A UV-sensitive hydrogel based combinatory drug delivery chip (UV gel-Drug Chip) for cancer cocktail drug screening†
Ying-Ting Chena,
Venkanagouda S. Goudara,
Ren-Guei Wua,
Hsin-Yi Hsieha,
Chung-Shi Yangb,
Hwan-You Changc,
Gwo-Bin Leed,
Chih-Ming Hoe and
Fan-Gang Tseng*af
aEngineering and System Science Dept., National Tsing Hua University, Hsinchu, Taiwan. E-mail: fangangtseng@gmail.com
bNational Health Research Institutes, Zhunan, Miaoli, Taiwan
cDepartment of Medical Science, National Tsing Hua University, Hsinchu, Taiwan
dDepartment of Power Mechanical Engineering, National Tsing Hua University, Hsinchu, Taiwan
eMechanical Engineering Department, University of California, Los Angeles, CA, USA
fApplied Science Centre, Academia Sinica, Taipei, Taiwan
First published on 1st April 2016
Abstract
The effective and efficient treatment of diseases, such as HIV, cancer or hereditary diseases, requires accurate and precise control of the combinatorial drug-dosage and their release. Herein, we introduce a simple photosensitive poly(ethylene glycol) diacrylate (PEGDA) hydrogel based platform for high dynamic range testing of combinatorial cocktail drug screening using three chemical and two protein drug treatments for colon cancer. UV cross linked PEGDA hydrogel droplet arrays on a Teflon patterned glass substrate enable a rapid yet accurate selection and dosage assignment of the drugs. Precisely loaded cocktails of the anticancer drugs were simultaneously released in-parallel with the PEGDA hydrogel chips into 2D or 3D cultured HCT-8 colon cancer cells for combinatorial drug screening. We demonstrate the functionality of our UV gel-Drug Chips 1000 fold range of concentrations for each of the five drugs in 30 seconds to find the optimized drug cocktail using a fractional factorial control system. Our device has low drug consumption, requiring only 12 nL per screening run per droplet. In addition, our UV gel-Drug Chips were employed for find the optimized drug cocktail using a fractional search algorithm. Our cocktail drug response results for both 2D (cell viability is 7.3%) and 3D (cell viability is 10.8%) colon cancer cells were analogous to those found by conventional method (6.8 and 9.3 respectively). In contrast to conventional method, our approach is faster, more effective, less time consuming and requires a lower amounts of drug volume.
Introduction
Conventional drug delivery techniques are time consuming, painful and can cause side effects.1 As such, there is a need for alternative, high throughput, cost effective and user friendly laboratory drug screening methods for highly efficient drug testing.2 Current trends in point-of-care, automatized and precise drug assay platforms are superior to conventional techniques. Because they can achieve higher accuracy with lower drug dosages.2 Microfluidic based platforms have emerged as an efficient chip based assay for cell line tests. With their in-built advantages of low reagent consumption, precise control and high throughput scalability will provide robust analytical tools to investigate complex biological processes at the cellular level.3–7 Depending on cancer type and location of cancer, anticancer drugs will have specific targeting sites. For example, 5-fluorouracil and capecitabine drugs are anti-metabolites, irinotecan is a plant alkaloid and acts as a topoisomerase inhibitor, folinic acid is a chemo-protectant, which causes folic acid deficiency in cancer cells, which leads to cell death, oxaliplatin is alkylating agent used for inhibiting cell cycles, bevacizumub is a genetically modified drug for regulating angiogenesis by inhibiting vascular endothelial factor and cetuximub is a protein based drug, which targets endothelial growth factor receptor proteins.58 These examples are some of the drugs approved by the US drug agency for chemotherapy against cancer. However, none of the abovementioned drugs, on their own merit, cannot eliminate cancer thoroughly and efficiently.57 Chemotherapies can reduce tumour burden within the sub-clones by eliminating highly proliferative cells by targeting their RNA, DNA, enzymes, protein receptors and cell cycle. But, when metastasis occurs, those cancer cells will mutate at metastasis site. Therefore, better and more effective drugs for treating metastatic cancer need to be developed. Sub-clonal diversity can be altered with chemotherapies, which allows for the selection of cells with additional genetic mutations to confer a survival advantage.8Therefore, if one drug is not enough to control these tumours, one may think of using a combination of several drugs as a cocktail to potentially obtain better treatment results.9 The synergistic effect of multiple combinations of drugs will allow lower dosage of the drugs. But, also increases the efficient activity of drugs against many types of cancer targets. Which in turn, decreases the drug resistance capabilities of cancer cells.10 Indeed, combinatory drugs have been reported to have higher efficacy and a lower individual drug dosage in the treatment of various diseases, including cancer.11 However, the dose of the drugs used in the combination can also critically affect the efficacy or toxicity. Conventional drug cocktail optimization methods are not as cost effective due to the high drug cost owing to the considerable volume ratio, time consuming routine process for large-scale assays and limitation of the dynamic ranges of the drug concentrations.12,13 In comparison, the feedback system control (FSC) technique is considerably less time consuming and cost effective in regard to the labour cost.14–17 This study demonstrates a novel lab-on-a-chip approach for efficient FSC based combinatorial drug screening.
To operate cocktail drugs in batches, a sequential microfluidic process was developed. For example, integrated microvalves were effectively employed to control and generate individual droplets of precise size and drug composition. The formed droplets can be selectively and sequentially used as drug cocktails.18 The other process, which uses a photopolymerization method in a microfluidic device to sequentially produce 5-fluorouracil loaded biocompatible poly(ethylene glycol) diacrylate (PEGDA) microspheres with monodisperse size distribution has been reported to be used successfully for sustained drug release.19 For instance, for protein drug delivery, microfluidic based polymeric capsules were prepared with high encapsulation efficiency20–28 for oral protein drug delivery and polymeric based drugs were produced.30 For intracellular drug delivery, polymer nano-capsules with different pH resistance were also prepared.31 However, despite the success of the abovementioned methods, sequential processing for batch drug testing showed some unsolved challenges, which include variations in the delivery time, drug combination complications and complex drug delivery networks/pathways to individual cell wells.32
On the other hand, a parallel process using a bead-based assay platform was recently developed for cell drug testing.33 However, the need for laser pulse actuation to break the bead shells for drug release posed an additional issue. Therefore, heating and chemical reactions are bio-incompatible during laser activation. Furthermore, once the capsules were randomly assigned with drugs, it is not easy to select several different drugs in a combination of various concentrations on demand.
To alleviate these challenges, we propose a combinatory drug assay platform for cocktail drug testing employing UV cross linked PEGDA hydrogel droplets to precisely release various cocktail ingredients from 5 different anticancer drugs for in-parallel drug testing against colon cancer cells. The platform incorporates the techniques of self-formation of drug/hydrogel microdroplets and UV selective curing to obtain the desired drug dosage from the obtained cocktails, which are bio-compatible with a high dynamic range (1000 folds) for cocktail drug assignation. These drug combinations can be simultaneously tested on both 2D and 3D cell cultures right after the drug cocktail preparation without any time lag.
The FSC approach
Because the efficacy and toxicity effects on the biological system, cells, organs and body are not only functions of the drugs but also they are strongly affected by the doses used. NM combinations formed by M drug and N doses present a very large test parameter space. By integrating the experiments and a search algorithm to form a feedback system control (FSC) loop, we have found that about 15 feedback loops can identify the optimal drug-dose cocktail out from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 possible combinations.14,17 Derived from the serial testing feedback loop concept, a much more efficient parallel search FSC technology has been demonstrated in cell line tests and in preclinical in vivo experiments.15,16 To make it more achievable for disease treatment purposes, differential evolutionary algorithms were proposed by Dr. F. Wei et al. to find out the optimized combinatory drug system for the best disease suppressing result in a very limited number of experiments.29 Moreover, they reported a study using the FSC technique to investigate a biological system with Herpes simplex virus type 1 against five antiviral drugs. The final searched combinations of drugs demonstrated a higher virus killing efficacy and lower individual drug dosage when compared to the effect of any individual drug at a much higher dosage, while only fewer than 15 cycles of experiments were performed to obtain the final result. By considering the aforementioned study, we employed a similar search algorithm to our UV gel-Drug Chip system for colon cancer cells drug screening. Initially, we set an IC50 value (half maximal inhibitory concentration) of each drug, as code 5, which was also the highest concentration of drug used in the cocktail. Furthermore, to setup different and lower concentrations, one third of the concentration of code 5 was assigned to code 4 as a reduced dose. The dose will further be reduced to one third from 4, 3, 2 to 3, 2, 1 respectively, in a similar way, Fig. 1 depicts the overall process for searching the optimized cocktail combinations. The system randomly picked up initial values for four parallel experiments, recorded as VG as shown in the first step. In the next step, the variation (also called mutation) was conducted according to the formula MGi = VGr1 + F(VGr2 − VGr3), so that VG can be converted to the MG.
000 possible combinations.14,17 Derived from the serial testing feedback loop concept, a much more efficient parallel search FSC technology has been demonstrated in cell line tests and in preclinical in vivo experiments.15,16 To make it more achievable for disease treatment purposes, differential evolutionary algorithms were proposed by Dr. F. Wei et al. to find out the optimized combinatory drug system for the best disease suppressing result in a very limited number of experiments.29 Moreover, they reported a study using the FSC technique to investigate a biological system with Herpes simplex virus type 1 against five antiviral drugs. The final searched combinations of drugs demonstrated a higher virus killing efficacy and lower individual drug dosage when compared to the effect of any individual drug at a much higher dosage, while only fewer than 15 cycles of experiments were performed to obtain the final result. By considering the aforementioned study, we employed a similar search algorithm to our UV gel-Drug Chip system for colon cancer cells drug screening. Initially, we set an IC50 value (half maximal inhibitory concentration) of each drug, as code 5, which was also the highest concentration of drug used in the cocktail. Furthermore, to setup different and lower concentrations, one third of the concentration of code 5 was assigned to code 4 as a reduced dose. The dose will further be reduced to one third from 4, 3, 2 to 3, 2, 1 respectively, in a similar way, Fig. 1 depicts the overall process for searching the optimized cocktail combinations. The system randomly picked up initial values for four parallel experiments, recorded as VG as shown in the first step. In the next step, the variation (also called mutation) was conducted according to the formula MGi = VGr1 + F(VGr2 − VGr3), so that VG can be converted to the MG.
Then, we crossover VG and MG, which means we randomly selected values from VG and MG for crossover and finally, we could obtain a new combination of values called TG (after crossover). Lastly, we compared the results of the VG and TG groups, and in a respective parallel trial we chose the better readout as the next input VG+1. We iterated the selection process for about five to eight cycles to obtain a convergence value of the testing result, which will finally lead to the final optimization value of the drug combination after several more feedback search cycles.
Experimental
Materials
Polyethylene glycol diacrylate (PEGDA) polymer (Fisher Scientific, Model B2200R-1, Pittsburgh, PA), 1× PBS buffer, pH 7.4 (gibco®, by life technologies), Irgacure 2959 (Sigma-Aldrich). 5-Fluorouracil (MW = 130.1), capecitabine (MW = 359.4), irinotecan (MW = 558.64), folinic acid (MW = 473.44), oxaliplatin (MW = 397.28) and rhodamine 6G (MW = 479) from Uni Ward Corp. Importantly, all the chemicals used in the UVGEL-DRUGCHIP preparation were dissolved and prepared in 1× PBS buffer solution.Preparation of hydrogel
Currently, hydrogels have emerged as among one of the best materials for controlled drug delivery and tissue engineering, because of their biocompatibility and biodegradability.34–36 There are natural and synthetic hydrogels and polyethylene glycol (PEG) based gels are synthetic hydrogels. PEG gels are bioinert hydrogels and their chemical structures can be modified conveniently by adding copolymers and natural polymers to the side chains.37–39 Polymeric hydrogels are also used in different stimuli responsive drug delivery systems, including ATP,40 temperature,41 magnetic field,42 mechanical signals,43 redox44 and ultrasound-triggered drug releasing systems.45 Out of all the PEG derivatives, PEGDA can be easily cross-linked via UV irradiation46 and has been widely used in many biomedical applications due to its cytocompatibility, non-toxicity and ease of use.47 In this study, we prepared a 5 wt% of poly(ethylene glycol) diacrylate (PEGDA, Mw = 700; Sigma-Aldrich) solution in 1× PBS buffer solution (the total volume was 100 mL), which was agitated in an ultrasonic bath (Fisher Scientific, Model B2200R-1, and Pittsburgh, PA) for 15 minutes. Once the PEGDA polymer was completely dissolved, 0.25 mg of a photoinitiator (Irgacure 2959, Sigma-Aldrich) was added to the same solution and further agitated using the ultrasonic bath and the pH adjusted to 7.4. Importantly, Irgacure®2959 was more biocompatible with many type of cell lines than other UV photoinitiators.47–49,51,52Preparation of the hydrogel + drug complex
The procedure described above was repeated to prepare 100 mL of hydrogel solution. Furthermore, the 100 mL solution was equally divided into 5 parts. In addition, according to the feedback system control (FSC) results, the respective concentration of each drug [5-fluorouracil, capecitabine, irinotecan, folinic acid, and oxaliplatin] has been weighed separately and mixed into the respective hydrogel solution. Furthermore, individual hydrogel + drug complexes were used for drop casting.Microdroplet formation and UV gel-Drug Chip preparation
We designed a wettability based contrast surface by arranging superhydrophilic defects on the superhydrophobic background, which can cause the spontaneous separation of liquids. First, piranha cleaned glass wafers were spin coated (60 s at 3000 rpm) with Teflon (0.5 wt% Teflon in 1× PBS buffer solution). Furthermore, the Teflon coated substrates were introduced to a reactive ion etching [RIE] (Ar/O2, 5 sccm/10 sccm, 80 W) cabinet to form hydrophilic windows with the help of a shadow mask (Fig. 2(A)). Moreover, the previously prepared hydrogel + drug solution were pipetted out and the solution was rolled across the superhydrophilic and superhydrophobic spots, as shown in (Fig. 2(C)); hydrogel + drug droplet arrays were rapidly formed with a volume variation of less than 5%. However, each hydrogel + drug patch measures around 0.9 cm2 (Fig. 2(E)), which comprises 5 different regions (Fig. 2(E)). Each region has 100 droplets (each droplet size was 200 μm in diameter with a 400 μm pitch to pitch distance) (Fig. 2(E) and (F)). Each region measured around 0.18 cm2. Therefore, these structures are visible with the naked eye. Five different hydrogel + drug complex solutions can be applied to the designated regions in a batch manner (Fig. 2(D)). The individual hydrogel + drug complex were manually loaded into the 5 separate regions, respectively, without cross contamination (Fig. 2(D)).Dosage selection by UV exposure
The hydrogel + drug complex were selectively photopolymerized to assign their corresponding dosages (Fig. 3(I) (A)–(E)) by exposure to UV radiation at 15 mW cm−2 intensity (using a UV transilluminator, Daihan Scientific, Korea) with a 10 s curing time using a shadow mask (Fig. 3(I) (C)). In the next step, the non-cured droplets (Fig. 3(I) (D)) were removed using tissue paper via capillary action, while the cured droplets remained on the chip (Fig. 3(I) (E)). Fig. 3(I) (E) represents the 5 different drug concentration regions in a single drug patch. The images in Fig. 3(II) show the array of hydrogel droplets and each droplet has a diameter of 200 μm and a volume of 12 nL. The volume of the droplets was measured by taking droplet images (top and cross-section views) under a contact angle measurement instrument and calculated using solid work software.Cell-Chip preparation
However, the same patterned design (as UV gel-Drug Chip) was used to prepare a multi-welled Cell-Chip. First, a fine smooth surfaced metal mould was prepared. Then, PDMS was prepared with standard procedure and poured onto the metal mould surface to obtain the multi-welled Cell-Chip, which was placed in an oven at 80 °C for 3 h to cure. After curing, the PDMS was peeled out carefully from the metal mould and bonded onto a glass wafer. Each well (1 cm diameter) was slightly larger than the hydrogel + drug patch (0.9 cm diameter), as shown in Fig. 4(C).Cell culture conditions
It is well known that many cancer cells lose some of their phenotypic properties when grown in vitro as 2D monolayers over time. Moreover, 2D cultures lack metabolic and proliferative gradients because of the strong affinity of the cells to the artificial surface. Formation of tumour-like 3D structures is highly inhibited in 2D monolayer cultures. In other words, 3D cultures closely mimic natural tissue and organs compared to cells grown in 2D. Moreover, there will be natural cell to cell interactions in the 3D cell cultures. In this study, HCT-8 cells were chosen to be the target cells and incubated as both 2D and 3D cultures for drug testing. For the 2D cultures, cells were initially cultured on a 96-well Petri dish covered with 300 μL of culture medium. The culture medium was made from DMEM in the presence of 10% Fetal Bovine Serum and 1% Penicillin–Streptomycin (Pen–Strep). The 96-well Petri dish was incubated overnight at 37 °C under 5% CO2. To begin with, some basic and conventional experiments were carried out using the 96-well plates. However, we mainly used the PDMS based multi-welled Cell-Chip for our drug screening experiments, which measured around 1 cm in diameter and holds approximately 155 μL of medium.For the 3D cultures, the hanging drop method was relatively simple and has been reported to have a reproducibility of almost 100% for producing one 3D spheroid per drop.50 To produce the 3D cultures, we used insphero gravity trap™ ULA and gravity plus™ plates with a cell stock containing 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL by pipetting 40 μL of the cell suspension into the top side of the Hanging Drop 3D cell culture platforms. To prevent medium evaporation, 4 mL of distilled water was added into the peripheral water reservoir. The growth media was exchanged every other day by removing 10 μL of solution from a drop and adding 20 μL of fresh growth medium into the drop. After the cells were incubated for 72 h, 200 μL of cell medium was added to the spheroid cells and then collected into the gravity trap plate. Furthermore, those cell spheroids were pipetted out and dropped into the multi-welled Cell-Chip and also into the 96-well chip for further respective investigations.
000 cells per mL by pipetting 40 μL of the cell suspension into the top side of the Hanging Drop 3D cell culture platforms. To prevent medium evaporation, 4 mL of distilled water was added into the peripheral water reservoir. The growth media was exchanged every other day by removing 10 μL of solution from a drop and adding 20 μL of fresh growth medium into the drop. After the cells were incubated for 72 h, 200 μL of cell medium was added to the spheroid cells and then collected into the gravity trap plate. Furthermore, those cell spheroids were pipetted out and dropped into the multi-welled Cell-Chip and also into the 96-well chip for further respective investigations.
Drug releasing process
Conventionally, to test all the combinations of the 5 drugs each with 5 different concentrations required at least 55 operations. Using the search algorithm,17,29 52 times can reach the optimal value. The current platform not only can quickly ensemble all 5 drugs with the desired concentration in each test but also can deliver in-parallel into each well for drug testing. To achieve a high throughput process, as we mentioned earlier, different areas on the glass substrate were patterned for cocktail drug delivery. Each area (0.18 cm2) contained hundreds of drug carrying hydrogel droplets with 5 different drug concentrations, which is called a hydrogel patch (Fig. 4(A)).As explained before, to define the dosage of each drug for a specific test, a shadow mask was employed to partially expose the selected area to cure the drug carrying hydrogel droplets. Due to the discrete nature of the hydrogel droplets, control over the volume and ratio can be very precise for the combination of 5 drugs. After hydrogel–drug droplet formation and cocktail dosage selection, the UV gel-Drug Chip was immersed into the corresponding multi welled Cell-Chip for cocktail drug delivery at a controllable time pace, as shown in (Fig. 4(A) and (C)). As mentioned before, the well size (1 cm) of the cell chip was designed slightly larger than that (0.9 cm) of the drug chip. Therefore, the area of one drug patch was large enough to be easily visible by the naked eye. So, we aligned these two chips manually without using any alignment keys and any other specific instruments.
Measurement of the released drug concentration
We used five anticancer drugs and one fluorescent molecule to define the releasing rate: 5-fluorouracil (MW = 130.1), capecitabine (MW = 359.4), irinotecan (MW = 558.64), folinic acid (MW = 473.44), oxaliplatin (MW = 397.28) and rhodamine 6G (MW = 479) (the data is reported in Fig. 2 of the ESI†). The released concentration of the drug was measured using a UV spectrophotometer. Initially, a standard calibration curve was obtained by measuring the absorbance from initial concentration of all 5 drugs. Furthermore, the drug released from the PEGDA hydrogel into the cell medium was measured to determine the released drug concentration. Fig. 6(B) shows the releasing rate of the different drugs from the hydrogel droplets. The releasing rate is defined as the percentage ratio of the released concentration to the loaded concentration and was dependent on the molecular weight, molecular charge, diffusion coefficient and solubility of each drug.In this study, we plotted graphs using Origin.8 and Microsoft Excel software.
Results and discussion
Cell viability tests
A detailed process and the results of the cell viability test are mentioned in the ESI (1).†Drug sensitivity test
Initially, the cytotoxicity was tested individually for all five anticancer drugs (5-fluorouracil, capecitabine, irinotecan, folinic acid and oxaliplatin) in a conventional manner (by directly pipetting the drugs into the 96 welled plates) against both the 2D and 3D cultures of HCT-8 cell lines. With this, we calculated and plotted the half maximal inhibitory concentration (IC50), as shown in Fig. 5, in which the IC50 values obtained for the anticancer drugs were significantly higher in the 3D culture systems than those found in the 2D culture systems. | ||
| Fig. 5 The half maximal inhibitory concentration (IC50) values between the 2D and 3D cultures of the individual drugs. | ||
This result is mainly due to the increased drug resistance from the dense multicellular/multilayer structures in the 3D cell spheroids. In this structure, the naturally synthesized extracellular matrix promotes strong cell-to-cell interactions, migration, ion transfer and cell-to-cell communication,53 which in turn may have caused a significant retardation of the drug penetration potential into the core region of the 3D cell spheroid. However, folinic acid and 5-FU are the two protein based drugs we have used in our experiments. Actually, folinic acid is marketed with the trade name Leucovorin, which does not absorb UV light and therefore it will not undergo further photolysis.59 Moreover, the experimental results (M. L. Pascu et al.) support that there will not be a structural change in 5-FU upon irradiation with a Hg lamp or N2 laser beam in the UV spectral range.60,61 Moreover, according to our experimental results, the IC50 values are similar (within 5% deviation) for all the 5 drugs by either the traditional pipetting or the UV hydrogel method, which suggests the similar effectiveness of both methods in drug delivery. However, in our approach we used significantly less chemicals (drugs and medium) and rapid drug release was achieved when compared to the conventional method.
Released drug concentration test
To understand the drug releasing rate from the UV-cured PEGDA hydrogels, we used 3 different concentrations (5, 10 and 20 wt%) of the PEGDA hydrogel with different UV curing times (15, 30 and 45 s). The graphical representation in Fig. 6(A) shows the releasing rate of the 3 different concentrations (5, 10 and 20 wt%) of PEGDA hydrogels with different UV curing times (15, 30 and 45 s). From the figure, one can easily design the releasing rate and time period on demand by adjusting the curing and concentration conditions of the hydrogels.54–56 Out of all the values, the 5 wt% hydrogels with curing times of 15, 30 and 45 s were found to have greater drug releasing capacity, i.e., nearly 80%. Therefore, we used the 5 wt% hydrogel and 15 s curing time as a standard condition to test the releasing rate of 5 different drugs and the result shown in (Fig. 6(B)). The drug releasing rate is defined as the number of molecules released per second from the hydrogel into the solution. The percentage ratio between the released concentration to the loaded concentration obtains the amount of drug released into the medium. The releasing rate depends on the molecular weight of each drug. These values will be a calibration base for assigning the drug releasing dose in the rest of the experiments in the cocktail designs. In addition, each drug has its own releasing rate (time constant) and effectiveness (IC50 value) and those basic properties have been carefully calibrated in Fig. 6 and Tables 1(A) and 2(B), respectively. Therefore, the final cocktail releasing was normalized using these two factors to ensure the correct final real releasing dose, as shown in Tables 1(B) and 2(B). Therefore, both the releasing speed and drug effectiveness have been considered at the same time in this study.Cocktail drug testing
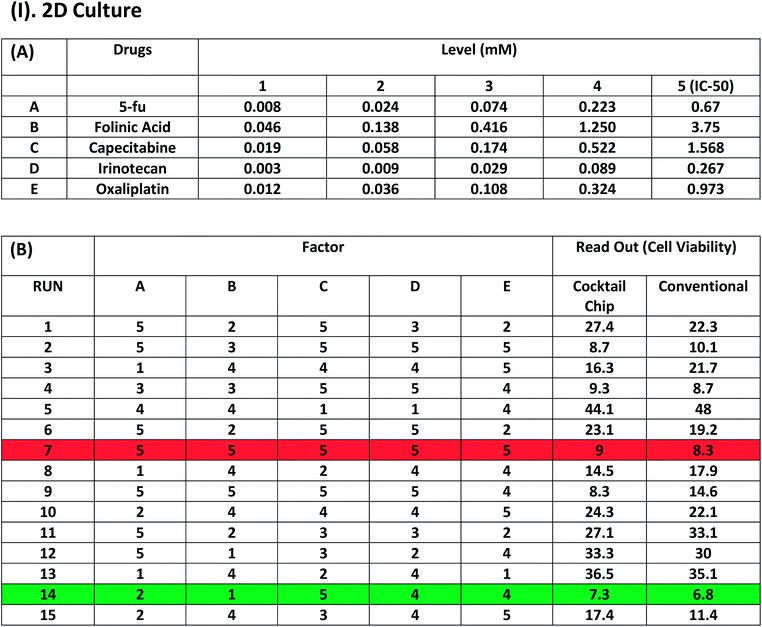
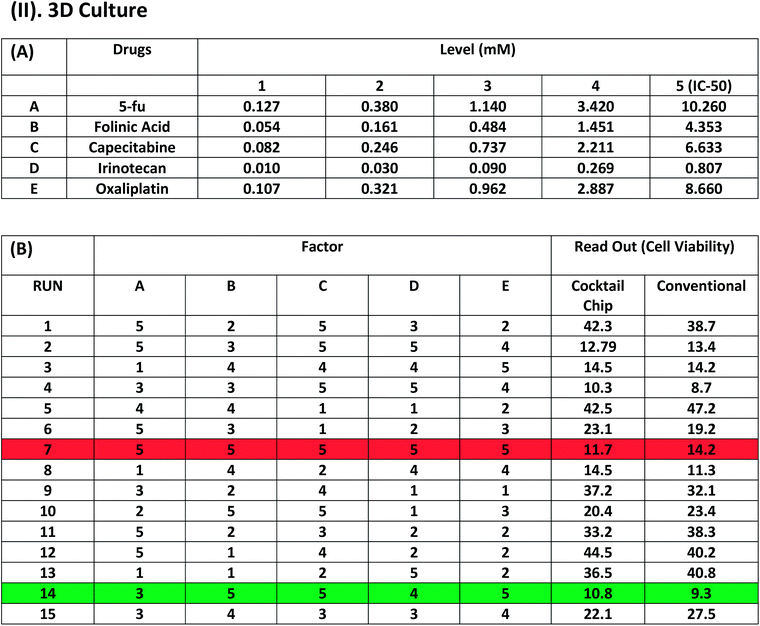
Once we have the IC50 value and releasing rate data for all 5 drugs, we can perform the cocktail fractional search algorithm to find out the optimized cocktail drug combination against colon cancer cells. Table 1(A) shows the cocktail drug testing on the 2D cell cultures. Each code (1, 2, 3, 4 and 5) on the top of the tables corresponds to the actual concentration of the 5 different drugs. Herein, code 5 represents the IC50 value of the individual drugs and the concentration of the next code will be one third of the previous one. Drugs consist of various concentrations corresponding to the five different codes used in further cocktail drug combinations for drug screening. Table 2(A) is also a replication of the same process for the 3D cell culture system. The remaining tables, Tables 1(B) and 2(B) illustrate the 15 experimental runs with different permutation combinations of cocktail drug concentrations to test both the 2D and 3D cell cultures, respectively. The values represent the cell viability for each cocktail testing result. Interestingly, run#7 (in italics) indicated the applied dose of each drug combinations are at their highest value, but its efficiency in terms of cell viability was worse than run#14 (in bold) drug combinations with much lower total dosages. We can observe the same results in both the 2D and 3D culture systems. In addition, the cell viability and concentration of drugs was slightly higher in the 3D cell culture than the 2D cell culture system because of the drug penetration/inhibition issue mentioned earlier for the 3D culture. We used the lowest survival rate (local minimum) of the drug testing result for defining the end point. To make sure this end point was correct, 5–6 more runs than the 14 runs were performed, which makes the end point clear at 14 runs. Therefore, we conclude that run#14 (in bold) was the best combination of drug cocktail to treat colon cancer in the 2D or 3D culture system. At the same time, we also compared the cell viability obtained by a conventional technique and our technique. The readout (conventional) represents the viability of cells treated with drugs alone in conventional microtiter plates via a direct drug pipetting process and served as reference data. Our results are very much analogous to those obtained using the conventional method. Importantly, 8–10 combinatorial assays can be performed using our single UV gel-Drug Chip. For more assays, several chips can be employed sequentially to accommodate the final desired numbers.Conclusions
In summary, we have developed and optimized a simple method employing UV selectable hydrogel droplets for cocktail drug screening (UV gel-Drug Chip) against colon cancer. With the advantage of the UV sensitive hydrogel, precise drug-loaded hydrogel droplet arrays were formed by simply rolling a drop over wettability contrast surface. The drug droplets can be further selected by a UV-curing process of the hydrogel droplets for high dynamic range dosage assignation. In other words, the method is rapid and compatible with both adhesive (2D) and suspension (3D) cells system and does not require expensive or complicated fabrication steps and operations. We combined 5 drugs with 1000 fold dynamic range in 30 s with minimal drug consumption. We also examined and confirmed that the cell viability of the 2D and 3D cultures searched using the employment of UV gel-Drug Chip is quite close to the cell viability obtained using the conventional drug screening method. The current platform provides a better operation for cocktail drug testing in three aspects over the traditional Petri dishes methods: (1) significantly, less wastage of drugs during the drug testing (a days drug usage using the conventional method is 1–10 μL. Our drug chip contains only 12 nL per drop. Therefore, which is a 10 fold reduction in drug usage), (2) more accurate drug releasing time/dosage control, (3) reduction in the individual dosage preparation and drug-well registration issues found in the pipetting process (10–15 min is needed for a conventional 96-well experiment, whilst 1–2 min is more than enough for our platform) and (4) the method is capable to produce a programmed delay-time releasing process for each drug. Although this chip is still in the early stage of development, with further advanced arrangements of the different curing times or concentrations, the chip may have the potential for multiple drug release not only in precise dosage control, but also releasing time pace assignation, which will open up a new opportunity for advanced drug screening processes.Acknowledgements
This study was financially supported by the Ministry of Science and Technology (MOST) of Taiwan under grant number MOST 104-2221-E-007-072-MY3, 104-2321-B-007-003, and 103-2321-B-007-004, and the Excellence University Centre Project for Biomedical Technology, NTHU, 2012–2015. We would like to thank Dr Helen Chang for grammatical correction.Notes and references
- N. T. Nguyen, S. A. M. Shaegh, N. Kashaninejad and D. T. Phan, Adv. Drug Delivery Rev., 2013, 35(11–12), 1403–1419 CrossRef PubMed.
- P. Neuzil, S. Giselbrecht, K. Lange, T. J. Haung and A. Manz, Nat. Rev. Drug Discovery, 2012, 11, 620–632 CrossRef PubMed.
- P. A. Auroux, D. Iossifidis, D. R. Reyes and A. Manz, Anal. Chem., 2002, 74(12), 2623–2636 CrossRef.
- P. S. Dittrich and A. Manz, Lab-on-a-Chip, 2006, 5, 210–218 CAS.
- P. S. Dittrich, K. Tachikawa and A. Manz, Anal. Chem., 2006, 78, 3887–3907 CrossRef CAS PubMed.
- D. R. Reyes, D. Iossifidis, P. A. Auroux and A. Manz, Anal. Chem., 2002, 74, 2623–2636 CrossRef CAS PubMed.
- Z. T. Yu, K. Kamei, H. Takahashi, C. J. Shu, X. Wang, G. W. He, R. Silverman, C. G. Radu, O. N. Witte, K. B. Lee and H. R. Tseng, Biomed. Microdevices, 2009, 11(3), 547–555 CrossRef CAS PubMed.
- A. Kresol and J. E. Dick, Cell Stem Cell, 2014, 14(3), 275–291 CrossRef PubMed.
- H. Podder, S. M. Stepkowski, K. L. Napoli, J. Clark, R. R. Verani, T. C. Chou and B. D. Kahan, J. Am. Soc. Nephrol., 2001, 12, 1059–1071 CAS.
- P. M. LoRusso, R. Canetta, J. A. Wagner, E. P. Balogh, S. J. Nass, S. A. Boerner and J. Hohneker, Clin. Cancer Res., 2012, 18(22), 6101–6109 CrossRef PubMed.
- L. Xu, E. Tomaso, D. Fukumura and R. Jain, Nature, 2002, 416, 279–280 CrossRef PubMed.
- A. Carnero, Clin. Transl. Oncol., 2006, 8, 482–490 CrossRef CAS PubMed.
- S. E. Chung, J. Kim, D. Y. Oh, Y. Song and S. Kwon, Nat. Commun., 2014, 5, 3468 Search PubMed.
- P. K. Wong, F. Yu, A. Shahangian, G. Cheng, R. Sun and C. M. Ho, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(13), 5105–5110 CrossRef CAS PubMed.
- H. Wang, D. K. Lee, K. Y. Chen, J. Y. Chen, K. Zhang, S. Silva, C. M. Ho and D. Ho, ACS Nano, 2015, 9(3), 3332–3344 CrossRef CAS PubMed.
- A. Weiss, R. H. Berndsen, X. Ding, C. M. Ho, P. J. Dyson, H. V. D. Bergh, A. W. Griffioen and P. N. Sliwinska, Sci. Rep., 2015, 5, 14508 CrossRef CAS PubMed.
- X. Ding, H. Xu, C. Hopper, J. Yang and C. M. Ho, Proc. Natl. Acad. Sci. U. S. A., 2012, 29, 299–304 Search PubMed.
- H. Zeng, B. Li, X. Su, J. Qin and B. C. Lin, Lab Chip, 2009, 9, 1340–1343 RSC.
- P. Xue, Y. Wu, N. V. Menon and Y. Kang, Microfluid. Nanofluid., 2015, 18, 333–342 CrossRef CAS.
- J. Pessi, H. A. Santos, I. Miroshnyk, J. Yliruusi, D. A. Weitz and S. Mirza, Int. J. Pharm., 2014, 472(1–2), 82–87 CrossRef CAS PubMed.
- F. Kong, X. Zhang, H. Zhang, X. Qu, D. Chen, M. Servos, E. Mäkilä, J. Salonen, H. A. Santos, M. Hai and D. A. Weitz, Adv. Funct. Mater., 2015, 25(22), 3330–3340 CrossRef CAS.
- R. M. Hernandez, G. Orive, A. Murua and J. L. Pedraz, Adv. Drug Delivery Rev., 2010, 62(7–8), 711–730 CrossRef CAS PubMed.
- G. Orive, M. De Castro, H. J. Kong, R. M. Hernandez, S. Ponce, D. J. Mooney and J. L. Pedraz, J. Controlled Release, 2009, 135(3), 203–210 CrossRef CAS PubMed.
- L. Li, A. E. Davidovich, J. M. Schloss, U. Chippada, R. R. Schloss, N. A. Langrana and M. L. Yarmush, Biomaterials, 2011, 32(20), 4489–4497 CrossRef CAS PubMed; C. A. Hoesli, K. Raghuram, R. L. J. Kiang, D. Mocinecova, X. Hu, J. D. Johnson, I. Lacik, T. J. Kieffer and J. M. Piret, Biotechnol. Bioeng., 2010, 108(2), 424–434 CrossRef PubMed.
- D. B. Seifert and J. A. Phillips, Biotechnol. Prog., 1997, 13(5), 562–568 CrossRef CAS.
- H. Zimmermann, S. G. Shirley and U. Zimmermann, Curr. Diabetes Rep., 2007, 7(4), 314–320 CrossRef CAS.
- M. R. Prausnitz, Adv. Drug Delivery Rev., 2004, 56(5), 581–587 CrossRef CAS PubMed.
- J. Wan, Polymers, 2012, 4(2), 1084–1108 CrossRef CAS.
- F. Wei, B. Bai and C. M. Ho, Biosens. Bioelectron., 2011, 30(1), 174–179 CrossRef CAS PubMed.
- S. Freiberg and X. Zhu, Int. J. Pharm., 2004, 282(1–2), 1–18 CrossRef CAS PubMed.
- M. Yan, J. Du, Z. Gu, M. Liang, Y. Hu, W. Zhang, S. Priceman, L. Wu, Z. H. Zhou, L. Zheng, T. Segura, Y. Tang and Y. F. Lu, Nat. Nanotechnol., 2010, 5, 48–53 CrossRef CAS PubMed.
- N. Romano, D. Sengupta, C. Chung and S. C. Hailshorn, Biochim. Biophys. Acta, 2011, 1810(3), 339–349 CrossRef CAS PubMed.
- Y. Song, T. Kwon, D. Lee, J. Kim, D. Ho, T. Park and S. Kwon, Encoding of liquid capped microcapsule and heterogeneous assembly for multiplexed assay, 2012, 978-0–9798064–5–2/μTAS Search PubMed.
- D. F. Williams, Biomaterials, 2009, 30, 5897–5909 CrossRef CAS PubMed.
- B. Balakrishnan and R. Banerjee, Chem. Rev., 2011, 111, 4453–4474 CrossRef CAS PubMed.
- H. Zimmermann, F. Ehrhart, D. Zimmermann, K. Mueller, A. Katsen-Globa, M. Behringer, P. J. Feilen, P. Gessner, G. Zimmermann, S. G. Shirley, M. M. Weber, J. Metze and U. Zimmermann, Appl. Phys. A: Mater. Sci. Process., 2007, 89(4), 909–922 CrossRef CAS.
- L. Yu and J. Ding, Chem. Soc. Rev., 2008, 37(8), 1473–1481 RSC.
- O. D. Krishna and K. L. Kiick, Biopolymers, 2010, 94(1), 32–48 CrossRef CAS PubMed.
- A. Sionkowska, Prog. Polym. Sci., 2011, 36(9), 1254–1276 CrossRef CAS.
- S. W. Choi, Y. Zhang and Y. Xia, Angew. Chem., Int. Ed., 2010, 49(43), 7904–7908 CrossRef CAS PubMed.
- H. Oliveira, E. P. Andres, J. Thevenot, O. Sandre, E. Berra and S. Lecommandoux, J. Controlled Release, 2013, 169(3), 165–170 CrossRef CAS PubMed.
- K. Y. Lee, M. Peters and D. Mooney, Adv. Mater., 2001, 13(11), 837–839 CrossRef CAS.
- W. Ong, Y. Yang, A. C. Cruciano and R. L. McCarley, J. Am. Chem. Soc., 2008, 130(44), 14739–14744 CrossRef CAS PubMed.
- N. Huebsch, C. J. Kearney, X. Zhao, J. Kim, C. A. Cezar, Z. Suo and D. J. Mooney, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(27), 9762–9767 CrossRef CAS PubMed.
- M. Ravi, V. Paramesh, S. R. Kaviya, E. Anuradha and F. D. Paul Solomon, J. Cell. Physiol., 2015, 230(1), 16–26 CrossRef CAS PubMed.
- R. Mo, T. Jiang, R. DiSanto, W. Tai and Z. Gu, Nat. Commun., 2014, 5, 3364–3373 Search PubMed.
- J. P. Mazzoccoli, D. L. Feke, H. Baskaran and P. N. Pintauro, J. Biomed. Mater. Res., Part A, 2010, 93(2), 558–566 Search PubMed.
- C. G. Williams, A. N. Malik, T. K. Kim, P. N. Manson and J. H. Elisseeff, Biomaterials, 2005, 26(11), 1211–1218 CrossRef CAS PubMed.
- S. Abhimanyu, R. Maham, C. Christopher and K. T. Nguyen, J. Biomed. Mater. Res., Part A, 2009, 91(1), 52–59 Search PubMed.
- R. Foty, J. Visualized Exp., 2011, 51, 2720 Search PubMed.
- J. P. Fouassier, Photoinitiation, photopolymerization, and photocuring: funda-mentals and applications, Carl Hanser Verlag, Munich, 1995 Search PubMed.
- C. G. Williams, A. N. Malik, T. K. Kim, P. N. Manson and J. H. Elisseeff, Biomaterials, 2005, 26(11), 1211–1218 CrossRef CAS PubMed.
- J. P. Kehrer, Crit. Rev. Toxicol., 1993, 23(1), 21–48 CrossRef CAS PubMed.
- D. J. Quick and K. S. Anseth, J. Controlled Release, 2004, 96(2), 341–351 CrossRef CAS PubMed.
- D. J. Quick, K. K. Macdonald and K. S. Anseth, J. Controlled Release, 2004, 97(2), 333–343 CrossRef CAS PubMed.
- J. Bryant, C. R. Nuttelman and K. S. Anseth, J. Biomater. Sci., Polym. Ed., 2000, 11(5), 439–457 CrossRef PubMed.
- American cancer society.
- http://www.chemocare.com.
- http://toxnet.nlm.nih.gov/cgi-bin/sis/search/a?dbs+hsdb:@term+@DOCNO+6544.
- M. L. Pascu, Biochem. Pharmacol., 2012, 1(2), 1–3 Search PubMed.
- M. L. Pascu, M. Brezeanu, L. Voicu, A. Staicu, B. Carstocea and R. A. Pascu, 5-fluorouracil as a phosensitiser, in vivo, 2005, 19, 215–220 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra01733a |
| This journal is © The Royal Society of Chemistry 2016 |