Strong base pre-treatment for colorimetric sensor array detection and identification of N-methyl carbamate pesticides†
Abstract
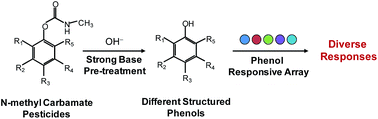
Colorimetric sensor arrays demonstrate numerous superior features in chemo- and bio-sensing, but they are generally not applicable to less-reactive analytes. Based on the findings that N-methyl carbamate pesticides could be decomposed into reactive phenols in basic media, herein, a novel strategy of strong base pre-treatment was developed and employed for the colorimetric sensor array detection and differentiation of the N-methyl carbamate pesticides in an indirect manner. With the use of five inexpensive and commercially available phenol responsive indicators, such a colorimetric sensor array can be facilely fabricated. Classification analysis (e.g. hierarchical clustering analysis (HCA) and principal component analysis (PCA)) reveals that the as-fabricated sensor array has an extremely high dimensionality and, consequently, exhibits excellence in discriminating a variety of N-methyl carbamates from other types of pesticides and potential interferants, and further identifying them exactly from each other. Moreover, semi-quantitative detection could also be achieved through combining HCA/PCA, recognition patterns, and corresponding fitting curves. Overall, the as-developed method exhibits high selectivity and sensitivity, good anti-interference, simultaneous detection and identification capability for each of the N-methyl carbamate pesticides, and potential applicability in real samples. Most importantly, this study demonstrates that pre-treatment strategies are very effective in expanding the range of applications of colorimetric sensor array methodology to less-reactive analytes.

 Please wait while we load your content...
Please wait while we load your content...