Interactions of 1-hydroxypyrene with bovine serum albumin: insights from multi-spectroscopy, docking and molecular dynamics simulation methods†
Jing Zhanga,
Weixiao Chenab,
Bowen Tangc,
Wei Zhanga,
Linfeng Chena,
Ying Duana,
Yuxiu Zhua,
Yaxian Zhud and
Yong Zhang*ae
aState Key Laboratory of Marine Environmental Sciences of China (Xiamen University), College of Environment and Ecology, Xiamen University, Xiamen, 361102, China. E-mail: yzhang@xmu.edu.cn; Fax: +86 592 2888685; Tel: +86 592 2888685
bCollege of Urban and Environmental Sciences, Peking University, Beijing 100871, China
cCollege of Pharmaceutical Sciences, Xiamen University, Xiamen 361102, China
dDepartment of Chemistry, College of Chemistry and Chemical Engineering, Xiamen University, Xiamen, 361005, China
eZhangzhou Institute of Technology, Zhangzhou, 363000, China
First published on 22nd February 2016
Abstract
The interaction between a typical PAH metabolite, 1-hydroxypyrene (1-OHP), and a transport protein, bovine serum albumin (BSA), has been investigated using fluorescence, UV-visible absorption (UV-vis), circular dichroism (CD) spectra, docking and molecular dynamics (MD) simulation methods under simulated physiological conditions (in Tris–HCl buffer, pH = 7.40). The experimental results suggested that the fluorescence quenching of BSA by 1-OHP occurred through a mixed static and dynamic quenching mechanism with a binding constant of 2.40 × 106 L mol−1 at 291 K. The thermodynamic parameters together with the docking and MD study revealed that van der Waals forces dominate the formation of the 1-OHP–BSA complex. Applying Förster's non-radiation energy transfer theory, the binding distance of 1-OHP to BSA was calculated to be 2.88 nm. In addition, as confirmed by time-resolved fluorescence, UV-vis, three-dimensional (3-D) fluorescence and CD spectra, high concentrations of 1-OHP induced conformational transitions of BSA, increasing the content of the α-helix of BSA and exposing its tryptophan residue to a more hydrophilic microenvironment. An inhibition test showed that 1-OHP strongly inhibits the binding constant of vitamin B2 with BSA. A molecular docking study visualized the binding mode of 1-OHP with BSA. 1-OHP inserted into the binding pocket IB of BSA, leaving its hydroxyl group outside. Based on that, the MD study further unveiled the stability of 1-OHP–BSA complex and their dynamic binding modes, and clarified the contributions of each binding force component and the key residues to the binding process.
Introduction
Polycyclic aromatic hydrocarbons (PAHs) are widespread environmentally persistent organic pollutants.1 Because of their highly hydrophobic and persistent characteristics, PAHs can accumulate in various organisms and pose a great potential hazard to human bodies and animal health on a global basis through exposure to various environmental phases, such as food chains, skin exposure and inhalation.2 Upon entry into a human body, inactive parent PAHs are primarily metabolized by cytochrome P450 enzymes, forming a number of more active oxy-derivatives, including epoxides and hydroxyl compounds. These metabolites can bind to biomacromolecules, such as DNA and proteins, causing DNA damage and eventually leading to mutations.3,4Serum albumin (ALB), the most abundant carrier protein in blood plasma, plays a fundamental role in the maintenance of the plasma colloid osmotic pressure and the binding and transportation of various endo- and exogenous compounds, such as fatty acids and drugs.5 As has been reported, PAH metabolites can bind and form stable adducts with ALB.6 These adducts can last about one month in human plasma and undergo no repair during the whole lifetime of the protein. Those PAH metabolites can then be transported to the target organs via blood circulation. As a result, the distribution, free concentration and disposition of PAH metabolites in vivo can be significantly affected by their binding to ALB. More importantly, the binding of PAH metabolites to ALB can result in adverse effects on the carrier protein, most likely by altering its structure or capturing its active binding sites. This may affect the normal biological functions of ALB and cause potential hazards to organisms. For decades, studies have focused on developing methods to detect the level of PAHs or PAH derivatives–ALB adducts in human plasma and the correlation between these levels and the total exposure of PAHs.7 However, to date, as an essential part to understand the disposition, transition and toxicity processes of PAH derivatives in vivo, the binding mechanism of PAH metabolites with ALB are not fully understood.
The interactions of ligands with ALB are of considerable interest for decades, especially in drugs and nanomaterials. There are plentiful literature precedents about their binding affinity with ALB, dominant binding forces, major binding sites on ALB, as well as the conformation transitions of ALB.8–10 In contrast, to date, only the equilibrium constants6 and binding site11,12 information of a few PAHs and their metabolites with ALB have been investigated. Some recent studies have sought a more detailed understanding on the mechanism of such a binding process. Wu et al. investigated the interactions of 1-naphthol and 2-naphthol with bovine serum albumin (BSA) using spectroscopy methods. Their results indicated that the two naphthol compounds form a complex with BSA through a hydrophobic interaction.13 Ouyang et al. studied the interaction between 1-hydroxypyrene (1-OHP) and BSA with the presence of a surfactant, sodium dodecyl benzene sulfonate (SDBS), by combining fluorescence spectroscopy with UV-visible absorption (UV-vis) spectra. Their findings suggested that the fluorescence of BSA was quenched by 1-OHP through a static quenching mechanism; the process was dominated by hydrogen bonding and van der Waals forces.14 However, it is plausible that the presence of SDBS can significantly affect the environment surrounding 1-OHP and BSA.15 As a result, the findings cannot be applied to reflect the actual binding behavior, as discussed in our previous work.16 Xu et al. studied the binding process of pyrene with BSA using spectroscopy methods and demonstrated that pyrene can form a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex with BSA with a high affinity constant mainly through van der Waals forces and hydrogen bonding, while inducing damage to BSA.17 Although some progress has been made in the field, studies focusing on thoroughly integrated interactions of PAH metabolites with ALB in a more bio-relevant environment are still lacking. Knowledge on the binding modes between PAH metabolites and ALB is scarce. More importantly, the impact of PAH metabolites on the structure and biological functions of ALB are still unclear.
1 complex with BSA with a high affinity constant mainly through van der Waals forces and hydrogen bonding, while inducing damage to BSA.17 Although some progress has been made in the field, studies focusing on thoroughly integrated interactions of PAH metabolites with ALB in a more bio-relevant environment are still lacking. Knowledge on the binding modes between PAH metabolites and ALB is scarce. More importantly, the impact of PAH metabolites on the structure and biological functions of ALB are still unclear.
Spectroscopy methods, as a result of their sensitivity, relative ease of use and informational properties, are widely used to investigate ALB–ligand interactions. In recent years, theoretical calculation methods, such as molecular docking, have been applied to simulate the binding process of a ligand into the active site of a protein. Combinational use of experimental and docking research can provide more information on the interactions between ligands and ALB, such as ref. 18 and 19. In our previous work, we also employed multi-spectroscopy and docking method to study the interaction between inorganic mercury(II) and catalase (CAT) and predict their binding mode.20 However, in most molecular docking studies, proteins are commonly treated as “rigid” molecules to save computational time, thus their conformations are not allowed to adjust during docking.21 However, ALB is a very flexible protein, and its conformational changes induced by binding to ligands have been reported.17,19 Thus, molecular dynamics (MD) simulations, considering the flexibility of ALB, are deemed necessary to carry out to further simulate the dynamic interactions of ligands with ALB, as has been successfully used in ref. 9 and 22.
Therefore, in this study, based on our previous work,16,23 1-OHP was selected as a typical PAH metabolite, and BSA was selected as a model transport protein because of its well characterized physical properties, good stability and high similarity to human serum albumin (HSA). Multi-spectroscopy, in combination with the docking method, was employed to study the binding parameters (quenching mechanism, quenching constant, number of binding sites, binding distance and binding mode) of 1-OHP with BSA and the effects of the binding to 1-OHP on the structure and biological functions of BSA. MD simulations were also innovatively employed here to elucidate the stability of 1-OHP–BSA complex, and reveal the dynamic binding modes of 1-OHP with BSA at the atomic level. Their binding free energies were decomposed, and the contributions of each interaction force and key residues to the binding were further deciphered. To the best of our knowledge, an in-depth understanding of the interaction between PAH metabolites and BSA and their adverse impact on BSA was investigated here for the first time. The results obtained here will provide a detailed basic data to clarify the interaction mechanism of 1-OHP with BSA in vitro at both the molecular and atomic level, which is helpful to understand the toxicity effects of 1-OHP on the biological activity of the transport protein during the blood transportation process in vivo.
Experimental
Materials
BSA (purity > 99.5%), 1-OHP (purity > 99%) and vitamin B2 (VB2) (purity = 98%) were purchased from Sigma Chemical Company and were used without further purification. Tris–HCl buffer (0.05 mol L−1, containing 0.10 mol L−1 NaCl) was used to keep the pH of the solution at 7.40. Stock solutions of 1-OHP (2.0 × 10−3 mol L−1 in ethanol) and BSA (4.0 × 10−5 mol L−1 in Tris–HCl buffer) were both kept in the dark at 4 °C for storage. All of the chemicals that were used were of analytical reagent grade. Milli-Q water was used throughout the study.Methods
For all of the spectroscopy measurements, samples with different concentrations of 1-OHP and BSA were prepared by the sequential addition of 1-OHP and BSA (and VB2 in the inhibition test) to a series of 10 mL colorimetric tubes and were diluted to a total volume of 10 mL with Tris–HCl buffer. The only exception was for circular dichroism (CD) measurements; these samples were diluted with Milli-Q water. All of the samples were performed in triplicate. After equilibration for 20 min, the spectra were measured.UV-visible (UV-vis) absorption spectra
UV-vis absorption spectra were measured in the range of 200 to 500 nm at room temperature (291 K) on a Cary60 UV spectrophotometer (Varian, USA). A quartz cuvette with a 1 cm path-length was used.Fluorescence measurements
Fluorescence measurements were carried out on a FLS920 steady/transient fluorescence spectrometer (Edinburgh, UK) that was equipped with a 150 W xenon lamp. Samples were contained in a 1 cm quartz cuvette.Steady-state quenching experiments were carried out with samples at 291, 308 and 318 K; these temperatures were maintained by placing the samples in a thermostatic water bath for 20 min for equilibration. Fluorescence measurements of each individual sample were taken shortly afterwards. The excitation wavelength was 282 nm, and the emission spectra were recorded from 290 to 400 nm. To correct the non-negligible inner filter effect (IFE) of 1-OHP on BSA fluorescence, all of the reported experimental fluorescence values were corrected by multiplying a factor of 10(Aex+Aem)/2, where Aex/Aem is the absorption value of the BSA and 1-OHP mixture at the excitation/emission wavelength of BSA.24
Three-dimensional (3-D) fluorescence spectra at 291 K were measured with the initial excitation wavelength set at 200 nm with an increment of 5 nm, and the emission wavelength was recorded from 200 to 500 nm with increments of 5 nm. Thirty-one scanning curves were obtained. The excitation and emission slits were both set at 1 nm.
Time-resolved fluorescence intensity decays were also measured on a FLS920 fluorescence spectrometer at 291 K using the time-correlated single-photon counting (TCSPC) method. The excitation and emission wavelengths were set at 282 and 340 nm, respectively. Both excitation and emission slits were set at 10 nm. The lamp trigger delay was adjusted to 32 ns. The instrumental response function (IRF) was obtained by measuring a colloidal silica (Ludox AM-30, 30 wt% suspension in water) solution. The fluorescence decay curves were analyzed with an iterative fitting program and a F900 that was provided by Edinburgh Instruments after deconvolution of the IRF. Typically, the reduced χ2 value approaching 1 and a random distribution of weighted residuals indicates a good fit.
CD spectra
CD spectra (190–250 nm) were recorded using a Jasco-810 spectropolarimeter (Japan Spectroscopic, Japan) at 291 K. Three scans were obtained and averaged for each system. SELCON3 programs from the CDPro software package were used to calculate the secondary-structure content of BSA (http://lamar.colostate.Edu/%7Esreeram/CDPro/).Molecular docking study
AutoDock 4.2.6 (ref. 25) implemented with a Lamarckian genetic algorithm (LGA) for a ligand conformational search was employed to perform the docking process. The 3-D structure of 1-OHP was drawn using Gauss View 5.0 and was optimized with Gaussian 09.26 The native structure of BSA was downloaded from the Protein Data Bank (PDB ID 4F5S).27 The docking process and docking parameters were set up as described in a previous literature.28 The docking results presented here were analyzed using PyMOL software.29MD simulations
MD simulations were performed on the 1-OHP–BSA complex structure from docking using Amber14 and AmberTools15 software package30 following the reported protocols.31 The atomic partial charges for 1-OHP were computed by the electrostatic potentials (ESP) method at the level of B3LYP and the 6-31G(d) basis set using Gaussian 09 package.26 The Amber ff14SB force field and the general Amber force field (GAFF)32 were used to establish force field parameters for BSA and 1-OHP, respectively. The complex was neutralized by adding 35 Na+ ions, and then solvated in a truncated octahedron box surrounded by no less than 8 Å TIP3P water molecules. The resultant system was composed of approximately 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 897 atoms. The system was then optimized by energy minimization using steepest descent and conjugated gradient method, and then gradually heated to 300 K for 200 ps. Finally, the system was equilibrated in an isothermal–isobaric (NPT) ensemble with unconstrained MD simulations for first 50 ps and unconstrained MD simulations for further 50 ns. During above steps, the long range electrostatic interactions were calculated by the particle mesh Ewald (PME) method using a 8 Å nonbonded cutoff, while the SHAKE algorithm was applied to constrain all covalent bonds involving hydrogen atoms. The MD simulation results were analyzed using the cpptraj program in the AmberTools 15 package, PyMOL software and the Visual Molecular Dynamics (VMD) 1.9.2. Based on the obtained 50 ns MD trajectories, the binding free energy between 1-OHP and BSA was decomposed for every 5.0 ns MD simulation using the molecular mechanics Poisson–Boltzmann solvent accessible surface area (MM–PBSA) method. The total binding free energy and their decomposition were calculated as described by Ghadari et al.,33 and Cui et al.34
897 atoms. The system was then optimized by energy minimization using steepest descent and conjugated gradient method, and then gradually heated to 300 K for 200 ps. Finally, the system was equilibrated in an isothermal–isobaric (NPT) ensemble with unconstrained MD simulations for first 50 ps and unconstrained MD simulations for further 50 ns. During above steps, the long range electrostatic interactions were calculated by the particle mesh Ewald (PME) method using a 8 Å nonbonded cutoff, while the SHAKE algorithm was applied to constrain all covalent bonds involving hydrogen atoms. The MD simulation results were analyzed using the cpptraj program in the AmberTools 15 package, PyMOL software and the Visual Molecular Dynamics (VMD) 1.9.2. Based on the obtained 50 ns MD trajectories, the binding free energy between 1-OHP and BSA was decomposed for every 5.0 ns MD simulation using the molecular mechanics Poisson–Boltzmann solvent accessible surface area (MM–PBSA) method. The total binding free energy and their decomposition were calculated as described by Ghadari et al.,33 and Cui et al.34
Results and discussion
Characterization of the interaction of 1-OHP with BSA
The intrinsic fluorescence of BSA is very sensitive to its local microenvironment and can be easily affected when binding to small molecules.35 The fluorescence quenching method was thus employed here to clarify the binding mechanism between BSA and 1-OHP and obtain their binding constant. The fluorescence spectra of BSA in the presence of 1-OHP at various concentrations are illustrated in Fig. 1. As shown in Fig. 1, BSA has an intrinsic fluorescence peak at approximately 340 nm, which is mainly attributed to its tryptophan (TRP), tyrosine (TYR) and phenylalanine (PHE) residues.24 With the addition of 1-OHP, a steady quenching of the fluorescence intensity of BSA can be observed. Besides, the quenching effect was mainly dependent on the concentration of 1-OHP.The fluorescence quenching of BSA by 1-OHP may be caused by a ground-state complex formation or the occurrence of collisional encounters during the excited state between them. To determine the quenching mechanism between BSA and 1-OHP, the fluorescence lifetimes of pure BSA and 1-OHP–BSA systems were measured separately. A decrease in the fluorescence lifetime (τf) of BSA with increasing 1-OHP concentration was observed (Fig. S1†), which offers evidence that the dynamic quenching process has occurred.24 Moreover, the Stern–Volmer plot (using eqn (S1)†) of the fluorescence intensities of BSA at 340 nm clearly curves toward the y-axis (Fig. 2), as observed in many instances;36 this result qualitatively insinuated a mixed quenching process was involved in the interaction of 1-OHP with BSA.24
To extract a more quantitative view of the mixed quenching mechanism, the data were analyzed using the Stern–Volmer equations (eqn (S1) and (S2)†) according to ref. 24 and 36; the dynamic quenching constant KD of 1-OHP and BSA was determined to be 4.14 × 104 L mol−1, with the static quenching constant KS equal to 7.48 × 104 L mol−1. The results suggest a mixed static and dynamic quenching mechanism for the quenching process of 1-OHP on BSA, while static quenching is dominant.
For the static quenching dominant process, the double-logarithm equation can be used to determine the number of binding sites (n) and the binding equilibrium constant (Kb) between 1-OHP and BSA, as previously described.37 As calculated from the plots of log(F0/F − 1) versus log[Q] for 1-OHP and BSA (Fig. S3†), the values of Kb and n at 291, 308, 318 K for the 1-OHP–BSA system are listed in Table 1.
As shown in Table 1, the Kb and n values decrease regularly with increasing temperature, indicating that higher temperature decreased the formation of the 1-OHP–BSA complex. Furthermore, at 291 K, the n value approximately equalled 1, and the large Kb value of 2.40 × 106 L mol−1 suggested that 1-OHP has a strong affinity to BSA with one specific binding site. The Kb value is on the same order of magnitude with that of other PAH–ALB, such as ref. 6 and 38. Meanwhile, the Kb value is as large as that obtained for the interaction of ALB with various bioactive substances, such as nobiletin (3.66 × 106 L mol−1), chrysin (1.20 × 106 L mol−1) and kaempferol (2.58 × 106 L mol−1).8 This indicated that 1-OHP can bind with the transport protein as strongly as these bioactive substances that are important for organism functioning. Moreover, the Kb value was similar or even larger compared to that of the pyrene–BSA complex reported by Xu et al. (2.63 × 106 L mol−1)17 and Tsukamoto and Hikida (1.00 × l06 L mol−1).39 This is in accordance with the report that with the increase in the electrophilic properties of PAH molecule caused by the hydroxyl group, the affinity of some PAHs metabolites towards biomacromolecules increases compared with their parent PAHs.6 It is known that the binding constant of ligands to albumin is an important indicator of the activity of these substances in vivo.40 Thus, the result obtained here warns that when considering the importance of the toxicity of hydrophobic pollutants and their structurally similar substances (i.e., drugs), it is crucial to take into account their metabolites.16
Types of interaction force
The interaction forces between biological macromolecules and small organic molecules mainly include hydrogen bonds, van der Waals' interactions, electrostatic forces and hydrophobic interactions.41 Enthalpic change (ΔH) together with entropic change (ΔS) can provide insights into the type of interaction forces dominated in the binding process. Thus, the thermodynamic measurements were employed to help determine the major interaction forces that were present in the binding process of 1-OHP with BSA. Assuming that ΔH does not vary over the experimental temperature range, thermodynamic parameters can be calculated using a Van't Hoff analysis and thermodynamic equations.42 The ΔH and ΔS values were calculated as the slope and ordinate from the ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kb vs. 1/T plot (Fig. S4†), respectively. All of the relevant thermodynamic parameters of BSA and 1-OHP are listed in Table 1. The negative value of free-energy change (ΔG) revealed that the interaction of 1-OHP and BSA was spontaneous. Because ΔH < 0 and ΔS < 0, van der Waals or hydrogen bonding forces must be the main interaction forces involved in the binding process of 1-OHP with BSA.41,43
Kb vs. 1/T plot (Fig. S4†), respectively. All of the relevant thermodynamic parameters of BSA and 1-OHP are listed in Table 1. The negative value of free-energy change (ΔG) revealed that the interaction of 1-OHP and BSA was spontaneous. Because ΔH < 0 and ΔS < 0, van der Waals or hydrogen bonding forces must be the main interaction forces involved in the binding process of 1-OHP with BSA.41,43
Binding distance from BSA to 1-OHP
Fluorescence resonance energy transfer (FRET) is primarily used to measure the molecular distance of the donor–acceptor complex. Because the absorption spectrum of 1-OHP provides sufficient overlap with the fluorescence emission spectrum of BSA (Fig. 3), the energy transfer from BSA to 1-OHP can occur with a high probability according to Forster's dipole–dipole non-radioactive energy transfer theory.44 The binding distance between the TRP residue of BSA and 1-OHP was thus investigated using FRET. After fitting data to eqn (S3)–(S5),† all of the parameters values related to energy transfer are calculated and listed in Table S1.† | ||
| Fig. 3 Overlap of UV-vis spectrum of 1-OHP (b) with the fluorescence emission spectrum of BSA (a). CBSA = C1-OHP = 5.0 × 10−6 mol L−1. | ||
As calculated, the energy transfer efficiency (E) of BSA to 1-OHP is 0.4. The distance from the TRP residue of BSA to 1-OHP (r) is 2.88 nm, which is less than 7 nm; it confirms that there is a high probability of energy transfer from BSA to 1-OHP. Furthermore, the presence of a static quenching mechanism for the interaction between 1-OHP and BSA is demonstrated again.45
Fluorescence lifetime measurements
The fluorescence lifetime measurements are highly sensitive for detecting the excited-state interactions and measuring the possible micro-environment changes of the protein.24 To confirm that the binding of 1-OHP induces the micro-environmental changes of BSA, the time-resolved fluorescence decays of BSA in the presence of 1-OHP were measured with the corresponding parameters displayed in Table 2.| C1-OHP (mol L−1) | τ1 (ns) | τ2 (ns) | τ3 (ns) | α1 (%) | α2 (%) | α3 (%) | τf (ns) | χ2 |
|---|---|---|---|---|---|---|---|---|
| 0 | — | 3.16 | 6.56 | — | 17.04 | 82.96 | 5.98 | 1.086 |
| 2.0 × 10−6 | 0.11 | 3.05 | 6.45 | 5.75 | 19.94 | 74.31 | 5.41 | 1.250 |
| 4.0 × 10−6 | 0.11 | 3.29 | 6.63 | 7.61 | 38.63 | 53.76 | 4.79 | 1.091 |
| 5.0 × 10−6 | 0.17 | 3.20 | 6.56 | 8.92 | 41.34 | 49.74 | 4.59 | 1.169 |
| 6.0 × 10−6 | 0.52 | 3.56 | 7.21 | 8.64 | 56.96 | 34.40 | 4.41 | 1.137 |
| 8.0 × 10−6 | 0.68 | 3.80 | 8.92 | 10.97 | 70.64 | 18.39 | 4.39 | 1.039 |
| 1.0 × 10−5 | 0.62 | 3.59 | 8.58 | 11.86 | 68.19 | 19.95 | 4.15 | 1.135 |
In Table 2, the observed curves for free BSA and the 1-OHP–BSA complex fit exponentially with good χ2 values. For free BSA, two lifetime components of 3.16 ns and 6.56 ns were obtained. In contrast, three lifetime components were detected with the addition of 1-OHP, which suggests a higher system complexity. The lifetimes of the three components remain stable at a low 1-OHP concentration and then increase slightly at 5.0 × 10−6 mol L−1 of 1-OHP and continue to grow with the addition of 1-OHP. The contribution intensities of the short lifetimes of τ1 and τ2 increase with increasing 1-OHP concentration, while the contribution of the long lifetime τ3 displays a decreasing trend.
Short lifetimes of BSA at 0.3–0.4 ns and 2–3.5 ns have been commonly considered to be associated with the intrinsic property of the TRP structure, while the longer lifetime at about 6 ns is attributed to the interaction between the TRP residue (s) and the surrounding environment. Additionally, the relative contribution of each component depends on the number of emitting TRP residues or/and on the type of interaction occurring between TRP residues and the surrounding environment.46 Based on both the increasing trend of longer lifetime τ3 and the changes for each component contributions, it is therefore evident that the interaction of 1-OHP and BSA altered the protein structure near the TRP residues of BSA and changed the micro-environment around the TRP residues. The continual decreasing of the lifetimes indicates that higher concentrations of 1-OHP can induce greater structural alteration of BSA. The same phenomenon is also reported in ref. 19.
Conformation transition of BSA with 1-OHP
The UV-vis absorption, 3-D fluorescence and CD spectroscopy results are discussed in the following sections, focusing on how the interaction impacts the structure or micro-environment of BSA. | ||
| Fig. 5 3-D fluorescence spectra of BSA (A), 1-OHP–BSA system (B) and the 3-D contour map counter plots of BSA (C), 1-OHP–BSA system (D). CBSA = C1-OHP = 5.0 × 10−6 mol L−1. | ||
| Peaks | BSA | 1-OHP–BSA | ||||
|---|---|---|---|---|---|---|
| Peak position λex/λem (nm nm−1) | Stokes shift Δλ (nm) | Intensity | Peak position λex/λem (nm nm−1) | Stokes shift Δλ (nm) | Intensity | |
| Peak 1 | 200/200–330/330 | 0 | 1202–16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
240/240–330/330 | 0 | 2021–19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 030 030 |
| Peak 2 | 282/341 | 59 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 620 620 |
284/344 | 60 | 5923 |
Peak 1 in Fig. 5 is the Rayleigh scattering peak (λex = λem). The peak intensity increases with the addition of 1-OHP, which suggests the formation of the 1-OHP–BSA complex, causing a larger macromolecule diameter and enhancing the scattering effect.50 Peak 2 (λex = 282 nm, λem = 341 nm) in Fig. 5 primarily reveals the intrinsic fluorescence of TRP residues.51 Its fluorescence intensity exhibits a drastic decrease from 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 620 to 5923 with the addition of 1-OHP and a red shift of about 3 nm at the maximum emission wavelength, suggesting that the TRP residues of BSA exposed to a less hydrophobic micro-environment.52
620 to 5923 with the addition of 1-OHP and a red shift of about 3 nm at the maximum emission wavelength, suggesting that the TRP residues of BSA exposed to a less hydrophobic micro-environment.52
 | ||
Fig. 6 CD spectra of the 1-OHP–BSA systems at different molar ratios of 1-OHP to BSA. C1-OHP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CBSA (a–c) = 0 CBSA (a–c) = 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; 1 1; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; 10 1; 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. 1. | ||
In Table 4, pure BSA is mainly α-helices, with a percentage of 50.9%, which is in good agreement with previously published literature.19 Compared with the results of pure BSA, the secondary structure contents of BSA exhibit a steady change in response to the increasing ratio of 1-OHP. With a molar ratio of 1-OHP to BSA at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, there is approximately a 7% increase of α-helices and a 11% increase of turn, with an accompanying decrease of β-sheets and random coil contents of 30% and 12%, respectively. Therefore, our results suggest that the addition of 1-OHP obviously leads to secondary structural alterations of BSA, mainly by increasing the helical stability of BSA and breaking down the β-sheet and random coils structure. This may be a result of the formation of the 1-OHP–BSA complex, as demonstrated in.54 In addition, the greater structural alteration of BSA that was induced by higher concentrations of 1-OHP was confirmed again.
1, there is approximately a 7% increase of α-helices and a 11% increase of turn, with an accompanying decrease of β-sheets and random coil contents of 30% and 12%, respectively. Therefore, our results suggest that the addition of 1-OHP obviously leads to secondary structural alterations of BSA, mainly by increasing the helical stability of BSA and breaking down the β-sheet and random coils structure. This may be a result of the formation of the 1-OHP–BSA complex, as demonstrated in.54 In addition, the greater structural alteration of BSA that was induced by higher concentrations of 1-OHP was confirmed again.
Inhibition effects of 1-OHP on the physiological function of BSA to transport vitamin B2
From the results above, 1-OHP can form strong interaction with BSA, and induce the structural changes of BSA. Thus the normal biological function of BSA may also be affected by the accumulation of 1-OHP, such as the binding and carrying capacity of BSA.55 To reveal the effects of 1-OHP on the transport functions of BSA, the inhibition test of 1-OHP on the binding ability of BSA with VB2 was performed using the fluorescence quenching method. Fitting to the double-logarithm equation,56 the calculated Kb and n values for the VB2–BSA systems are listed in Table 5.With the addition of 1-OHP, the Kb and n values of BSA and VB2 decrease significantly compared with those without 1-OHP. 1-OHP at 1.0 × 10−5 mol L−1 reduces the binding constant of VB2 with BSA by nearly two orders of magnitude and decreases the number of binding sites of VB2 in BSA from 1.23 to 0.65. To the best of our knowledge, the binding sites of VB2 on BSA haven't been clearly clarified yet.57–60 Even so, the competition interaction between 1-OHP and VB2 with BSA cannot be excluded. Moreover, according to Zhang et al.61 and Chen et al.,62 small molecules can change the conformation of BSA after binding to BSA, making the conformation unfavorable for the binding of VB2 to BSA. Since the addition of 1-OHP alters the secondary structure of BSA and changes the microenvironment near Trp residues, the conformations of BSA after damaged by 1-OHP may also become unfavorable for binding to VB2.61 Thus the effect of the conformational changes of BSA on the binding of VB2 on BSA should not be neglected. In summary, the accumulation of 1-OHP could damage the physiological function of BSA possibly by altering its conformation and occupying its active binding sites, which may lead to potential danger for organisms.
Binding site for 1-OHP in BSA: molecular docking study
In order to know more information about the binding site location and binding mode, theoretical calculation methods could be used here. Because of the promising results in searching for the binding location of ligands on BSA,20,28 the AutoDock blind docking method has been employed to seek the preferred binding location of 1-OHP in BSA and to corroborate the experimental observations. Out of the 25 conformers that were obtained, the conformer with the lowest binding free energy (−28.55 kJ mol−1) was used for further analysis. | ||
| Fig. 7 Docking results of the 1-OHP–BSA system: (A) binding site and distance from TRP residues to 1-OHP, (B) binding mode, (C) neighboring amino acids within a distance of 4 Å approximately 1-OHP (the hydrogen bond between 1-OHP and GLU182 is shown as yellow dots) (read more about the colors in ESI†). | ||
The distances between 1-OHP and the two TRP residues, TRP134 and TRP213, are measured to be 1.2 nm and 2.5 nm, respectively. Such close proximity highly supports the possible energy transfer from TRP residues to the 1-OHP molecule, as revealed earlier in the results of the energy transfer study.
Molecular dynamic simulations
In order to gain deeper understanding of the interaction process, the lowest energy structure of 1-OHP–BSA complex resulting from the docking process was selected as the initial structure for 50 ns MD simulations. | ||
| Fig. 8 Time evolutions of the backbone RMSD of BSA (blue line) and 1-OHP (red line) in MD simulations. | ||
Physicochemical parameters such as the total energy and potential energy were also calculated and presented in Fig. S5.† The stable total energy and potential energy with smooth curves also indicate that the system reaches stability.68 Meanwhile, the evolution of the conformation of the complex over time was displayed using VMD software, which shows that 1-OHP is always immersed in the IB subdomain of BSA. These results further confirm that the molecular docking result for the binding location of 1-OHP in BSA is credible.
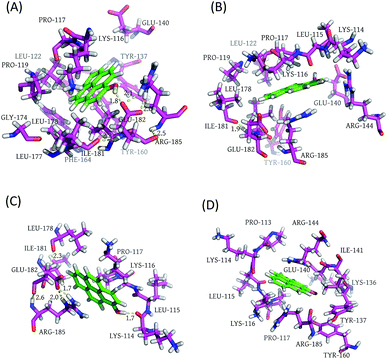 | ||
| Fig. 9 Binding modes of 1-OHP with BSA at 15 ns (A), 20 ns (B), 30 ns (C), and 40 ns (D) in the MD simulation (the hydrogen bonds between 1-OHP and GLU182, GLU182 and other residues are shown as yellow dots). Read more about the colors in ESI.† | ||
The position shifts and the rotations of the –OH group of 1-OHP may induce different interaction modes of 1-OHP with the nearby residues overtime, such as the different hydrogen bonds and cation–π interactions formed overtime. For instance, in Fig. 9, 1-OHP forms a hydrogen bond with GLU182 and LYS114, at 15 ns and 30 ns, respectively. Whereas no hydrogen bond is detected at 20 ns and 40 ns.
Since the hydrogen bond is closely related to the electrostatic interactions of 1-OHP to BSA,34 the occurrence and geometry of the hydrogen bonds between 1-OHP and BSA during the simulation time were further determined and analysed using H bond program from AmberTools15.
As listed in Table S3,† in agreement with the docking result, two hydrogen bonds are formed between the phenol OH of 1-OHP and OE1, OE2 group of GLU182 with an average distance of 2.63 Å and 2.64 Å. However, the two hydrogen bonds only occupy 34.42% and 17.24% of the simulation time, respectively. Meanwhile, other hydrogen bonds are formed between 1-OHP with GLU140, TYR160, LYS114, LEU115, PRO117 and PRO113 residues with low occupancies of all below 3%. The results confirm that the hydrogen bonds between 1-OHP and residues of BSA are not stable. The low occupancy of hydrogen bonds may be caused by that the solvent molecules penetrating into the binding site may attack on the hydrogen bonds, resulting in the weakness of hydrogen bond stability, as reported previously.31 Thus the hydrogen bond forces cannot play an important role in the binding process. Moreover, as illustrated in Fig. 9, during the most simulation time, there exit strong cation–π interactions between the large π-system from 1-OHP and the positively charged nitrogens from the side chain of nearby residues; the cation–π interactions formed between 1-OHP and ARG185 at 20 ns and 30 ns, or ARG144 at 40 ns (Fig. S7†). Comparing to the former hydrogen bonds forces, the cation–π interactions may be more important for 1-OHP binding to BSA. Moreover, in the simulations, some hydrophobic amino acid residues (TRP160, PRO117 and ILE181) and charged/polar residues (GLU182, ARG185 and LYS114, LYS116) always appear within 5 Å of 1-OHP. This result indicates that both hydrophobic and polar forces play an important role in the binding of 1-OHP to BSA.
Besides the interactions between 1-OHP and residues, the conformation of BSA at four snapshots were also calculated and lists in Table S4.† The secondary structures of the BSA during each 500 frames show no obvious changes, because of BSA being in the equilibrium state. Even so, it is worth to note the changes of the residues induced by the binding to 1-OHP. As shown in Fig. 9, during the simulation, after 1-OHP come into this binding site, the nearby residues change its conformation and position for more stable state of complex system. For instance, compared the snapshot at 30 ns to that at 15 ns, it is obvious to see that at 15 ns the OE1, OE2 atom of GLU182 formed a hydrogen bond with the HE, HH2 atom of ARG185, with corresponding bond lengths of 2.0 Å and 2.1 Å. Whereas, at 30 ns, the orientation of GLU182 changes significantly; the OE2 atom of GLU182 formed two hydrogen bonds with the HE, HH2 atomic of ARG185, with closer distance of 2.0 Å and 1.7 Å. Thus the conformation of GLU182 becomes more stable, and its surrounding microenvironment also changes. The position, orientation and microenvironment changes of the residues induced by binding to 1-OHP further confirm the UV-vis and CD spectra result that the interactions with 1-OHP induce the structural and microenvironmental changes of BSA.
Binding free energy analysis
To gain further insight into the forces involved in the binding process of 1-OHP with BSA, the total binding free energy was decomposed and analysed using MM–PBSA methods. The MM–PBSA binding free energy calculations were carried out for the 500 snapshots of every 5 ns MD simulation. The contributions of each component are shown in Fig. 10, with the data listed in Table S5.† In Table S5,† the total calculated binding energy for 1-OHP–BSA complex was (−17 ± 4) kcal mol−1. The difference from the aforementioned experimental ΔG value (about −8.18 kcal mol−1) was largely due to the ignorance of the entropy contributions in PBSA method. When the experimental ΔG also ignored the TΔS (−5.145 kcal mol−1) contribution, the calculated and experimental results were very close, indicating the accuracy of MM–PBSA method. In Fig. 10, the components of the binding free energy (ΔEbind) show that the van der Waals energy (ΔEvdw) makes the greatest contribution to the binding free energy, and the electrostatic energy (ΔEele) and the nonpolar solvation free energy (ΔEnonpolar) are also beneficial to the binding, while the polar solvation free energy (ΔEpolar) shows unfavorable contributions. In addition, compared to the value of sum of the polar interaction energies (∑Epolar), the larger negative value of the sum of the nonpolar interaction energies (∑Enonpolar) indicated that the hydrophobic forces played the dominant role in stabilizing the binding of 1-OHP with BSA. This result is in agreement with that obtained from the experimental results. It is worth to note that the magnitude of ΔEele for the complex fluctuates greatly. Since hydrogen bonds between the ligand and protein are closely related to the electrostatic energy, the fluctuations of ΔEele are possibly related to the aforementioned formation and breakdown of the hydrogen bonds.34Overall, the binding free energy decomposition results are consistent with the docking study and the experimental analysis. Combining the MD simulation, docking, and thermodynamic analysis, it is evident that hydrophobic interaction, especially the van der Waals dominates the binding of 1-OHP to BSA, despite the existence of hydrogen bonds and electrostatic interactions.
Conclusions
This work regarding the combination of multi-spectroscopy, docking and MD simulation methods has realized an in-depth understanding of not only the systematic interaction mechanism between 1-OHP and BSA but also the adverse effects of the binding on BSA. 1-OHP has been shown to form a strong 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex with BSA. Their quenching mechanism, binding constant, dominant binding forces and binding distance have been determined in a more realistic condition than the previous research reported by Ouyang et al.14 The structural transitions and transport functional inhibition of BSA induced by the binding process are corroborated here. The specific binding location of 1-OHP in BSA and their dynamic binding mode have also been revealed and are visually represented. The stability of the 1-OHP–BSA complex is corroborated. The contributions of each interaction force to the binding process have been clarified. The key residues contributing most to the binding energy have also been verified. These results reveal the interaction mechanism of 1-OHP with BSA, and warn of the great potential toxicity of 1-OHP on the transport protein in organisms.
1 complex with BSA. Their quenching mechanism, binding constant, dominant binding forces and binding distance have been determined in a more realistic condition than the previous research reported by Ouyang et al.14 The structural transitions and transport functional inhibition of BSA induced by the binding process are corroborated here. The specific binding location of 1-OHP in BSA and their dynamic binding mode have also been revealed and are visually represented. The stability of the 1-OHP–BSA complex is corroborated. The contributions of each interaction force to the binding process have been clarified. The key residues contributing most to the binding energy have also been verified. These results reveal the interaction mechanism of 1-OHP with BSA, and warn of the great potential toxicity of 1-OHP on the transport protein in organisms.
Besides, PAH metabolites can interact with other important biomacromolecules in vivo (e.g., glutathione peroxidase, catalase, superoxide dismutase, DNA, estrogen receptor, etc.), which can cause potential health hazards, such as oxidative damage, DNA damage, and endocrine disruption. Future work should be performed to further study the interactions of PAH metabolites with these important targets based on the established methods, so as to help further understand the toxicity mechanism of PAH metabolites at both molecular and atomic level. Such insights are well worth extending to the toxicity assessment aspects of other environmental contaminants, emerging nanoparticles and drugs.
Acknowledgements
We gratefully acknowledge the financial support of the National Natural Science Foundation of China (No. 21075102, 21177102, 21577110) and the Xiamen University Innovative Research Foundation (No. DC2013035). We also thank the College of Pharmaceutical Sciences, Xiamen University and Department of Chemistry, College of Chemistry and Chemical Engineering, Xiamen University for providing us with the amber14 and Gaussian 09 software. We also express our thanks to Elsevier Language Editing Services for their assistance with English. We also thank the reviewers and editors for their suggestions that are very helpful in improving the quality of this work.References
- S. Augusto, C. Máguas, J. Matos, M. J. Pereira and C. Branquinho, Environ. Pollut., 2010, 158, 483–489 CrossRef CAS PubMed.
- X. T. Wang, Y. Miao, Y. Zhang, Y. C. Li, M. H. Wu and G. Yu, Sci. Total Environ., 2013, 447, 80–89 CrossRef CAS PubMed.
- G. Prodi, S. Grilli, M. Mazzullo, A. Colacci and G. Arfellini, Toxicol. Pathol., 1984, 12, 185–188 CrossRef CAS PubMed.
- B. Moorthy, C. Chu and D. J. Carlin, Toxicol. Sci., 2015, 145, 5–15 CrossRef CAS PubMed.
- K. Yamasaki, V. T. G. Chuang, T. Maruyama and M. Otagiri, BBA, Biochim. Biophys. Acta, Gen. Subj., 2013, 1830, 5435–5443 CrossRef CAS PubMed.
- K. Skupińska, M. Zylm, I. Misiewicz and T. Kasprzycka-Guttman, Acta Biochim. Pol., 2006, 53, 101–112 Search PubMed.
- Y. Y. Qin, C. K. M. Leung, C. K. Lin, A. O. W. Leung, H. S. Wang, J. P. Giesy and M. H. Wong, Environ. Sci. Technol., 2011, 45, 1630–1637 CrossRef CAS PubMed.
- S. Naveenraj and S. Anandan, J. Photochem. Photobiol., C, 2013, 14, 53–71 CrossRef CAS.
- S. L. Zhuang, H. F. Wang, K. K. Ding, J. Y. Wang, L. M. Pan, Y. L. Lu, Q. J. Liu and C. L. Zhang, Chemosphere, 2016, 144, 1050–1059 CrossRef CAS PubMed.
- Z. Q. Xu, Q. Q. Yang, J. Y. Lan, J. Q. Zhang, W. Peng, J. C. Jin, F. L. Jiang and Y. Liu, J. Hazard. Mater., 2016, 301, 242–249 CrossRef CAS PubMed.
- A. M. Saletskii, A. G. Mel’Nikov, A. B. Pravdin, V. I. Kochubei and G. V. Meln’ikov, J. Appl. Spectrosc., 2008, 75, 402–406 CrossRef CAS.
- P. Brunmark, S. Harriman, P. L. Skipper, J. S. Wishnok, S. Amin and S. R. Tannenbaum, Chem. Res. Toxicol., 1997, 10, 880–886 CrossRef CAS PubMed.
- T. Q. Wu, Q. Wu, S. Y. Guan, H. X. Su and Z. J. Cai, Biomacromolecules, 2007, 8, 1899–1906 CrossRef CAS PubMed.
- Y. Ouyang, Y. Wang and L. Guiying, Phys. Test. Chem. Anal., Part B, 2011, 47, 253–256 CAS.
- R. T. Liu, W. S. Zong, K. Jin, X. T. Lu, J. H. Zhu, L. J. Zhang and C. Z. Gao, Spectrochim. Acta, Part A, 2008, 70, 198–200 CrossRef PubMed.
- J. Zhang, W. X. Chen, W. Zhang, Y. Duan, Y. X. Zhu, Y. X. Zhu and Y. Zhang, Chem. J. Chin. Univ., 2015, 36, 1511–1516 CAS.
- C. B. Xu, J. L. Gu, X. P. Ma, T. Dong and X. L. Meng, Spectrochim. Acta, Part A, 2014, 125, 391–395 CrossRef CAS PubMed.
- D. Wu, Y. M. Zhai, J. Yan, K. L. Xu, Q. Wang, Y. Z. Li and H. Li, RSC Adv., 2015, 5, 11036–11042 RSC.
- F. Zhang, J. Zhang, C. L. Tong, Y. D. Chen, S. L. Zhuang and W. P. Liu, J. Hazard. Mater., 2013, 263, 618–626 CrossRef CAS PubMed.
- L. F. Chen, J. Zhang, Y. X. Zhu and Y. Zhang, RSC Adv., 2015, 5, 79874–79881 RSC.
- J. Shen, W. Q. Zhang, H. Fang, R. Perkins, W. D. Tong and H. X. Hong, BMC Bioinf., 2013, 14, S6 CrossRef PubMed.
- W. Peng, F. Ding and Y. K. Peng, RSC Adv., 2016, 6, 1826–1843 RSC.
- L. F. Chen, J. Zhang, Y. X. Zhu and Y. Zhang, Chem. J. Chin. Univ., 2015, 36, 2394–2401 CAS.
- J. R. Lakowicz, Principles of fluorescence spectroscopy, Springer Science & Business Media, New York, 3rd edn, 2006 Search PubMed.
- G. M. Morris, D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew and A. J. Olson, J. Comput. Chem., 1998, 19, 1639–1662 CrossRef CAS.
- M. Frisch, G. Trucks, H. B. Schlegel, G. Scuseria, M. Robb, J. Cheeseman, G. Scalmani, V. Barone, B. Mennucci and G. Petersson, Gaussian 09, Revision A. 02, Gaussian, Inc., Wallingford, CT, 2009, p. 200 Search PubMed.
- A. Bujacz, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2012, 68, 1278–1289 CrossRef CAS PubMed.
- A. Ganguly, B. K. Paul, S. Ghosh, S. Dalapati and N. Guchhait, Phys. Chem. Chem. Phys., 2014, 16, 8465–8475 RSC.
- W. L. DeLano, DeLano Scientific, San Carlos, CA, 2002, p. 452 Search PubMed.
- D. A. Case, J. T. Berryman, R. M. Betz, D. S. Cerutti, T. E. Cheatham III, T. A. Darden, R. E. Duke, T. J. Giese, H. Gohlke, A. W. Goetz, N. Homeyer, S. Izadi, P. Janowski, J. Kaus, A. Kovalenko, T. S. Lee, S. LeGrand, P. Li, T. Luchko, R. Luo, B. Madej, K. M. Merz, G. Monard, P. Needham, H. Nguyen, H. T. Nguyen, I. Omelyan, A. Onufriev, D. R. Roe, A. Roitberg, R. Salomon-Ferrer, C. L. Simmerling, W. Smith, J. Swails, R. C. Walker, J. Wang, R. M. Wolf, X. Wu, D. M. York and P. A. Kollman, Amber, University of California, San Francisco, 2015 Search PubMed.
- K. K. Ding, H. X. Zhang, H. F. Wang, X. Lv, L. M. Pan, W. J. Zhang and S. L. Zhuang, J. Hazard. Mater., 2015, 299, 486–494 CrossRef CAS PubMed.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS PubMed.
- R. Ghadari, F. S. Alavi and M. Zahedi, Comput. Biol. Chem., 2015, 56, 33–40 CrossRef CAS PubMed.
- F. C. Cui, K. C. Yang and Y. Q. Li, PLoS One, 2015, 10, e0125848 Search PubMed.
- X. Peng, X. C. Wang, W. Qi, R. X. Su and Z. M. He, Food Chem., 2016, 192, 178–187 CrossRef CAS PubMed.
- B. P. Pahari, S. Chaudhuri, S. Chakraborty and P. K. Sengupta, J. Phys. Chem. B, 2015, 119, 2533–2545 CrossRef CAS PubMed.
- K. Lou, Z. H. Zhu, H. M. Zhang, Y. Q. Wang, X. J. Wang and J. Cao, Chem.-Biol. Interact., 2016, 243, 54–61 CrossRef CAS PubMed.
- N. A. Carmona, B. Cohen, J. A. Organero and A. Douhal, J. Photochem. Photobiol., A, 2012, 234, 3–11 CrossRef CAS.
- T. Tsukamoto and T. Hikida, J. Photochem. Photobiol., A, 1996, 95, 271–273 CrossRef CAS.
- S. Endo and K. U. Goss, Chem. Res. Toxicol., 2011, 24, 2293–2301 CrossRef CAS PubMed.
- P. D. Ross and S. Subramanian, Biochemistry, 1981, 20, 3096–3102 CrossRef CAS PubMed.
- S. Huang, F. W. Zhu, Q. Xiao, Q. Zhou, W. Su, H. N. Qiu, B. Q. Hu, J. R. Sheng and C. S. Huang, RSC Adv., 2014, 4, 36286–36300 RSC.
- X. R. Li and S. Wang, New J. Chem., 2015, 39, 386–395 RSC.
- T. Förster, Delocalized excitation and excitation transfer, Florida State University, 1965 Search PubMed.
- X. Peng, W. Qi, R. L. Huang, R. X. Su and Z. M. He, PLoS One, 2015, 10(4), e0118274 Search PubMed.
- N. Tayeh, T. Rungassamy and J. R. Albani, J. Pharm. Biomed. Anal., 2009, 50, 107–116 CrossRef CAS PubMed.
- O. S. Wolfbeis, M. Leiner, P. Hochmuth and H. Geiger, Berichte der Bunsengesellschaft für physikalische Chemie, 1984, 88, 759–767 CrossRef CAS.
- A. S. Sharma, S. Anandakumar, M. Ilanchelian and S. Anandakumar, RSC Adv., 2014, 4, 36267–36281 RSC.
- X. Chen, K. Qian and Q. Chen, Eur. J. Med. Chem., 2015, 93, 492–500 CrossRef CAS PubMed.
- Y. Z. Zhang, B. Zhou, Y. X. Liu, C. X. Zhou, X. L. Ding and Y. Liu, J. Fluoresc., 2008, 18, 109–118 CrossRef CAS PubMed.
- C. Jash and G. S. Kumar, RSC Adv., 2014, 4, 12514–12525 RSC.
- S. Huang, H. N. Qiu, S. Y. Lu, F. W. Zhu and Q. Xiao, J. Hazard. Mater., 2015, 285, 18–26 CrossRef CAS PubMed.
- N. J. Greenfield, Nat. Protoc., 2006, 1, 2876–2890 CrossRef CAS PubMed.
- X. Y. Xie, W. J. Lü and X. G. Chen, J. Hazard. Mater., 2013, 248, 347–354 CrossRef PubMed.
- F. Chiti and C. M. Dobson, Annu. Rev. Biochem., 2006, 75, 333–366 CrossRef CAS PubMed.
- Y. Shu, W. W. Xue, X. Y. Xu, Z. M. Jia, X. J. Yao, S. W. Liu and L. H. Liu, Food Chem., 2015, 173, 31–37 CrossRef CAS PubMed.
- X. J. Guo, X. D. Sun and S. K. Xu, J. Mol. Struct., 2009, 931, 55–59 CrossRef CAS.
- H. W. Zhao, M. Ge, Z. X. Zhang, W. F. Wang and G. Z. Wu, Spectrochim. Acta, Part A, 2006, 65, 811–817 CrossRef PubMed.
- M. Voicescu, D. G. Angelescu, S. Ionescu and V. S. Teodorescu, J. Nanopart. Res., 2013, 15, 1–10 CrossRef.
- F. Wang, W. Huang and Z. X. Dai, J. Mol. Struct., 2008, 875, 509–514 CrossRef CAS.
- X. Zhang, L. Chen, X. C. Fei, Y. S. Ma and H. W. Gao, BMC Mol. Biol., 2009, 10, 16 CrossRef PubMed.
- J. B. Chen, X. F. Zhou, Y. L. Zhang, Y. J. Qian and H. P. Gao, Amino Acids, 2012, 43, 1419–1429 CrossRef CAS PubMed.
- S. Neelam, M. Gokara, B. Sudhamalla, D. G. Amooru and R. Subramanyam, J. Phys. Chem. B, 2010, 114, 3005–3012 CrossRef CAS PubMed.
- A. Selva Sharma, S. Anandakumar and M. Ilanchelian, J. Lumin., 2014, 151, 206–218 CrossRef CAS.
- F. Zsila, Mol. Pharmaceutics, 2013, 10, 1668–1682 CrossRef CAS PubMed.
- H. Z. Shen, Y. Huang, R. Wang, D. Zhu, W. Li, G. F. Shen, B. Wang, Y. Y. Zhang, Y. C. Chen, Y. Lu, H. Chen, T. C. Li, K. Sun, B. G. Li, W. X. Liu, J. F. Liu and S. Tao, Environ. Sci. Technol., 2013, 47, 6415–6424 CAS.
- M. Shahlaei, B. Rahimi, M. R. Ashrafi-Kooshk, K. Sadrjavadi and R. Khodarahmi, J. Lumin., 2015, 158, 91–98 CrossRef CAS.
- S. Y. Liao, G. Q. Mo, J. C. Chen and K. C. Zheng, J. Mol. Model., 2014, 20, 1–9 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra00981f |
| This journal is © The Royal Society of Chemistry 2016 |





