A comparative antimicrobial and toxicological study of gold(iii) and silver(i) complexes with aromatic nitrogen-containing heterocycles: synergistic activity and improved selectivity index of Au(iii)/Ag(i) complexes mixture†
Abstract
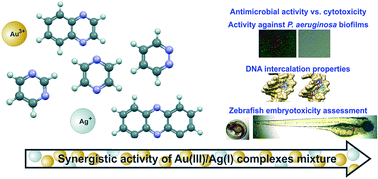
Five aromatic nitrogen-containing heterocycles, pyridazine (pydz, 1), pyrimidine (pm, 2), pyrazine (pz, 3), quinoxaline (qx, 4) and phenazine (phz, 5) have been used for the synthesis of gold(III) and silver(I) complexes. In contrast to the mononuclear Au1–5 complexes all having square-planar geometry, the corresponding Ag1–5 complexes have been found to be polynuclear and of different geometries. Complexes Au1–5 and Ag1–5, along with K[AuCl4], AgNO3 and N-heterocyclic ligands used for their synthesis, were evaluated by in vitro antimicrobial studies against a panel of microbial strains that lead to many skin and soft tissue, respiratory, wound and nosocomial infections. All tested complexes exhibited excellent to good antibacterial activity with minimal inhibitory (MIC) values in the range of 2.5 to 100 μg mL−1 against the investigated strains. The complexes were particularly efficient against pathogenic Pseudomonas aeruginosa (MIC = 2.5–30 μg mL−1) and had a marked ability to disrupt clinically relevant biofilms of strains with high inherent resistance to antibiotics. Moreover, the Au1–4 and Ag1–5 complexes exhibited pronounced ability to competitively intercalate double stranded genomic DNA of P. aeruginosa, which was demonstrated by gel electrophoresis techniques and supported by molecular docking into the DNA major groove. Antiproliferative effect on the normal human lung fibroblast cell line MRC5 has also been evaluated in order to determine therapeutic potential of Au1–5 and Ag1–5 complexes. Since the investigated gold(III) complexes showed much lower negative effects on the viability of the MRC5 cell line than their silver(I) analogues and slightly lower antimicrobial activity against the investigated strains, the combination approach to improve their pharmacological profiles was applied. Synergistic antimicrobial effect and the selectivity index of 10 were achieved for the selected gold(III)/silver(I) complexes mixtures, as well as higher P. aeruginosa PAO1 biofilm disruption activity, and improved toxicity profile towards zebrafish embryos, in comparison to the single complexes. To the best of our knowledge, this is the first report on synergistic activity of gold(III)/silver(I) complexes mixtures and it could have an impact on development of new combination therapy methods for the treatment of multi-resistant bacterial infections.

 Please wait while we load your content...
Please wait while we load your content...