Specific interaction of platinated DNA and proteins by surface plasmon resonance imaging†
Xiao Wangab,
Jiying Xu*a,
Chanjuan Liuab and
Yi Chen*abc
aKey Laboratory of Analytical Chemistry for Living Biosystems, Institute of Chemistry, Chinese Academy of Sciences, Beijing 100190, China. E-mail: xujy@iccas.ac.cn; chenyi@iccas.ac.cn
bUniversity of Chinese Academy of Sciences, Beijing 100049, China
cBeijing National Laboratory for Molecular Science, Beijing 100190, China
First published on 17th February 2016
Abstract
The DNA-targeting platinum complex cisplatin is one of the most successful drugs for clinical treatment of solid tumors, and several new analogues of transplatin have shown cytostatic activity recently. The specific recognition of platinated DNA with cellular proteins is of great interest for better understanding of the pharmaceutical mechanisms. Herein, a surface plasmon resonance imaging (SPRi) method to differentiate the interaction between the protein human high mobility group box 1 (HMGB1) and DNAs, and human nuclear protein positive cofactor 4 (PC4) and DNAs has been developed. Four kinds of DNAs were immobilized on the gold films including platinated-DNA adducts (cisplatin and trans-[PtCl2(NH3)(thiazole)] (trans-PtTz) damaged DNAs; referred to as cisPt–DNA1 and transPtTz–DNA2, respectively) and native DNAs (DNA1 and DNA2, as controls). The validation of the method has been proven by the specific recognition of HMGB1 and cisPt–DNA1 first. The results obtained indicated that the PC4 was more likely to bind to platinated DNAs (cisPt–DNA1 and transPtTz–DNA2) than the native DNAs. Temperature-dependent kinetics and thermodynamics revealed that the recognition behavior was not affected by a temperature changefrom 15 °C to 42 °C. This label-free method provides authentic results, as the controls can be simultaneously determined by a single chip under the same conditions, and this makes it suited for other high throughput analyses of interactions between drug-damaged DNAs and proteins to better understand the activity/inactivity mechanisms of drugs and drug screening/discovery.
Introduction
cis-Diamminedichloroplatinum(II) (cisplatin), one of the most successful anticancer drugs in clinical treatment of various human malignancies, has been demonstrated to bind to the target DNA to form cisPt–DNA adduct(s) or damaged DNA and then induce the cells into apoptosis.1–4 The NMR analysis of the interaction between the cisplatinated DNA and high-mobility-group (HMG) domain protein revealed that the DNA dodecamer duplex containing a d(GpG) intrastrand cross-link was the major adduct of the cisplatin, and cisplatin binding altered the DNA native conformation to facilitate HMG-domain protein recognition.5 Furthermore, the X-ray crystal characterization of the cisplatinated DNA and the HMG1 complex indicated that the protein HMG1 was likely to be bound to the widened minor groove of the 16-base-pair DNA duplex containing a site-specific cis-[Pt(NH3)2] adduct.2 S. Müller reported that there were significant differences in the binding characteristics of full HMGB1 and truncated HMGB1 suggesting an important role of the acid tail in modulating DNA binding.6 Cisplatin has high antitumor activity but its trans-isomer (transplatin) is inert. However, several new analogues of transplatin containing a planar amine (L) ligand in the general structure of trans-[PtCl2(NH3)(L)] have shown cytostatic activity against cisplatin-resistant tumor cells recently. In addition, human nuclear protein positive cofactor 4 (PC4)7–10 was also shown, using mass spectrometry combined with nano-HPLC, to bind selectively to trans-[PtCl2(NH3)(thiazole)] (trans-PtTz) damaged DNA,9 while trans-PtTz is a kind of trans-platinum in which one amino group is substituted by organic molecular thiazole amine group.HMGB1 and PC4 are a group of p53-interacting proteins, enhancing the sequence specific DNA binding ability and transactivation functions of p53, while p53 is a well-known tumor suppressor for the maintenance of genomic integrity and cellular homeostasis.11 HMGB1 is a nuclear protein which activates p53 binding to its cognate sites by providing a bent DNA for p53 recruitment, as well as p53-dependent transcription. It inhibits the repair of cisPt–DNA in vitro and in vivo and has been implicated in transcription, replication, and cellular differentiation.4,12–14 It has been confirmed that HMGB1 could recognize 1,2-intrastrand cross-link DNA damaged by platinum-based drugs. The endogenous PC4 is a highly abundant and multifunctional nuclear protein binding to DNA, which plays a vital role in several processes such as transcription, replication, DNA repair, and normal cellular growth. It stimulates activator-dependent class II gene transcription in vitro, and has been found to accumulate at DNA damage sites induced by either chemical agents or laser micro-irradiation.8,10 It has been reported that the transPtTz–DNA afforded mono-functional, bi-functional intra- and inter-stranded adducts in roughly equal proportions.12 However, differences in HMGB1/PC4 binding to the cis/trans-platinated DNA have not yet been well studied or remain unclear.
Surface plasmon resonance (SPR) is a promising method to study the mechanism of molecule–molecule interaction and a label-free technique to research the kinetics of molecular interaction, especially in physiology-compatible conditions.15–19 The SPR method has been used to research the binding affinity of platinum-free DNA with HMG1 and HMG2; a stronger binding ability of HMG1 with DNA than that of HMG2 was monitored.17 However, the presently adopted SPR methods can monitor only one interaction with a single flow cell, rather than multiple interactions. To overcome this weakness, SPR imaging (SPRi)15,16,20–23 has been shown promising to resolve such problems. SPRi extends SPR measurement to microarrays and enables direct measurement of a series of molecular binding kinetics.
In this paper, we present a SPRi method to differentiate the protein–DNA interaction reactions between HMGB1 and DNAs, & PC4 and DNAs. Four kinds of DNAs were immobilized on the gold films including platinated-DNA adducts (cisPt–DNA1 and transPtTz–DNA2) and native DNAs (DNA1 and DNA2, as controls). With a gold sensing chip array-dotted with a 1,2-cisplatin-cross-linked DNA motif (cisPt–DNA1, refer to Table 1), a 1,3-transPtTz-cross-linked DNA motif (transPtTz–DNA2, refer to Table 1) and two native DNAs (DNA1 and DNA2, refer to Table 1, as controls), the interactions and related kinetics between HMGB1 and DNAs & PC4 and DNAs were monitored under simulated physiological conditions, respectively, by our laboratory-built SPRi (Scheme 1). The differences of HMGB1/PC4 binding to the cis/trans-platinated DNA have not yet been well studied or remain unclear. The impact of temperature on the interactions was also inspected and some unexpected variations were observed and discussed thereafter.
Experimental
Materials and reagents
HS-oligonucleotides DNA1 and DNA2, platinated HS-oligonucleotides cisPt–DNA1 and transPtTz–DNA2, and proteins HMGB1 and PC4 (refer to Table 1) were provided by Dr Fuyi Wang (Key Laboratory of Analytical Chemistry for Living Bio-systems, Institute of Chemistry, Chinese Academy of Sciences). Bovine serum albumin (BSA) and pepsin were purchased from Amresco (Pkwy, Solon, Ohio, USA). Hemoglobin, cytochrome c, cysteamine hydrochloride and 6-mercapto-1-hexanol (MCH) were from Sigma (St. Louis, Mo, USA). NaCl, EDTA, MgCl2, HCl, ethanol, acetone, ethanolamine (EOA), Tween-20 and tris-(hydroxylmethyl) aminomethane (Tris) were all of analytical reagent grade from Beijing Chemical Works (Beijing, China). All chemicals were used as received. Aqueous solutions were all prepared with pure water (>18.2 MΩ cm) generated from a Milli-Q academic system (Billerica, MA, USA).Preparation of microarray chips
Glass slides (1.0 cm × 1.0 cm from Shitai Co. Ltd., China) were deposited with 2 nm-thickness Cr underlayer and 48 nm-thickness gold film by vapor-deposition in a JEE-420 vacuum evaporator (JEOL Ltd., Tokyo, Japan) and sequentially rinsed with acetone, pure water, ethanol and pure water again, dried under a nitrogen gas stream. They were activated by plasma cleaning treatment (PDC-MG, Mingheng, China) for 3 min, then array-spotted with thiolated DNA probes dissolved in a TME buffer at pH 7.8 composed of 10 mM Tris, 100 mM NaCl, 10 mM MgCl2 and 1 mM EDTA by a laboratory-built spotter, which can deliver a 15 nL solution drop onto the gold surface to form a spot of 250 μm in diameter with Au–S bonding.24,25 The spotted chips were incubated at room temperature in a humid environment for 2 h. The chips were frozen in liquid nitrogen for 3 min, then inserted into a TME buffer containing 20 mM cysteamine hydrochloride and 1 mM MCH immediately and kept for 2 h to block the unused active regions and nonspecific adsorption sites (Scheme 1). The chips were then washed thoroughly with TME buffer and pure water for at least three times. After drying under a nitrogen gas stream, the chips were used directly or could be stored in dark at −20 °C for at least one and a half months.SPRi measurement
All SPRi measurements were performed on our laboratory-built SPR imager, model TX7100, which was fabricated based on the Kretschmann configuration. Briefly, a p-polarized light beam was collimated and directed at a certain angle from one side of a prism into its bottom attached to a gold sensing chip via a refraction index matching oil (i.e. cedar oil) and sealed in a 40 μL flow cell. The light beam reflected from the other side was filtered through a narrow bandpass filter (λ 645 nm) and imaged onto a CCD camera (WAT-902B, Watec Co., Ltd., Japan). The temperature of the flow cell was controlled by a Peltier cooling and heating system and the analysis temperature can be adjusted from 2 °C to 95 °C. Whenever required, the images or video signals were acquired at 5 frames per second, then saved and analyzed with a laboratory-edited imaging workstation.To monitor interaction events, an array-dotted gold sensing chip was assembled onto the prism bottom and equilibrated with TME buffer containing 0.005% Tween-20 (v/v, TMETween), pumped at a flow rate of 30 μL min−1. Once the monitored signal became stable, the TMETween buffer was replaced by a protein solution dissolved in the same buffer, pumped at a flow rate of 10 μL min−1, then kept incubation at 20 °C (otherwise noted) for 10 min, the chip surface was washed by pumping the TMETween buffer into the flow cell again. The interacted gold chip was regenerated by pumping 2 M NaCl through the flow cell for 3 min, and then pumped the TMETween buffer until the recorded signal returned to the baseline and kept stable (Scheme 1).
Results and discussion
The interaction and reaction between the soluble protein (A) of either HMGB1 or PC4 with the immobilized DNA probes (B) was assumed to be at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry.16–18,21 Here, we carried out the following kinetic analysis:
1 stoichiometry.16–18,21 Here, we carried out the following kinetic analysis:
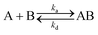 | (1) |
The Gibbs free energy change (ΔG) is a criterion to determine whether the reaction is spontaneous or balanced under constant temperature and pressure conditions. It can be obtained as follows:18
ΔG = RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kd Kd
| (2) |
Chip optimization
It showed a considerably lower amount of non-specific protein adsorption compared with the other two commonly used blocking solutions, 0.1% BSA (w/v) in TME buffer and 1 M EOA aqueous solution (Fig. S2 in the ESI†). In the buffer of pH 7.8 used here, BSA is negatively charged while HMGB1 is positively charged, for the pI values of two proteins are 4.85 and 10.1, respectively. Positively charged HMGB1 was easily absorbed onto negatively charged BSA, so BSA was not appropriate to be used here owing to serious electrostatic adsorption. However, cysteamine28 is a small hydrophilic molecule containing a mercapto group at one end which can facilitate the monolayer self-assembled on the gold surface and an amino group at the other end, and exhibits a positive charge in neutral solution. Thus we thought cysteamine would be a suitable blocking reagent to be used here due to its property of repelling proteins of the same charge. Meanwhile, in order to prevent DNA lodging on the sensor chip surface, 1 mM MCH was added into the blocking solution to keep the DNA chain in vertical extension and improve the space configuration of DNA to make the interaction more effective.24,25 This step reduces duplex density and is a key factor in increasing protein accessibility to specific recognition sequences within individual areas.29 In addition, BSA has been described as an ellipsoidal molecule with hydrodynamic dimensions of 14 × 4 × 4 nm;30 the portrait height is the same as the used DNAs with 16 bases per single strand (the theoretical chain length of the DNA is about 5 nm). So, BSA blocking may enclose the protein binding site of DNAs. Meanwhile the smaller molecules cysteamine and MCH will not affect the protein binding site of DNAs, which minimize the steric hindrance and improve the molecular accessibility.
Method development and verification
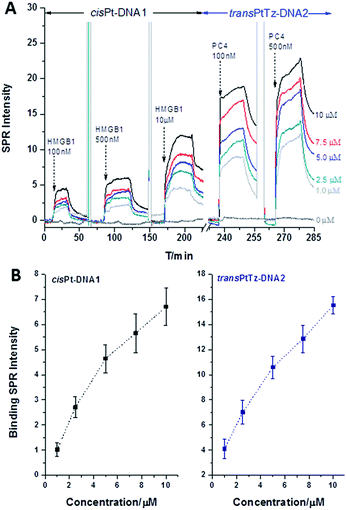
A method of SPR/SPRi was developed and validated under simulated physiological conditions. The DNA–protein interaction signals exhibited a probe-type-dependent behaviour, as Fig. 2 shows. From these results, it was obvious that HMGB1 had a higher affinity to the cisPt–DNA1 than to DNA1, DNA2 and transPtTz–DNA2, and the same result was shown in the SPRi image (interior Fig. 2). The binding signal of HMGB1 to the transPtTz–DNA2 was a little stronger than that of the DNA2. The binding signal of HMGB1 to no DNA confirmed that there was little non-specific adsorption. In the study of the binding characteristics of different concentrations of HMGB1 with the four DNAs (Fig. 3), we knew that the binding SPR intensities of the cisPt–DNA1 and DNA1 to HMGB1 increased in a HMGB1 concentration dependent manner, and the binding ability of HMGB1 to the cisPt–DNA1 was stronger than that of the control native DNA1. The signals of HMGB1 binding to the transPtTz–DNA2 and DNA2 increased a little when HMGB1 concentration increased, and that of the transPtTz–DNA2 was a little higher than DNA2. So, we deduced that the HMGB1 recognized the cisplatinated DNAs specifically.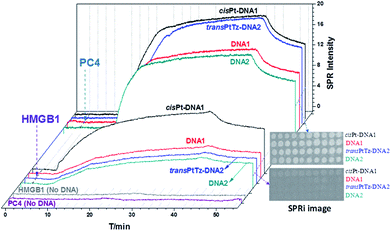 | ||
| Fig. 2 Typical SPRi sensorgram of 10 μM HMGB1/PC4 binding to cisPt–DNA1/DNA1 and transPtTz–DNA2/DNA2. The inset is the SPRi subtraction image. The spotting concentrations of all probes were 10 μM. | ||
Combining SPRi detection and data analysis, the Kd and ΔG at room temperature were obtained as: (4.09 ± 0.25) × 10−6 M and (−30.24 ± 0.29) kJ mol−1, respectively, between HMGB1 and cisPt–DNA1; and (2.22 ± 0.79) × 10−5 M and (−26.12 ± 0.73) kJ mol−1, respectively, between HMGB1 and DNA1; which confirmed that HMGB1 was more likely to link to cisPt–DNA1 than DNA1. The Kd and ΔG were (1.72 ± 0.19) × 10−5 M and (−26.85 ± 0.36) kJ mol−1, respectively, between HMGB1 and transPtTz–DNA2; (1.93 ± 0.43) × 10−5 M and (−26.52 ± 0.53) kJ mol−1, respectively, between HMGB1 and DNA2. Comprehensive comparison of these four sets of data, confirmed that HMGB1 was more likely to link to cisPt–DNA1 than DNA1, transPtTz–DNA2 and DNA2, indicating the specific recognition of HMGB1 with cisPt–DNA1. The specific recognition of HMGB1 with cisplatinated DNAs was also in good agreement with the result reported by Dr Fuyi Wang9 that HMGB1 bound preferentially to 1,2-cisplatin-cross-linked DNA than native DNAs, as HMGB1 can recognize the 16-base-pair DNA duplex containing a site-specific cis-[Pt(NH3)2] adduct with a widened minor groove. These results demonstrated that the developed SPRi method was reliable and applicable to characterize the affinity interaction between platinated DNAs and proteins, realizing the verification of this method by SPRi.
Interactions of PC4 and platinated DNAs
In the same as HMGB1, the interaction and recognition behaviors between the four immobilized kinds of DNAs and PC4 were inspected. The SPRi image (internal Fig. 2) showed that the cisPt–DNA1, DNA1, transPtTz–DNA2 and DNA2 all exhibited strong binding SPR signals with similar intensities. The typical SPRi kinetics curves (Fig. 2) showed that the binding signals of PC4 to cisPt–DNA1 and transPtTz–DNA2 were a little stronger than those of native DNA1 and DNA2. In addition, the binding signal of PC4 to no DNA confirmed that there was little non-specific adsorption. The study of the interactions between PC4 at different concentrations (0.625, 1.25, 2.5, 5.0, 10 μM) and four different DNAs (Fig. 3) showed that the binding intensity of all four kinds of DNAs increased when the PC4 concentration increased, and the binding signals of the cisPt–DNA1 and transPtTz–DNA2 were stronger than the native DNA1 and DNA2, respectively. Consequently, the affinity interactions between the platinated DNAs (cisPt–DNA1 and transPtTz–DNA2) and PC4 are stronger than those between native DNAs and PC4. This result was also confirmed by kinetic and thermodynamic analysis. At room temperature, Kd and ΔG were: (3.35 ± 0.27) × 10−7 M and (−36.34 ± 0.26) kJ mol−1, respectively, between PC4 and transPtTz–DNA2; and (8.39 ± 0.36) × 10−7 M and (−34.10 ± 0.31) kJ mol−1, respectively, between PC4 and DNA2; which confirmed that PC4 was more likely link to transPtTz–DNA2 than DNA2. Similarly, the Kd and ΔG were: (4.22 ± 0.25) × 10−7 M and (−35.78 ± 0.14) kJ mol−1, respectively, between PC4 and cisPt–DNA1; and (9.02 ± 0.16) × 10−7 M and (−33.92 ± 0.05) kJ mol−1, respectively, between PC4 and DNA1; which confirmed that PC4 was more likely linked to cisPt–DNA1 than DNA1. As we know, the high antitumor activity of cisplatin and the inert of its trans-isomer (transplatin) have long been considered as a classic example of antineoplastic QSAR. However, recent studies showed that when one amino group of trans-platinum was substituted by an organic molecule such as a thiazole amine group, transplatin compounds showed varying degrees of inhibition activity towards a variety of solid tumor cells, including cisplatin-resistant cells.9 The reverse structure–activity relationship aroused great interest in researchers, but the detailed molecular mechanism is still unclear. The SPR and dynamic results indicated that the PC4 could specifically recognize the transPtTz–DNA2, but the selectivity of the PC4 binding to transPtTz–DNA2 and DNA2 herein was not obvious; as reported by Dr Fuyi Wang,9 it may be due to the different protein structure or sequence length. The detailed recognition mechanism between PC4 and transPtTz–DNA2 needs to be investigated further, and this research lays the foundation for the further study of the transplatin antitumor compounds.Temperature-dependent kinetics and thermodynamicss
The interaction or reaction kinetics of DNA-targeting drugs were usually investigated at room temperature or below, far from a human body temperature, which is normally maintained at about 37 °C (but may be elevated up to 42 °C during fever). To gain an insight into its impact, a wide range of temperatures from 15 °C to 42 °C was studied by taking advantage of simulated human physiological conditions via SPRi. The calculated Kd at different temperatures (Fig. 4) showed that the affinity interactions between HMGB1/PC4 with the platinated DNAs were all stronger than the native DNAs and all the interactions were maintained under simulated human physiological conditions (37 °C or 42 °C). HMGB1 can recognize specifically cisPt–DNA1 at 15 °C to 42 °C with a Kd 10 times less than transPtTz–DNA2, native DNA1 and DNA2. PC4 was more likely to link to cisPt–DNA1 and transPtTz–DNA2 than native DNA1 and DNA2 at 15 °C to 42 °C with a Kd 2–3 times less than native DNAs. The corresponding ΔG at different temperatures were also calculated (Table S1 in ESI†). The results confirmed that the interaction or recognition behavior was not affected by a temperature change from 15 °C to 42 °C.Interaction with other proteins
In order to demonstrate further the specificity and selectivity of interaction between the platinated DNAs and proteins, the interactions between four kinds of DNAs and BSA/pepsin/hemoglobin/cytochrome c were analyzed (Fig. S4 in the ESI†). In a buffer at pH 7.8, BSA (pI = 4.85) and hemoglobin (pI = 7.23) are negatively charged, while pepsin (pI = 8.1) and cytochrome c (pI = 10.2) are positively charged. For BSA, pepsin and cytochrome c, no obvious interaction signals were observed. Meanwhile, for hemoglobin, though the interaction signals were clearly shown, there was no selective recognition behavior. This result further confirmed the selective or specific recognition between the platinated DNAs and HMGB1/PC4 proteins.Conclusions
In conclusion, an SPRi method to study the interactions between platinated DNAs and HMGB1/PC4 under simulated human physiological conditions with label-free, high throughput, in situ and real-time advantages has been developed. Its validation was proved by the specific recognition of HMGB1 and cisplatin damaged DNA1. The results indicated that PC4 was more likely to link to platinated DNAs (cisPt–DNA1 and transPtTz–DNA2) than the native DNAs (DNA1 and DNA2); the selectivity needs to be investigated further. The maintenance of these interactions under simulated human physiological conditions at normal body temperature (37 °C) and fever (42 °C) also have been revealed. The method provided authentic results such that controls could be simultaneously obtained by a single chip under the same conditions. Furthermore, this method is suited for the high throughput analyses of interactions between other drug-damaged DNAs and proteins to better understand the activity/inactivity mechanisms of drugs and for drug screening/discovery.Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 21235007 and 21135006) and the Chinese Academy of Sciences (No. KJCX2-EW-N06-01 and 2016ga03).References
- J. B. Kašpárková and V. Brabec, Biochemistry, 1995, 34, 12379–12387 CrossRef.
- U. M. Ohndorf, M. A. Rould, Q. He, C. O. Pabo and S. J. Lippard, Nature, 1999, 399, 708–712 CrossRef CAS PubMed.
- Y. Jung and S. J. Lippard, Chem. Rev., 2007, 107, 1387–1407 CrossRef CAS PubMed.
- E. R. Jamieson and S. J. Lippard, Chem. Rev., 1999, 99, 2467–2498 CrossRef CAS PubMed.
- A. Gelasco and S. J. Lippard, Biochemistry, 1998, 37, 9230–9239 CrossRef CAS PubMed.
- S. Müller, M. E. Bianchi and S. Knapp, Biochemistry, 2001, 40, 10254–10261 CrossRef.
- K. Batta and T. K. Kundu, Mol. Cell. Biol., 2007, 27, 7603–7614 CrossRef CAS PubMed.
- M. Liao, Y. Zhang, J. H. Kang and M. L. Dufau, J. Biol. Chem., 2011, 286, 7681–7691 CrossRef CAS PubMed.
- Z. Du, Q. Luo, L. Yang, T. Bing, X. Li, W. Guo, K. Wu, Y. Zhao, S. Xiong, D. Shangguan and F. Wang, J. Am. Chem. Soc., 2014, 136, 2948–2951 CrossRef CAS PubMed.
- O. Mortusewicz, W. Roth, N. Li, M. C. Cardoso, M. Meisterernst and H. Leonhardt, J. Cell Biol., 2008, 183, 769–776 CrossRef CAS PubMed.
- S. Banerjee, B. R. P. Kumar and T. K. Kundu, Mol. Cell. Biol., 2004, 24, 2052–2062 CrossRef CAS PubMed.
- V. Marini, P. Christofis, O. Novakova, J. Kašpárková, N. Farrell and V. Brabec, Nucleic Acids Res., 2005, 33, 5819–5828 CrossRef CAS PubMed.
- D. B. Zamble, D. Mu, J. T. Reardon, A. Sancar and S. J. Lippard, Biochemistry, 1996, 35, 10004–10013 CrossRef CAS PubMed.
- E. R. Jamieson and S. J. Lippard, Biochemistry, 2000, 39, 8426–8438 CrossRef CAS PubMed.
- L. K. Wolf, Y. Gao and R. M. Georgiadis, J. Am. Chem. Soc., 2007, 129, 10503–10511 CrossRef CAS PubMed.
- F. Pillet, A. Sanchez, C. Formosa, M. Severac, E. Trévisiol, J. Y. Bouet and V. A. Leberre, Biosens. Bioelectron., 2013, 43, 148–154 CrossRef CAS PubMed.
- A. Yamamoto, Y. Ando, K. Yoshioka, K. Saito, T. Tanabe, H. Shirakawa and M. Yoshida, J. Biochem., 1997, 122, 586–594 CrossRef CAS PubMed.
- B. Dey, S. Thukral, S. Krishnan, M. Chakrobarty, S. Gupta, C. Manghani and V. Rani, Mol. Cell. Biochem., 2012, 365, 279–299 CrossRef CAS PubMed.
- F. Zhang, S. Wang, L. Yin, Y. Yang, Y. Guan, W. Wang, H. Xu and N. Tao, Anal. Chem., 2015, 87, 9960–9965 CrossRef CAS PubMed.
- G. J. Wegner, H. J. Lee and R. M. Corn, Anal. Chem., 2002, 74, 5161–5168 CrossRef CAS PubMed.
- G. Krishnamoorthy, J. B. Beusink and R. B. M. Schasfoort, Anal. Methods, 2010, 2, 1020–1025 RSC.
- A. J. Thiel, A. G. Frutos, C. E. Jordan, R. M. Corn and L. M. Smith, Anal. Chem., 1997, 69, 4948–4956 CrossRef CAS.
- F. Pillet, C. Romera, E. Trévisiol, S. Bellon, M. P. Teulade-Fichou, J. M. François, G. Pratviel and V. A. Leberre, Sens. Actuators, B, 2011, 157, 304–309 CrossRef CAS.
- R. L. L. A. B. Steel, T. M. Herne and M. J. Tarlov, Biophys. J., 2000, 79, 975–981 CrossRef PubMed.
- R. L. T. M. H. M. J. T. S. K. Satija, J. Am. Chem. Soc., 1998, 120, 9787–9792 CrossRef.
- P. R. Edwards and R. J. Leatherbarrow, Anal. Biochem., 1997, 246, 1–6 CrossRef CAS PubMed.
- S. Hoebel, D. Vornicescu, M. Bauer, D. Fischer, M. Keusgen and A. Aigner, Anal. Chem., 2014, 86, 6827–6835 CrossRef CAS PubMed.
- O. R. Bolduc and J. F. Masson, Langmuir, 2008, 24, 12085–12091 CrossRef CAS PubMed.
- J. C. O'Brien, J. T. Stickney and M. D. Porter, J. Am. Chem. Soc., 2000, 122, 5004–5005 CrossRef.
- N. N. Mamedova, N. A. Kotov, A. L. Rogach and J. Studer, Nano Lett., 2001, 1, 281–286 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra27719a |
| This journal is © The Royal Society of Chemistry 2016 |



![[G with combining low line]](https://www.rsc.org/images/entities/i_char_0047_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif)