3423 cm−1 in the IR spectrum of 4 were attributed to carbonyl and hydroxyl ester groups. 1H and 13C NMR spectroscopic data (Table 2) suggested that 4 was a derivative of compound 2, mainly differing in terms of carbon resonances in a group of acetoxy signals at δC 22.0 and 170.1; a group of angeloyl signals at δC 15.9, 21.0, 128.5, 138.5, 167.7; and a methoxy signal at δC 52.3. The presence of D-galactose, L-arabinose, and D-glucuronic acid could be verified by the method described for sugar analysis. The HMBC correlation (Fig. 2) of the anomeric proton of glucuronic acid at δH 4.88 with C-3 (δC 89.9) of the aglycone (barringtogenol C) confirmed the attachment of this sugar unit to the aglycone. The connections of the acetoxy group to the C-16 position, as well as that of two angeloyl groups to the C-21 and 22 positions, were also confirmed by the HMBC experiment, which showed long-range correlations between H-16 (δH 5.56) and carboxyl at δC 170.1, between H-21 (δH 6.01) and carboxyl at δC 167.7, and between H-22 (δH 6.31) and the carboxyl of angeloyl at δC 167.2 (Fig. 2). The HMBC correlation of the proton resonance of methoxy at δH 3.77 with the C-6′ of glucuronic acid at δC 170.0 indicated that the methoxy was linked at the C-6′ position. An additional NOESY experiment (Fig. 3) confirmed the stereochemistry of the aglycone, which showed substitutes configurations identical to those of compound 2. Finally, compound 4 was determined to be 3-O-[α-L-arabinofuranosyl(1→3)]-β-D-galactopyranosyl(1→2)-β-D-6′-methyl-glucuronic acid-16-O-acetyl-21,22-O-diangeloyl-3β,16α,21β,22α,28-pentahydroxy-olean-12-ene and subsequently named 16-O-acetyl-aesculioside G12.
Compound 5 was assigned a molecular formula of C66H102O27 with 16 degrees of unsaturation, as deduced from its HR-ESI-MS (positive-ion model) data ([M + Na]+ m/z 1349.6426, calcd for C66H102O27Na, 1349.6506). The IR spectrum of 5 showed the absorption bands of carbonyl (1723 cm−1) and hydroxyl (3427 cm−1) groups. The 1H-NMR (600 MHz, pyridine-d5) and 13C-NMR (150 MHz, pyridine-d5) spectral data (Table 2) suggested that structure 5 presents an aglycone similar to that of compound 8. The presence of four sugar units could be verified by carbon signals at δC 71.2, 76.7, 78.6, 85.4, 105.1, 170.0 (for D-glucuronic acid); δC 61.8, 70.0, 73.4, 75.1, 76.7, 104.8 (for D-galactose); δC 62.3, 77.6, 83.5, 85.9, 111.0 (for L-arabinose) and δC 16.3, 69.4, 69.7, 70.9, 74.3, 106.2 (for D-fucose). Two angeloyl groups (δC 15.9, 20.2, 127.8, 138.4, 167.3; δC 15.9, 20.7, 128.0, 138.8, 167.3), an acetyl group (δC 20.7 and 170.7), and a methoxy group (δC 52.2) were found. Direct connections between the protons and carbons of compound 5 were identified from its HSQC spectrum. In the HMBC spectrum (Fig. 2), the linkage between two angeloyl groups with D-fucose units at C-3′′′′ and C-4′′′′ could be verified by the long-range correlations between the proton resonance at δH 5.54 (Fuc-H-3′′′′) and δC 167.3 (Ang) and between δH 5.63 (Fuc-H-4′′′′) and δC 167.3 (Ang); moreover, the anomeric proton signal of fucose moiety (δH 4.92, H-1′′′′) revealed a correlation with C-21 (δC 92.0). The HMBC experiment showed long-range correlations between the methoxyl group at δH 3.71 (–OCH3) and a carbonyl carbon at δC 170.0 (GlcA-6′), as well as between the protons δH 4.21 and 4.32 (each 1H, d, J = 10.9 Hz, Ha,b-28) and carbonyl at δC 170.7. The similarity of locations and linkages sequences for three other sugar unites to those of compound 5 could be verified by HMBC correlations between the anomeric proton signal of glucuronic acid (δH 4.83) and the C-3 carbon signal at δC 90.0, between δH 5.28 (Gal-H-1′′) and δC 78.6 (C-2′), and between δH 5.97 (Ara-H-1′′′) and δC 85.4 (C-3′) (Fig. 2). Thus, 5 was determined to be 3-O-[α-L-arabinofuranosyl(1→3)]-β-D-galactopyranosyl(1→2)-β-D-6′-methyl-glucuronic acid-21-O-(3′′′′,4′′′′-O-diangeloyl)-β-D-fucopyranosyl-28-O-acetyl-3β,16α,21β,22α,28-pentahydroxy-olean-12-ene.
The positive HR-ESI-MS spectrum of compound 6 showed a pseudomolecular ion peak [M + Na]+ at m/z 1193.5719 (calcd for C58H90O24Na, 1193.5720), hence suggesting the molecular formula C58H90O24 with 14 degrees of unsaturation. The IR spectrum of 6 showed absorption bands of carbonyl (1720 cm−1) and hydroxyl (3400 cm−1) groups. Analysis of the carbon and proton resonance in the NMR [1H-NMR (600 MHz, C5D5N) and 13C-NMR spectra (150 MHz, C5D5N)] of compound 6 revealed the presence of three sugars [D-glucuronic acid: δC 71.5, 76.4, 78.2, 86.3, 104.7, 169.8; D-glucose: δC 61.3, 69.5, 75.5, 77.8, 78.2, 103.9; L-arabinose: δC 62.3, 77.5, 83.6, 85.4, 111.1], two angeloyls (δC 15.7, 21.2, 128.9, 136.5, 167.7 and δC 15.7, 20.6, 129.1, 137.4, 168.1), and a methoxyl at δC 52.3. A total of 30 carbon signals, including carbons for six methyls (δC 15.7, 17.3, 20.2, 21.0, 22.2, 29.5), two for an olefinic bond [δC 125.2 (C-12), δC 143.7 (C-13)], two oxygenated methylenes [δC 63.1 (C-24), δC 63.3 (C-28)], four oxygenated methines (δC 67.5, 73.3, 73.5, 78.4, for C-15, 16, 21, 22, respectively), and other alkyl carbons, could be clearly assigned to the aglycone 24-hydroxy-R1-barrigenol by the direct connections observed between protons and carbons in HSQC spectrum. In the HMBC spectrum (Fig. 2), the linked positions of two angeloyl groups were determined by a long-range correlation between H-21 (δH 6.76) and a carbonyl carbon at δC 167.7 (Ang) and between H-22 (δH 6.39) and a carbonyl carbon at δC 168.1 (Ang). Additionally, long-range correlations between the anomeric proton signal of glucuronic acid [δH 4.90 (1H, d, J = 7.3 Hz)] and a C-3 carbon signal at δC 91.5, between the anomeric proton signal at δH 5.51 [(1H, d, J = 7.5 Hz)] (Glc-H-1′′) and δC 78.2 (C-2′) and between the anomeric proton signal at δH 6.06 (Ara-H-1′′′) and δC 86.3 (C-3′) (Fig. 2) were observed in HMBC spectrum. Correlation of the proton resonance of methoxy at δH 3.81 with the C-6′ of glucuronic acid at δC 169.8 indicated that the methoxy group was linked at the C-6′ position. Consequently, compound 6 was determined to be 3-O-[α-L-arabinofuranosyl(1→3)]-β-D-glucopyranosyl(1→2)-β-D-6′-methyl-glucuronic acid-21,22-O-diangeloyl-3β,15α,16α,21β,22α,24β,28-heptahydroxy-olean-12-ene and named 6′-methylester-O-xanifolia-Y2.
Compound 7 was obtained as a white powder (CH3OH), and its molecular formula was deduced to be C58H90O23 (14 degrees of unsaturation) by the pseudomolecular ion peak [M + Na]+ at m/z 1177.5759 (calcd for C58H90O23Na, 1177.5771) in its HR-ESI-MS spectrum. The IR spectrum of 7 showed absorption bands of carbonyl (1720 cm−1) and hydroxyl (3400 cm−1) groups. 1H-NMR (300 MHz, C5D5N) and 13C-NMR (150 MHz, C5D5N) spectra (Table 1) demonstrated signal patterns similar to those of compound 6, although an oxygenated methylene carbon signal at δC 34.8 (C-15) in 7 was observed instead of the oxygenated methine at δC 67.5 in 6. HSQC correlations for two proton signals at δH 1.60 (1H, m, Ha-15) and 1.88 (1H, overlap, Hb-15) with C-15 were also observed. Based on the HMBC correlations (Fig. 2), compound 7 was determined to be 3-O-[α-L-arabinofuranosyl(1→3)]-β-D-glucopyranosyl(1→2)-β-D-6′-methyl-glucuronic acid-21,22-O-diangeloyl-3β,16α,21β,22α,24β,28-hexahydroxy-olean-12-ene and named 6′-methylester-O-xanifolia-Y8.
The structures of three known triterpenoids were elucidated to be xanifolia Y (8),11 xanifolia ACH-Y (9),10 and xanifolia Y2 (10)8 on the basis of their spectroscopic data compared with published values. The NMR spectral data of compounds 8–10 were listed in extraction and isolation part.
Inhibition of U87MG, HepG2 and HCT-116 cell growth by compounds 1–10
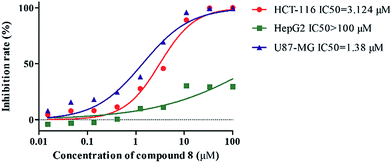
Considering that many barrigenol-like triterpenoids containing the angeloyl group show significant anti-cancer activity,1,8 all of the extracted compounds were evaluated for tumor cell proliferation inhibition against three human tumor cell lines (U87MG, HCT-116 and HepG2) (Table 3 and Fig. 4). New compounds 3, 6, and 7 exhibited potent inhibitory activities against the tumor cells with the IC50 values under 20 μM for each cell line; known compound 8 also showed very significant inhibition of HCT-116 and U87MG cells with IC50 values of 3.12 and 1.38 μM, respectively (Fig. 5). Known compound 10 showed moderate anti-cancer activities against three human malignant neoplasm cell lines (10 μM < IC50 < 30 μM for each cell line). Compared with other compounds, compound 1 showed no inhibitory activity. Analysis of the relationship between structure and activity indicated that angeloyl groups at the C-21 or/and C-22 positions and carbohydrates at C-3 may be the key factors responsible for the anti-tumor activity of barrigenol-like triterpenoids.| Compd | IC50 (μM) | ||
|---|---|---|---|
| HCT-116 | HepG2 | U87-MG | |
| 1 | >100 | >100 | >100 |
| 2 | >100 | 27.7 | >100 |
| 3 | 11.06 | 3.83 | 14.22 |
| 4 | 97.6 | 48.53 | >100 |
| 5 | 36.15 | >100 | 54.01 |
| 6 | 8.24 | 4.16 | 5.64 |
| 7 | 6.11 | 3.02 | 11.71 |
| 8 | 3.124 | >100 | 1.38 |
| 9 | 11.19 | >100 | >100 |
| 10 | 29.83 | 11.71 | 17.73 |
| Doxorubicin hydrochloride | 0.87 | 0.39 | 0.27 |
 | ||
| Fig. 4 The inhibition ratio of compounds treatment on HCT-116, HepG2 and U87-MG cells at the concentration of 11.11 μM. | ||
Apoptotic effect of barrigenol-like triterpenoid-treatment on U87-MG cells
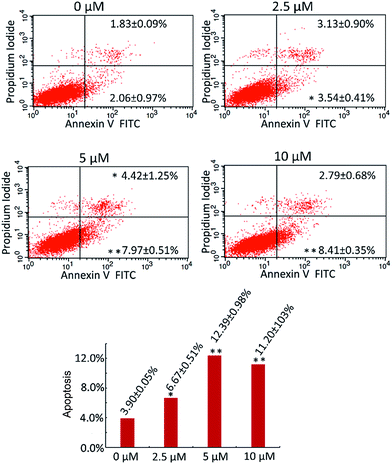
A study on the apoptotic effect of compound 8 as an example of a highly active barrigenol-like triterpenoid on U87-MG cells was carried out. Cell cycle analysis of compound 8-treated U87-MG cells was performed through propidium iodide (PI) staining and flow cytometry (Fig. 6). G0/G1 phase arrest was observed to increase from 46.53% in the control to 60.98% after addition of compound 8 (10 μM). Similar results at three other concentrations (5, 2.5, and 1.25 μM) were observed.The apoptosis-mediated cytotoxicity of compound 8 was further determined by double-staining assay. Annexin V is a calcium-dependent phospholipid-binding protein that presents a strong affinity with phosphatidylserine (PS). PS externalization is an important characteristic of the early stages of cell apoptosis; annexin V-FITC can detect PS externalization to the cell surface, which indicates apoptosis. PI could also pass through the cellular membrane of late apoptotic cells and mark the nucleus.14
When U87-MG cells were incubated with compound 8 for 24 h, different levels of apoptosis, especially early apoptosis, were observed (Fig. 7). Total apoptosis rates of 6.67%, 12.39% and 11.2% (control 3.9%) were observed in cells treated with compound 8 at concentrations of 2.5, 5, and 10 μM, respectively. These findings suggested that barrigenol-like triterpenoids induced apoptosis in a dose-dependent manner.
Conclusion
Among the secondary metabolites of X. sorbifolia Bunge, barrigenol-like triterpenoids are the most remarkable constituents. The n-butanol extract of X. sorbifolia Bunge exhibited excellent antitumor activities toward the HCT-116, HepG2 and U87-MG cell lines with IC50 values of 22.66, 6.741, and 16.31 μg ml−1, respectively. Detection of this active extract by thin-layer chromatography suggested that it contained a number of triterpenoid compounds. Chemical studies resulted in the isolation of seven new and three known barrigenol-type compounds from the bioactive portion of the extract. The aglycone moieties of the triterpenoids were classified into three types (compound 1, 16-deoxybarringtogenol C type; compounds 3, 6, 8, 9, and 10, barrigenol R1; and compounds 2, 4, 5, and 7, barringtogenol C). Compounds 3, 6, 7, 8, and 10, with featured angeloyl groups at the C-21 and C-22 positions and a sugar chain at the C-3 position, displayed noteworthy inhibition effects against the proliferation of malignancy cells (HCT-116, HepG2, and U87-MG). In this study, G0/G1 phase arrest and apoptosis in compound 8-treated U87-MG cells were confirmed by flow cytometry assay. Research on barrigenol triterpenoids from X. sorbifolia Bunge and their biological activities may not only yield more novel active structures, but also contribute to the development of this plant as a novel source of potential anti-tumor agents.Experimental
Plant materials
Husks of X. sorbifolia were collected in October 2011 at Chifeng City Inner-Mongolia autonomous region, China, and identified by Associate Prof. Jiu-Zhi Yuan of Shenyang Pharmaceutical University. A voucher specimen (ZB-11-XS001A) was deposited in the Department of Natural Products Chemistry, Shenyang Pharmaceutical University, Shenyang, China.Apparatus and instruments
NMR spectra were obtained by Bruker ARX-300, AV-400, and AV-600 spectrometers (Bruker, Billerica, MA, USA) using tetramethyl silane as an internal standard. HRESI-MS spectra were obtained using a Bruker micro-TOF-Q mass spectrometer, IR spectra were obtained with a Bruker IFS-55 Fourier transform infrared (FT-IR) spectrometer (Bruker), and a PerkinElmer Spectrum 100 FT-IR Spectrometer (PerkinElmer, USA). Rotation spectra were obtained using a JASCO PU-4100 pump and JASCO ORD-4090 ORD spectrophotometric detector with an NH2P-50 4E column (20 × 250 mm), and specific rotation were obtained by Anton-Paar MCP 200 polarimeter (Anton-Paar, AT). Open-column chromatography was performed using silica gel (200–300 mesh, Qingdao Marine Chemical Co., Ltd.), macroporous adsorption resin D101 (Langfang Nanda resin Co., Ltd.), and Sephadex LH-20 (Pharmacia Biotech, USA). Preparative RP-HPLC was conducted on an Agela P1050 pump and Agela UV1000D UV spectrophotometric detector at 210 nm using a Mightysil RP-18 GP 250-20 column (5 μm, 20 × 250 mm) eluted with gradient CH3OH–H2O or CH3OH–H2O–CF3COOH solvent systems. Flash chromatography was carried out on an Agela Cheetah Flash System using an ODS Flash Column (spec: C-18, 80 g, 120 g, 20–45 μm; Tianjin Agela Technologies Co., Ltd.). The cytotoxicity assay was performed on a microplate spectrophotometer (Gemini EM; Molecular Devices). Cell cycle and apoptosis were analyzed using Becton Dickinson FACS Calibur System.Extraction and isolation
Husks of X. sorbifolia (15.0 kg, dry weight) were pulverized and extracted three times with 70% ethanol (v/v) under reflux for 2 h. Solvents were removed in a vacuum to obtain a crude extract (1050 g), which was suspended in water and partitioned with EtOAc and n-BuOH to yield an EtOAc part (185 g), an n-BuOH part (390 g) and an aqueous extract (382 g). The n-BuOH part (350 g) was subjected to silica gel column chromatography using gradient CH2Cl2–CH3OH–H2O (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–1
1–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to obtain 12 fractions based on TLC analysis. Fraction 6 (8 g) was submitted to silica gel column chromatography and eluted with a CH2Cl2–CH3OH system (15
1) to obtain 12 fractions based on TLC analysis. Fraction 6 (8 g) was submitted to silica gel column chromatography and eluted with a CH2Cl2–CH3OH system (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–1
1–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to yield six sub-fractions. Subfraction 6-5 (2.8 g) was further separated by Sephadex LH-20 column chromatography (CH2Cl2–CH3OH, 1
1) to yield six sub-fractions. Subfraction 6-5 (2.8 g) was further separated by Sephadex LH-20 column chromatography (CH2Cl2–CH3OH, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and the resultant fractions were further purified by RP-18 HPLC (CH3OH–H2O, 78
1), and the resultant fractions were further purified by RP-18 HPLC (CH3OH–H2O, 78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22) to obtain compounds 2 (11 mg) and 5 (13 mg). Fraction 8 (4.2 g) was separated by flash ODS column chromatography (CH3OH
22) to obtain compounds 2 (11 mg) and 5 (13 mg). Fraction 8 (4.2 g) was separated by flash ODS column chromatography (CH3OH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, 30
H2O, 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70–80
70–80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) to yield six subfractions. Sub-fraction 8-4 (600 mg) was further purified by RP-18 HPLC (CH3OH
20) to yield six subfractions. Sub-fraction 8-4 (600 mg) was further purified by RP-18 HPLC (CH3OH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, 70
H2O, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) to yield compounds 3 (13 mg), 4 (9 mg), 6 (12 mg), and 7 (11 mg). Compound 1 (8 mg) was also obtained from subfraction 8-6 (800 mg) by RP-18 HPLC (CH3OH
30) to yield compounds 3 (13 mg), 4 (9 mg), 6 (12 mg), and 7 (11 mg). Compound 1 (8 mg) was also obtained from subfraction 8-6 (800 mg) by RP-18 HPLC (CH3OH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, 65
H2O, 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35). Fraction 11 (75.9 g) was subjected to flash ODS column chromatography (CH3OH
35). Fraction 11 (75.9 g) was subjected to flash ODS column chromatography (CH3OH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, 10
H2O, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90–100
90–100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) to afford nine subfractions, and subfraction 11-5 (150 mg) was purified by RP-18 HPLC with CH3OH–H2O (75
0) to afford nine subfractions, and subfraction 11-5 (150 mg) was purified by RP-18 HPLC with CH3OH–H2O (75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25) solvent system to yield compound 9 (10 mg). Compound 8 (3 g) was recrystallized from subfraction 11-7 (10 g) in methanol–dichloromethane system. Fraction 12 (7.9 g) was subjected to silica gel column chromatography and eluted with a CH2Cl2–CH3OH system (100
25) solvent system to yield compound 9 (10 mg). Compound 8 (3 g) was recrystallized from subfraction 11-7 (10 g) in methanol–dichloromethane system. Fraction 12 (7.9 g) was subjected to silica gel column chromatography and eluted with a CH2Cl2–CH3OH system (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5–100
5–100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70) to obtain eight subfractions. Subfraction 12-2 (190 mg) was purified with using Sephadex LH-20 column chromatography (CH3OH
70) to obtain eight subfractions. Subfraction 12-2 (190 mg) was purified with using Sephadex LH-20 column chromatography (CH3OH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, 65
H2O, 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35), and the resultant fractions were further purified using RP-18 HPLC with a CH3OH
35), and the resultant fractions were further purified using RP-18 HPLC with a CH3OH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O
H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CF3COOH (70
CF3COOH (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.03) solvent system to obtain compound 10 (13 mg).
0.03) solvent system to obtain compound 10 (13 mg).
Acid hydrolysis
Solutions of compounds 1–7 (2 or 4 mg) in 2 M HCl (5.0 ml) were heated under reflux for 5 h at 90 °C. The reaction mixture was extracted with CHCl3 (8 ml × 3), and the aqueous layer was evaporated repeatedly to pH = 7 and subsequently concentrated to dryness to obtain a residue. The residue were subjected to HPLC analysis using an NH2P-50 4E column (4.6 × 250 mm, Showa Denko K.K., Japan) and an optical rotation detector (JASCO ORD-4090, JASCO International Co. Ltd., Japan). D-Glucose, L-rhamnose, D-galactose, L-arabinose, D-fucose were confirmed by comparison of their retention times with those of authentic samples [mobile phase: acetonitrile–water (75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25, v/v); flow rate: 0.8 ml min−1, tR: 11.4 min (L-arabinose, positive optical rotation, Tokyo Chemical Industry Co. Ltd., Lot. LQAGA-A1); tR: 15.6 min (D-galactose, positive optical rotation, Tokyo Chemical Industry Co. Ltd., Lot. ISDKF-DH); tR: 15.3 min (D-glucose, positive optical rotation, ChromaDex Inc., Lot. 00007265-1JA); tR: 8.7 min (L-rhamnose, negative optical rotation, Tokyo Chemical Industry Co. Ltd., Lot. T5P7E-GA); tR: 9.7 min (D-fucose, positive optical rotation, Aladdin Co. Ltd., Lot. # D1503126)].
25, v/v); flow rate: 0.8 ml min−1, tR: 11.4 min (L-arabinose, positive optical rotation, Tokyo Chemical Industry Co. Ltd., Lot. LQAGA-A1); tR: 15.6 min (D-galactose, positive optical rotation, Tokyo Chemical Industry Co. Ltd., Lot. ISDKF-DH); tR: 15.3 min (D-glucose, positive optical rotation, ChromaDex Inc., Lot. 00007265-1JA); tR: 8.7 min (L-rhamnose, negative optical rotation, Tokyo Chemical Industry Co. Ltd., Lot. T5P7E-GA); tR: 9.7 min (D-fucose, positive optical rotation, Aladdin Co. Ltd., Lot. # D1503126)].
Cell culture
HCT-116 and HepG2 cells were purchased from Stem Cell Bank, Chinese Academy of Science, China. The cells were grown in Dulbecco's Modified Eagle's Medium (DMEM) (Gibco, USA) containing 10% heat-inactivated fetal bovine serum (FBS) (Gibco) and 0.5% penicillin/streptomycin at 37 °C in a 5% CO2 humidified atmosphere.15,16 U87-MG cells (purchased from Stem Cell Bank, Chinese Academy of Science, China) were grown in minimum essential medium (MEM) (Gibco) supplemented with 10% FBS (Gibco), 1% sodium pyruvate (Gibco), 1% GlutaMAX (Gibco), and 0.5% penicillin/streptomycin at 37 °C in a humidified atmosphere containing 5% CO2.17Cytotoxicity assay
The antiproliferative activities of compounds (1–10) and doxorubicin hydrochloride (positive control) toward HCT-116, HepG2 and U87-MG were assessed using a cell counting kit-8 (CCK-8, Tianjin Biolite Biotech Co., Ltd., China).18 Briefly, cells (5000 per well) were cultured at 37 °C, 100% relative humidity, and 5% CO2 and dispersed in replicates in 96-well plates for 24 h. The test compounds and extracts in DMSO (0–100 μM or 0–400 μg ml−1) were added to the wells, which were further incubated for 72 h under the same conditions. The medium was removed, and complete medium contained 10% CCK-8 was added to each well. The plates were incubated once more in the same conditions for 2–6 h. Finally, the absorbance of the solutions was measured at 450 nm, and 650 nm for comparison. The percentage of cell inhibition was calculated as follows: cell death (%) = [A450(negative control) − A450(test)]/[A450(negative control) − A450(positive control)] × 100%. IC50 results were obtained by curve-fitting using Graphpad Prism 5 software.Cell cycle analysis
U87-MG cells (4 × 105 per well) were seeded into 6-well plates and incubated with four concentrations (10, 5, 2.5, and 1.25 μM) for 24 h. The cells were collected through centrifuging (1000 × g, 5 min), and the supernatant was discarded. The remaining cells were rinsed with PBS, fixed in 70% (v/v) ice-cold ethanol at 4 °C for 12 h, and then centrifuged (1000 × g, 5 min). After supernatant removal, cells were resuspended in 535 μl of stain solution [Cell Cycle and Apoptosis Analysis Kits (Beyotime Biotechnology Co. Ltd., China); 500, 25, and 10 μl of staining buffer, 20× propidium iodide solution, and 50× RNase A, respectively] at 37 °C for 30 min and analyzed using a Becton Dickinson FACS Calibur System. The excitation and emission wavelengths of PI were 488 and 585 nm, respectively, and a total of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were required for each sample.19,20
000 events were required for each sample.19,20
Annexin V-FITC/PI assay
U87-MG cells (4 × 105 per well) were treated with compound 8 (10, 5, and 2.5 μM) for 24 h. After incubation, the cells were collected, pelleted by centrifugation (1000 × g, 5 min), and washed with PBS; about 5 × 105 cells were collected for testing. The apoptotic effect of barrigenol-like triterpenoids on U87-MG cells was determined by an annexin V-FITC apoptosis detection kit (Gen-View Scientific Inc., USA). Cells were resuspended in 500 μl of binding buffer. Then, 5 μl of annexin V-FITC and 5 μl PI were added to this mixture in order, and the suspension was incubated in darkness at room temperature for 15 min before analysis. Cells were analyzed using a Becton Dickinson FACS Calibur System. The excitation wavelength of annexin V-FITC and PI was 488 nm and their emission wavelength were 520 and 585 nm, respectively. A total of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were required for each sample.14
000 events were required for each sample.14
Statistical analysis
Results were expressed as mean ± standard deviation. IC50 values were obtained by curve-fitting using Graphpad Prism 5 software. One-way analysis of variance (ANOVA) followed by Dunnett's test was performed using SPSS 19.0 software.Acknowledgements
This work was supported by the Project of Innovation Team (LT2015027) of Liaoning of P. R. China, and Project of Key Laboratory of Structure-Based Drug Design & Discovery of Ministry of Education of Shenyang Pharmaceutical University.Notes and references
- W. Yuan, P. Wang, G. R. Deng and S. Y. Li, Phytochemistry, 2012, 75, 67–77 CrossRef CAS PubMed.
- P. Mangala Gowri, S. V. S. Radhakrishnan, S. Jeelani Basha, A. V. S. Sarma and J. Madhusudana Rao, J. Nat. Prod., 2009, 22, 791–795 CrossRef PubMed.
- G. S. Wan, X. B. Wang, L. J. Wu and H. Y. Gao, Chin. Tradit. Herb. Drugs, 2013, 44, 1842–1851 CAS.
- S. Aslan Erdem, A. C. Mitaine-Offer, T. Miyamoto, M. Kartal and M. A. Lacaille-Dubois, Phytochemistry, 2015, 110, 160–165 CrossRef CAS PubMed.
- X. Li, G. P. Chen, L. Li, K. J. Wang, B. Q. Zhang and S. J. Hu, Microvasc. Res., 2010, 79, 63–69 CrossRef CAS PubMed.
- J. Li, Y. G. Zu, M. Luo, C. B. Gu, C. J. Zhao, T. Efferth and Y. J. Fu, Food Chem., 2013, 138, 2152–2158 CrossRef CAS PubMed.
- Menghejirigala and Saqier, J. Med. Pharm.Chin. Minorities, 2014, 11, 40–42 Search PubMed.
- P. K. Chan, M. Zhao, C. T. Che and E. Mak, J. Nat. Prod., 2008, 71, 1247–1250 CrossRef CAS PubMed.
- Y. J. Chen, T. Takeda, Y. Ogihara and Y. Iitaka, Chem. Pharm. Bull., 1984, 32, 3378–3383 CrossRef CAS PubMed.
- Y. J. Chen, T. Takeda, Y. Ogihara and Y. Iitaka, Chem. Pharm. Bull., 1985, 33, 127–134 CrossRef CAS.
- Z. L. Li, B. Z. Yang, X. Li, S. J. Wang, N. Li and Y. Wang, J. Asian Nat. Prod. Res., 2006, 8, 361–366 CrossRef CAS PubMed.
- Z. L. Li, X. Li, D. Y. Li, D. Li, D. L. Meng, W. Li and Y. Sha, J. Asian Nat. Prod. Res., 2007, 9, 387–392 CrossRef CAS PubMed.
- S. Nakamura, X. Z. Li, H. Matsuda and M. Yoshikawa, Chem. Pharm. Bull., 2008, 56, 536–540 CrossRef CAS PubMed.
- M. Eskandani, H. Hamishehkar and J. Ezzati Nazhad Dolatabadi, Food Chem., 2014, 153, 315–320 CrossRef CAS PubMed.
- D. K. Man, L. Casettari, M. Cespi, G. Bonacucina, G. F. Palmieri, S. C. Sze, G. P. Leung, J. K. Lam and P. C. Kwok, Mol. Pharm., 2015, 12, 2112–2125 CrossRef CAS PubMed.
- B. C. He, J. L. Gao, X. Luo, J. Luo, J. Shen, L. Wang, Q. Zhou, Y. T. Wang, H. H. Luu, R. C. Haydon, C. Z. Wang, W. Du, C. S. Yuan, T. C. He and B. Q. Zhang, Int. J. Oncol., 2011, 38, 437–445 CrossRef CAS PubMed.
- S. Banerjee, A. K. Sahoo, A. Chattopadhyay and S. S. Ghosh, RSC Adv., 2014, 4, 39257 RSC.
- Y. Y. Guan, H. J. Liu, X. Luan, J. R. Xu, Q. Lu, Y. R. Liu, Y. G. Gao, M. Zhao, H. Z. Chen and C. Fang, Phytomedicine, 2015, 22, 103–110 CrossRef CAS PubMed.
- S. K. Lam and T. B. Ng, Food Chem., 2011, 126, 595–602 CrossRef CAS.
- O. Y. Zolotarskaya, L. Xu, K. Valerie and H. Yang, RSC Adv., 2015, 5, 58600–58608 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: 1D and 2D NMR, HR-ESI-MS, and IR spectrum of compounds 1–10. See DOI: 10.1039/c6ra02706g |
| This journal is © The Royal Society of Chemistry 2016 |