Ultrahigh-throughput generation and characterization of cellular aggregates in laser-ablated microwells of poly(dimethylsiloxane)†
Abstract
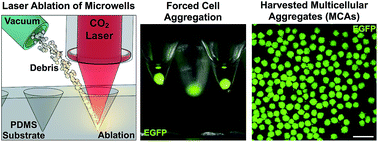
Aggregates of cells, also known as multicellular aggregates (MCAs), have been used as microscale tissues in the fields of cancer biology, regenerative medicine, and developmental biology for many decades. However, small MCAs (fewer than 100 cells per aggregate) have remained challenging to manufacture in large quantities at high uniformity. Forced aggregation into microwells offers a promising solution for forming consistent aggregates, but commercial sources of microwells are expensive, complicated to manufacture, or lack the surface packing densities that would significantly improve MCA production. To address these concerns, we custom-modified a commercial laser cutter to provide complete control over laser ablation and directly generate microwells in a poly(dimethylsiloxane) (PDMS) substrate. We achieved ultra rapid microwell production speeds (>50 000 microwells per h) at high areal packing densities (1800 microwells per cm2) and over large surface areas for cell culture (60 cm2). Variation of the PDMS substrate distance from the laser focal plane during ablation allowed for the generation of microwells with a variety of sizes, contours, and aspect ratios. Casting of high-fidelity microneedle masters in polyurethane allowed for non-ablative microwell reproduction through replica molding. MCAs of human bone marrow derived mesenchymal stem cells (hMSCs), murine 344SQ metastatic adenocarcinoma cells, and human C4-2 prostate cancer cells were generated in our system with high uniformity within 24 hours, and computer vision software aided in the ultra-high-throughput analysis of harvested aggregates. Moreover, MCAs maintained invasive capabilities in 3D migration assays. In particular, 344SQ MCAs demonstrated epithelial lumen formation on Matrigel, and underwent EMT and invasion in the presence of TGF-β. We expect this technique to find broad utility in the generation and cultivation of cancer cell aggregates, primary cell aggregates, and embryoid bodies.

 Please wait while we load your content...
Please wait while we load your content...