PEG functionalized selenium nanoparticles as a carrier of crocin to achieve anticancer synergism
Thottumugathu Ancil Mary†
b,
Krishnamurthy Shanthi†b,
Karuppaiya Vimala†a and
Kannan Soundarapandian*a
aProteomics and Molecular Cell Physiology Laboratory, Department of Zoology, Periyar University, Salem-636 011, Tamil Nadu, India. E-mail: sk_protein@buc.edu.in; skperiyaruniv@gmail.com; Tel: +91-0427-2345766 Tel: +91-0427-2345520
bDepartment of Zoology, Bharathiar University, Coimbatore-641 046, Tamil Nadu, India
First published on 9th February 2016
Abstract
Selenium nanoparticles (SeNPs) have garnered a great deal of attention as potential cancer therapeutic payloads. However, the in vivo targeting drug delivery has been challenging. Herein, we describe the synthesis of PEG functionalized SeNPs (PEG-SeNPs) and its use as a cancer-targeted delivery vehicle to achieve enhanced anticancer efficacy. Crocin, a major active product of saffron (dried stigmas of Crocus sativus), having many therapeutic properties was conjugated to PEG-SeNPs. PEG-SeNPs showed pH-mediated crocin release for potential cancer therapy. Crocin conjugated PEG-SeNPs exhibits unprecedented enhanced cytotoxicity toward lung cancer cells through induction of apoptosis. Furthermore, crocin conjugated PEG-SeNPs significantly inhibited in vivo tumor growth in nude mice model. This cancer-targeted design of SeNPs opens a new path for synergistic treating of cancer with higher efficacy and decreased side effects.
1. Introduction
Cancer is one of the most feared diseases globally and there has been a sustained rise in its incidence in both developing and developed countries. It is one of the major non-communicable diseases posing a threat to world health. Despite the growing therapeutic options for patients with cancer, their efficacy is time-limited and also their curability is limited.1 The current cancer treatments often kill healthy cells and thus show significant toxicity and unavoidable side effects.2 Therefore, the discovery of novel, selective, efficient, and safe drugs for cancer chemotherapy remains an urgent and high priority for medicinal research.3 Currently, nanoparticle-based drugs are emerging as an important class of therapeutics.4 The most promising aspects of utilizing nanoparticles as therapeutics are their potential to localize (or be targeted) in a specific manner to the site of disease and reduce or eliminate the possible numerous untoward side effects. The nanometric size of these materials precludes them from being readily cleared through the kidneys, thereby extending circulation in the blood pool depending on their surface-functionalization characteristics.5 Also, when considering novel cancer treatments, blood vessels in many tumor types are irregular in shape, dilated, leaky, and can present fenestrations in endothelial cells. Due to the altered anatomy of tumor vessels, nanosized particles can easily extravasate from the blood pool into tumor tissues and be retained due to poor lymphatic drainage. This phenomenon of selective accumulation of nanosized particles near tumor tissues is termed the enhanced permeability and retention (or EPR) effect.6,7 Additionally, nanoparticles have high surface area-to-volume ratios, yielding high loading capacities. Thus, nanoparticles can be loaded with therapeutic drugs and imaging agents; they may also be surface-functionalized with targeting ligands and cloaking agents like poly(ethyleneglycol) (PEG), with the goal of reducing systemic toxicity.A number of nanosystems with different structure and compositions, such as metals, polymers, oxides, and semi-conductors, have been designed and prepared to carry anticancer drugs.8–11 Among these nanomaterials, selenium nanoparticles (SeNPs) have garnered a great deal of attention as potential cancer therapeutic agents and drugs carriers.12–16 Selenium (Se) is an essential trace element with important physiological functions and extensive pharmacological actions. It is a structural component of the active centre of many antioxidant enzymes and functional proteins.17 SeNPs possess potent effects both on scavenging various free radicals and on protecting DNA from oxidation damage in vitro. SeNPs could efficiently increase the activity of selenoenzymes, including glutathione peroxidase, phospholipid hydroperoxide glutathione peroxidase thioredoxin reductase and deiodinase.18 Cellular Se plays an important role in the reduction of oxidative stress in the body.19 It also regulates the function of the thyroid gland and helps in the proper functioning of the immune system.20 It plays an important role to prevent various diseases, such as diabetes, hypercholesterolemia,21 cardiovascular disease.22,23 Many studies have shown that the supplementation of Se could prevent cancer and reduce cancer incidence.24–26 Moreover, recent studies have indicated that SeNPs express important anticancer activity by inhibiting the growth or triggering the apoptosis of different types of cancer cells containing human hepatocyte cells (HepG2),27 human breast-cancer cells (MCF-7, MDA-MB-231),28 human melanoma cells (A375),29 human cervical carcinoma cells (HeLa).30 Despite the cytotoxicity toward cancer cell lines, SeNPs could enhance the cell viability and minimize the DNA damage caused by UV exposure on human lymphocytes.31 Although clinical trials with Se are currently limited to cancer chemoprevention, recent evidence strongly showed the potential for utilization of Se in a new way, to overt cancer through a combination with well-established chemotherapeutic and hormonal agents. Many studies showed that Se could sensitize cancer cells to conventionally used anticancer drugs.32,33 Over the past decade, SeNPs have attracted increasing attention because of their antioxidant activities and low toxicity.34,35 Compared to other nanoparticles that are currently most often studied, such as gold nanoparticles, SeNPs are superior, because Se is degradable in vivo. Degraded Se can be used as a nutrient for many kinds of normal cells or as an antiproliferative agent for many kinds of cancer cells.36 Abundant evidence supports the better biocompatibility, bioefficacy and lower toxicity of SeNPs by comparing with inorganic and organic selenocompounds.37 In addition, SeNPs, by nature, display desired biological activities and can be used as drug carriers as well. In this study, we report the use of SeNPs as carriers of crocin to enhance their anticancer outcome.
Crocin, a major active product of saffron (dried stigmas of Crocus sativus), has many therapeutic properties such as antitumoral,38,39 antioxidant,40 anxiolytic,41,42 neuronal protective,43 anti-ischemic44 and protective against DNA damage45 activities. Crocin are also effective agents as antidepressant, anticonvulsant, memory enhancer and sedative in treatment of central nervous system disorders.46 Owing to the therapeutic potential of both SeNPs and crocin, we look forward to design a synergistic system by conjugating crocin to the surface of the SeNPs, which could enhance the cure rate and lower their toxicity. By altering the surface chemistry of SeNPs using PEG, crocin could be conjugated to the nanoparticles, and this drug delivery system can be utilized to target cancer. Herein, we describe the synthesis of PEG functionalized SeNPs (PEG-SeNPs) and its use as a cancer-targeted drug delivery system for crocin to achieve enhanced anticancer efficacy against lung cancer. The in vivo anticancer activity of crocin conjugated PEG-SeNPs and the underlying molecular mechanisms were also investigated in this study.
2. Experimental
2.1. Materials
Human normal lung epithelial cell lines L-132 and lung cancer A549 cell were procured from National Centre for Cell Science (Pune, India). Certified dried saffron stigma sample were purchased from Coimbatore, Tamil Nadu. Sodium selenite (Na2SeO3), poly(ethylene glycol) (200k) (PEG), 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT), 4′-6-diamidino-2-phenylindole (DAPI), acridine orange/ethidium bromide AO/EtBr Dulbecco's Modified Eagles medium (DMEM) were purchased from Sigma-Aldrich (Bangalore). Analytical grade reagents were purchased from Sigma-Aldrich (Bangalore). All the samples were prepared in Milli-Q water.2.2. Extraction of crocin from saffron stigmas
Crocin was isolated from saffron by previously described method.47 Saffron stigmas powders (10 g) were suspended in 25 mL ethanol (80%) at 0 °C and shaked by vortex for 2 min. After centrifugation at 4000 rpm for 10 min the supernatant was separated. 25 mL of ethanol (80%) was added to sediment and extraction was repeated again. This step was repeated 6 other times. The total volume of solvent consumption for 10 g saffron stigmas in extraction process was 200 mL (8 × 25 mL). The resulting solution was kept in a thick walled glass container at −5 °C for 24 days in darkness. The container was sealed in this period. The obtained crystals were separated from solution and washed with acetone to remove remaining water. The yielded amount of crystals was 1.7 g. In the next step, the obtained crystals were dissolved in 120 mL ethanol (80%) and kept at −5 °C in darkness for 20 extra days for re-crystallization. The final amount of yielded crystals was 1.02 g.2.3. HPLC analysis
For HPLC analysis, we used a Varian 9012 liquid chromatographic system equipped with a Varian 9050 UV detector (Walnut Creek, CA). The separations were carried out on a Phenomenex Lichrosphere 5 RP C18 column (250 × 4.6 mm, 5 μm) (Torrance, CA). The precolumn was a Phenomenex C18 column (30 × 4 mm). The detector was set at 442 nm with a spectral acquisition rate of 1.25 scans per s. For the mobile phase, solvent A (methanol) and solvent B [1% (v/v) aqueous acetic acid in water] were used. The mixing of the gradient solvent eluting system was as follows: initial 30% A and 70% B; 0–5 min, linear change to 40% A; 5–10 min, change to 55% A; 10–25 min, change to 68% A; 25–27 min, change to 90% A; 27–30 min, 90% A; 30–33 min, change to 30% A; 33–40 min, 30% A. The flow rate of the mobile phase was 0.8 mL min−1, and the injection volume was 20 μL. All solutions were filtered through a 0.2 μm hydrophilic polypropylene membrane (Merck Millipore, Billerica, MA) before use. Separation was accomplished at 25 °C. Five different concentrations of crocin solutions were prepared to determine the calibration curve. The calibration curve was constructed with crocin content versus peak area (y = 0.0002x + 1.0422; R2 = 0.9993; linear range: 0.01–0.2 mg mL−1). The content of crocin was calculated using the standard curve of crocin, and determinations were repeated 3 times.2.4. Preparation of PEG-SeNPs
PEG-SeNPs was synthesized using a previously reported method with slight modification.48 A stock solution of 5 mM sodium selenite (Na2SeO3) was prepared by dissolving 8.7 mg of Na2SeO3 powder in 10 mL of Milli-Q water. A 5 mL aliquot of Na2SeO3 stock solution was mixed with 10 mL PEG200 solution at 210–220 °C for 15–20 minutes, under magnetic stirring. The product was then mixed with water in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. The solution was centrifuged at 10
1 ratio. The solution was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 minutes and then washed with Milli-Q water five times to remove excess PEG. The obtained products were characterized by various spectroscopic methods.
000 rpm for 10 minutes and then washed with Milli-Q water five times to remove excess PEG. The obtained products were characterized by various spectroscopic methods.
2.5. Preparation of crocin conjugated PEG-SeNPs
Crocin was conjugated onto PEG-SeNPs by a previously reported method with slight modification.49 A 5 mL aliquot of PEG-SeNPs was mixed with 5 mL of 32.5 mg mL−1 crocin solution. The mixture was reconstituted to a final volume of 25 mL with Milli-Q water. Then the mixed solution was stirred for 24 h at room temperature. Excess crocin were removed by dialysis against Milli-Q water overnight. Se concentration was determined by ICP-AES analysis.The drug loading efficacy was calculated by two ways, first based on indirect method by estimating the crocin content of the supernatant and second based on direct estimation of the crocin content present in the pellet obtained after centrifugation. The drug concentration in supernatant and redispersed pellets was determined by measurements of its UV absorbance at 470 nm using UV/visible spectroscopy and the percentage loading of crocin onto nanoparticles were estimated by the following formula.
2.6. In vitro drug release of crocin conjugated PEG-SeNPs
Two copies of crocin conjugated PEG-SeNPs (10 mg) were respectively suspended in 10 mL PBS solution at pH 5.3 and pH 7.4 with constantly shaking in dark tubes at 37 °C. At specific intervals, a certain volume of buffer was taken out from tubes and same volume of fresh buffer was replaced. For the measurement of released crocin concentration, the absorbance of the release medium at 475 nm was recorded on a Shimadzu UV-vis absorption spectrophotometer.2.7. Cell viability assay
Cell viability was determined by measuring the ability of cells to transform MTT to a purple formazan dye.50 Cells were seeded in 96-well tissue culture plates at 2.5 × 103 cells per well for 24 h. The cells were then incubated with crocin conjugated PEG-SeNPs at different concentrations for different periods of time. After treatment, 20 μL per well of MTT solution (5 mg mL−1 phosphate buffered saline) was added to the well and incubated for another 5 h. To dissolve the formazan salt formed, the medium was aspirated and replaced with 150 μL per well DMSO. The cell growth condition was reflected by the color intensity of the formazan solution. Absorbance at 570 nm was taken on a 96-well microplate reader (MD VERSA max).2.8. Synergy analysis
Isobologram method was conducted to analyze the synergistic effect between PEG-SeNPs and crocin. Briefly, line segment between the IC50 value of PEG-SeNPs and crocin on the x- and y-axes respectively represented the additive line. The data point near or on the additive line represented an additive treatment effect, while the data point below or above the additive line remarked the synergism or antagonism respectively. In addition, the extent of synergism or antagonism was evaluated by combination index (CI). CI value of 1 meant an additive effect between two drugs, while CI value <1 represents synergism, CI value >1 indicates antagonism. The extent of CI value below or above 1 is positively related to the extent of synergism and antagonism respectively.2.9. Fluorescence microscopic studies
2.10. Western blotting analysis
Crocin conjugated PEG-SeNPs treated cells were washed in PBS and lysed in 100 μL of buffer containing 50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 10 μg mL−1 pepstatin, and 10 μg mL−1 leupeptin. After 20 min, extracts were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min at 4 °C and supernatants were stored at −80 °C until further use. Proteins (30 μg per lane) were separated using 10% SDS-PAGE, and then transferred to polyvinylidene difluoride (PVDF) membranes. Afterwards, the membranes were blocked in TBST solution containing 5% (w/v) non-fat milk for 2 h, followed by overnight incubation at 4 °C with primary antibodies such as bax, bcl-2, caspase 9 and 3 and cytochrome c. β-Actin was used as an internal control. After being washed with TBST buffer, the membranes were incubated for 1 h with the secondary antibody, horseradish peroxidase-conjugated goat anti-rabbit IgG. Antibody-bound proteins were detected using enhanced chemiluminescence reagents. Blots were washed with washing buffer and incubated with secondary antibodies conjugated with horseradish peroxidase for 1 h at room temperature.
000 rpm for 10 min at 4 °C and supernatants were stored at −80 °C until further use. Proteins (30 μg per lane) were separated using 10% SDS-PAGE, and then transferred to polyvinylidene difluoride (PVDF) membranes. Afterwards, the membranes were blocked in TBST solution containing 5% (w/v) non-fat milk for 2 h, followed by overnight incubation at 4 °C with primary antibodies such as bax, bcl-2, caspase 9 and 3 and cytochrome c. β-Actin was used as an internal control. After being washed with TBST buffer, the membranes were incubated for 1 h with the secondary antibody, horseradish peroxidase-conjugated goat anti-rabbit IgG. Antibody-bound proteins were detected using enhanced chemiluminescence reagents. Blots were washed with washing buffer and incubated with secondary antibodies conjugated with horseradish peroxidase for 1 h at room temperature.
2.11. Hemolytic assay
Ethylenediaminetetraacetic acid (EDTA)-stabilized human blood samples were freshly collected. A sample of whole blood (4 mL) was added to phosphate-buffered saline (8 mL, PBS: pH 7.4). The red blood cells (RBCs) were isolated by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 016g for 5 min and further washed five times with sterile PBS solution. Following the last wash, the RBCs were diluted with PBS (40 mL). Then diluted RBC suspension (0.2 mL) was added to crocin conjugated PEG-SeNPs solutions at systematically varied concentrations and mixed by vortexing. All the sample tubes were kept in static condition at room temperature for 3 h. Finally, the mixtures were centrifuged at 10
016g for 5 min and further washed five times with sterile PBS solution. Following the last wash, the RBCs were diluted with PBS (40 mL). Then diluted RBC suspension (0.2 mL) was added to crocin conjugated PEG-SeNPs solutions at systematically varied concentrations and mixed by vortexing. All the sample tubes were kept in static condition at room temperature for 3 h. Finally, the mixtures were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 016g for 3 min, and 100 μL of supernatant of all samples was taken, and its absorbance was recorded on a spectrophotometer (Shimadzu UV-vis spectrophotometer) at 545 nm. The percentage hemolysis was calculated using the following relationship.
016g for 3 min, and 100 μL of supernatant of all samples was taken, and its absorbance was recorded on a spectrophotometer (Shimadzu UV-vis spectrophotometer) at 545 nm. The percentage hemolysis was calculated using the following relationship.Herein, RBC incubation with deionized water and PBS were used as the positive and negative controls, respectively.
2.12. Assessment of anti-tumor activity in vivo
The anti-tumor efficiency of crocin conjugated PEG-SeNPs was assessed in tumor-induced mice. Briefly, the subcutaneous dorsa of male nude mice were inoculated with A549 cells (1 × 107) in 100 mL of normal saline. When the volume of the xeno-graft tumor reached approximately 50–75 mm3 the mice were randomly divided into 3 groups and a control group with six mice in each group. Crocin conjugated PEG-SeNPs at dosages of 3.0, 6.0 and 9.0 mg per kg per day was injected intravenously every 2 days, and the mice were then observed for 16 days. The tumor diameters were measured every 3 days interval for each group. The tumor volumes (V) and body weight were calculated using the formula V = [length × (width)2]/2. For the assessment of toxicity, organs such as, liver, kidney and lung were collected, fixed in 4% paraformaldehyde solution and made into 4 mm sections which were stained with hematoxylin and eosin (H&E) and observed under a microscope.2.13. Statistical analysis
All the measurements were made in triplicate and all values were expressed as the mean ± standard error. The results were subjected to an analysis by Student's t-test. The results were considered statistically significant if the p-value was ≤0.05.2.14. Live subject statement
The authors state that all experiments were performed in compliance with the relevant laws and institutional guidelines (Animal Ethical Committee, Periyar University, Salem) and this work has been approved by the IAEC (Institutional Animal Ethical Committee) constituted as per the Rules and Regulations of Ministry of Animal Husbandry, Government of India. The authors also state that informed consent was obtained for any experimentation with human subjects and Animal Ethical Committee, Periyar University, Salem is committed to the protection and safety of human subjects involved in research.3. Results and discussion
3.1. Identification of active compound
The GC-MS spectrum revealed the presence of various compounds present in saffron extracts (Fig. 1). Sixteen major compounds from GC/MS results were listed along with their retention indices and molecular weight (Table 1). The qualitative analysis of crocin was further confirmed with the assistance of HPLC. Chromatogram of high-performance liquid chromatographic analysis of commercially available crocin (used as standard) and crocin isolated from saffron extract were shown in (Fig. 2). Chromatogram of crocin isolated from saffron extract (Fig. 2b) showed peaks between 14–18 minutes consistent with the standard crocin51 (Fig. 2a). The purity of crocin was 96%.| S. no. | Compound | Rate time | Formula | Molecular weight |
|---|---|---|---|---|
| 1 | Propyl acetate | 4.192 | C5H10O2 | 102.13 |
| 2 | Methyl (2R)-2-hydroxypropanoate | 4.325 | C4H8O3 | 104.10 |
| 3 | Ethylene glycol | 4.655 | C2H6O2 | 62.06 |
| 4 | Methyl eugenol | 6.886 | C11H14O2 | 178.22 |
| 5 | Methyl 2-methylbutyrate | 7.392 | C6H12O2 | 116.15 |
| 6 | Alloocimene | 11.508 | C10H16 | 136.23 |
| 7 | Isophorone | 11.787 | C9H14O | 138.20 |
| 8 | Ketoisophorone | 12.202 | C9H12O2 | 152.19 |
| 9 | 1,4-Cyclohexanedione, 2,2,6-trimethyl | 12.656 | C9H14O2 | 154.20 |
| 10 | Crocine | 13.319 | C44H64O24 | 976.96 |
| 11 | 2,4-Cycloheptadien-1-one, 2-phenyl- | 13.694 | C13H12O | 184.23 |
| 12 | Mintlactone | 15.255 | C10H14O2 | 166.21 |
| 13 | Hexadecanoic acid, methyl ester | 17.152 | C17H34O2 | 270.45 |
| 14 | 9,12-Octadecadienoic acid | 26.265 | C18H32O2 | 280.44 |
| 15 | 8,11,14-Docosatrienoic acid | 26.333 | C23H40O2 | 348.56 |
| 16 | Octadecanoic acid, methyl ester | 26.604 | C19H38O2 | 298.50 |
| 17 | Ethyl linoleate | 27.026 | C20H36O2 | 308.49 |
| 18 | 1-(4-Undecylphenyl)ethanone | 30.654 | C19H30O | 274.44 |
| 19 | 2-Fluoroadenine | 32.225 | C5H4FN5 | 153.11 |
 | ||
| Fig. 2 (a) Chromatogram of HPLC analysis of commercially available crocin (used as standard), the insert shows the structure of crocin and (b) crocin isolated from saffron extract. | ||
3.2. Preparation and characterization of NPs
To validate the synthesis of PEG-SeNPs, UV-visible spectroscopy was performed (Fig. 3). The spectrum of PEG-SeNPs exhibited absorption maxima at 395 nm. Similar absorption maxima were observed for SeNPs synthesized using lemon leaf extract.52 In addition previous reports have shown that the SeNPs contribute to the absorption maximum at around 200–400 nm in the UV-visible spectra.53 Besides the insert of Fig. 3 represents change in color during the nanoparticles synthesis. Initially the colloidal solution appeared colorless but after reduction with PEG, it turned to red color. This color change may be due to the surface plasma resonance (SPR) with a broad peak. Similar color chances were noted by Estevez et al.54 during the formation of chitosan-stabilized selenium nanoparticles and Zheng et al.55 during the formation of polyamidoamine-modified selenium nanoparticles. Thus color change from colorless selenious acid to red color (SeNPs), having absorption maximum (λmax) at 390 nm clearly indicates the formation of SeNPs using PEG. Further the conjugation of crocin onto the NPs was confirmed by the appearance of additional peaks at 470 and 475 nm related to crocin.56 These results support the successful conjugation of crocin to PEG-SeNPs.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–O–) groups which are due to constituents of alcohol groups found in crocin.
C–O–) groups which are due to constituents of alcohol groups found in crocin.
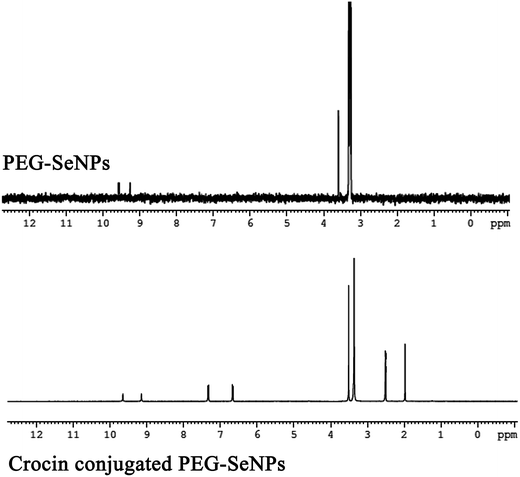
The NMR spectra depicted in Fig. 8 authenticate the presence of PEG-SeNPs. The respective chemical shifts peaks had been noticed at 9.61, 9.22, 3.61 and 3.34 ppm. The peak at 3.61 ppm is related to the principle proton peaks from PEG.65 The incorporation of PEG in SeNPs was thus confirmed by observing the proton peaks from PEG (CH2 at 3.61 ppm) in the PEG-SeNPs. Interestingly the crocin conjugated PEG-SeNPs accentuated the characteristic peaks at 9.65, 9.15, 3.50 and 3.36 ppm. The conjugation of crocin was confirmed by the appearance of principle peaks of crocin at 1.97, 2.40, 6.50 and 7.40 ppm.66,67 The Chemical shift timing may slightly be varied for a complex when it is structurally further modified with other compounds or molecules. Thus the data presented in the form of NMR spectra are more convening to confirm the nanoformulation of crocin conjugated PEG-SeNPs.
3.3. pH-Mediated release of crocin in vitro
The crocin release behavior from PEG-SeNPs was investigated in PBS solution at pH 7.4 and pH 5.3 to intimate the blood and lysosome environments in vivo. As shown in Fig. 10, the cumulative release amount of crocin from the nanoparticles at pH 5.3 was 47.0% within 1 h and 91.0% for 48 h, whereas the release rate at pH 7.4 was 11.6% in 1 h and finally reached 34.5% for 48 h. The results demonstrated that the release process at pH 7.4 was much slower than that at pH 5.3. One of possible reasons was the low solubility of crocin at pH 7.4 than that at pH 5.3. Thus PEG-SeNPs hold a promise as a pH-mediated release delivery vehicle for potential cancer therapy.3.4. In vitro cytotoxicity
The in vitro cytotoxic effects of crocin conjugated PEG-SeNPs was evaluated against human lung cancer and normal cell lines by MTT assay (Fig. 11). As shown in Fig. 11a, crocin conjugated PEG-SeNPs inhibited A549 cell growth in a time- and dose-dependent manner. Despite this potency, the toxicity of crocin conjugated PEG-SeNPs toward human normal cells (L-132) showed no appreciable deduction in cell viability in both 24 h and 48 h incubation, indicating that crocin conjugated PEG-SeNPs is highly biocompatible (Fig. 11b). The IC50 concentration of crocin conjugated PEG-SeNPs was found to be 18.6 μM for 24 h and 7.9 μM for 48 h. These results indicate that, crocin conjugated PEG-SeNPs is efficient in reducing the toxicity in normal cells without sacrifice of its anticancer activity. Similar cytotoxicity was reported by Yanyu Huang et al.37 in MCF-7 cells incubated with DOX-loaded Tf-conjugated SeNPs (Tf-SeNPs). More recently, Wen et al.2 demonstrated that 5-fluorouracil-SeNPs (5FU-SeNPs) exhibited a broad spectrum inhibition against A375, MCF-7, HepG2, Colo201, and PC-3 cancer cells. Despite this potency, 5FU-SeNPs showed much lower cytotoxicity toward human normal cells (Hs68 human fibroblasts, HK-2 proximal tubular cells, and MCF-10A human mammary epithelial cells). Interestingly, MCF-10A cells were also used as a model to examine the effects of 5FU-SeNPs on normal breast cells as compared to human breast cancer cells (MCF-7 cells). The results of their study showed that 5FU-SeNPs exhibited lower cytotoxicity toward MCF-10A than MCF-7 cells. Consistently our results also showed no appreciable toxicity toward human normal cells (L-132) when compared to human lung cancer (A549). These suggest that, the effects of crocin conjugated PEG-SeNPs on the human cells are cell-type specific. This selectivity could be partly due to the different protein and gene expression profiles of different cells which resulted in activation of different intracellular signaling pathways after exposure to crocin conjugated PEG-SeNPs. Taken together, our results suggested that crocin conjugated PEG-SeNPs possess great selectivity between cancer and normal cells and displays potential application in cancer chemotherapy.To understand the synergistic interaction between PEG-SeNPs and the conjugated crocin, the growth inhibition of crocin conjugated PEG-SeNPs were analyzed by isobologram examination. The IC50 values for crocin conjugated PEG-SeNPs, crocin and PEG-SeNPs, were found at 6.2, 153.0, and 243.9 μM, respectively (Fig. 11c). The results of the isobologram analysis revealed that the growth inhibitory effects between crocin and PEG-SeNPs in the crocin conjugated PEG-SeNPs system was strongly synergistic, as evidenced by the location of the data point in the isobologram being far below the line defining an additive effect.37 The combination index (CI) of the crocin conjugated PEG-SeNPs was found at 0.024, which further confirmed the strong synergistic effects between crocin and the PEG-SeNPs. Taken together, our results clearly demonstrate that the strategy to use a SeNP as a carrier of crocin could be a highly efficient way to enhance its anticancer efficacy.
3.5. AO/EtBr staining for detection of apoptotic cells
The induction of apoptosis, after the treatment with IC50 concentrations of crocin conjugated PEG-SeNPs for 24 and 48 h was assessed by fluorescence microscopy after staining with acridine orange/ethidium bromide (AO/EtBr). The images of untreated and crocin conjugated PEG-SeNPs treated A549 cells are presented in Fig. 12a (Upper panel). The fluorescence microscopic analysis demonstrated that untreated A549 cells were stained with a uniform green fluorescence. Because AO can penetrate the normal cell membrane, the cells without treatment were observed as green fluorescence. In contrast the apoptotic cells formed as a result of nuclear shrinkage, blebbing were observed as orange colored bodies due to their loss of membrane integrity when viewed under fluorescence microscope.583.6. HOECHST 33342 staining for nuclear apoptosis
The characterization of the cell death induced by crocin conjugated PEG-SeNPs was further examined with the help of fluorescent DNA binding agent, HOECHST 33342. HOECHST 33342 is known to form fluorescent complexes with natural double-stranded DNA and is useful to find out the apoptotic nuclei. As seen from the images in Fig. 12a (Middle panel) untreated A549 cells had normal morphology with intact round nucleus emitting a weak fluorescence. However, cells treated with crocin conjugated PEG-SeNPs showed apoptotic nuclei, identified by reduced nuclear size, condensed chromatin gathering at the periphery of the nuclear membrane and a total fragmented morphology of nuclear bodies. Shanyuan Zheng et al.48 explained the apoptosis of PEG-SeNPs treated HepG2 cells by means of similar morphological characteristics such as DNA fragmentation and nuclear condensation using staining techniques. Therefore, the anti-proliferation effect of crocin conjugated PEG-SeNPs would be associated with their potential to induce apoptosis in A549 cancer cells.3.7. Analysis of mitochondrial membrane potential (Δψm) by rhodamine 123 staining
The mitochondrial membrane potential (Δψm) loss of cancer cells was analyzed using the dye, Rh-123 [Fig. 12a (Lower panel)]. As can be seen from the image, a decrease in mean fluorescence intensity was observed following the treatment of cells with crocin conjugated PEG-SeNPs. The fluorescence images demonstrated the loss of mitochondrial membrane potential (Δψm) due to mitochondrial membrane depolarization, which was considered to be an initial and irreversible step of apoptosis.68 The data indicated that the induction of apoptosis in cells by crocin conjugated PEG-SeNPs was accompanied by alterations in the mitochondrial membrane potential (Δψm). It was reported that mitochondria played an important role in an intrinsic apoptotic pathway by releasing cytochrome c, leading to the activation of the caspase cascade.69 The results demonstrated that crocin conjugated PEG-SeNPs could disrupt the functions of mitochondria at the early stages of apoptosis, subsequently coordinate caspase 3 activation through the cleavage of caspases by the release of cytochrome c. Fig. 12b shows that the total number of apoptotic cells increases when the incubation time increases.3.8. Effect of crocin conjugated PEG-SeNPs on markers of intrinsic apoptotic gene expression
Apoptotic signaling pathway regulated by a complex network of molecules, involves the expression changes of distinct apoptotic proteins.70 To elucidate the apoptotic pathways activated by crocin conjugated PEG-SeNPs, Western blot analyses were carried out to measure the expression of mitochondrial mediated apoptotic genes. It has been reported that bcl-2 members (e.g., bcl-2) protect against multiple signals that lead to cell death whereas bax members (e.g., bax) induce apoptosis.71–73 Previous studies demonstrated that down regulation of anti-apoptotic protein bcl-2 leads to release of cytochrome c from the mitochondria to cytosol, which is an essential step in the induction of apoptosis. Cytochrome c release from mitochondria to cytosol in turn leads to the activation of the caspase cascade such as caspase-3 and 9 which is critical in executing apoptosis, as it is either partially or totally responsible for the proteolytic cleavage of many key proteins.58 Thus it is remarkable to speculate the analysis of bax, bcl-2, cytochrome c, and caspases-3 and 9 gene expressions. The results (Fig. 13) revealed a significant decrease in the expression of bcl-2 and with a significant increase in the expression of bax, cytosolic cytochrome c and caspase-3 in cells treated with crocin conjugated PEG-SeNPs compared to untreated control. Thus, the induction of apoptosis was closely associated with the down-regulation of bcl-2, up-regulation of bax, loss of mitochondrial membrane potential, release of cytochrome c into cytosol, and subsequent activation of caspase cascades.3.9. Blood compatibility
Determination of hemolytic properties is one of the most common tests in studies of NPs interactions with blood components.58 Hemoglobin release analysis (Fig. 14) shows the hemolytic activity of crocin conjugated PEG-SeNPs. Hemolysis of crocin conjugated PEG-SeNPs at all the tested concentrations were found to be <5%. It has been reported that up to 5% hemolysis is permissible for biomaterials.74 The largest percentage hemolysis obtained was 0.68 ± 0.012% for crocin conjugated PEG-SeNPs at 9 mg mL−1. Since this is much lower than 5%, it indicates that crocin conjugated PEG-SeNPs are hemocompatible for drug delivery applications. Fig. 15a shows photographs of the hemolytic test on the nanoparticle samples. When water is added to RBCs, hemolysis takes place and the released hemoglobin emits red color. This serves as a positive control and represents 100% hemolysis. RBCs incubated with PBS were used as negative controls and represents 0% hemolysis. The supernatant from crocin conjugated PEG-SeNPs at different concentrations is achromatic, and is comparable to that suspended in PBS. Thus, crocin conjugated PEG-SeNPs at the tested concentration exhibited no significant hemolysis. The cell morphology analysis (Fig. 15b) indicated that incubation of RBCs with 9 mg mL−1 crocin conjugated PEG-SeNPs did not result in hemolysis or change in morphology of red blood cells when compared to control, thus implying the biocompatibility of the NPs. Yu-Shen Lin et al.75 showed the influence of PEG surface coating on hemolytic activity of mesoporous silica nanoparticle (MS NPs). The authors report that contrary to bare MS NPs, no apparent hemolysis was observed for PEG-coated MS NPs after 3 h blood incubation. In our study, the absence of hemolysis maybe due to biocompatible polymer PEG coating which prevented the adhesion of both the NPs to red blood cell membrane. Thus this simple surface modification stratagem is critical to ensure the safety of crocin conjugated PEG-SeNPs in biomedical applications.3.10. In vivo anticancer activity of crocin conjugated PEG-SeNPs
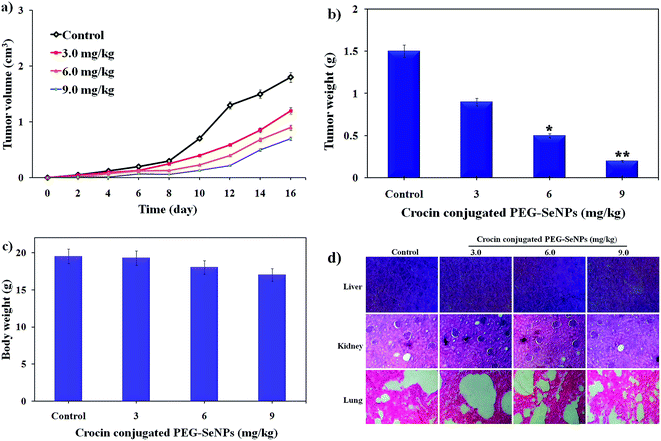
In vivo therapeutic efficacy of crocin conjugated PEG-SeNPs is a crucial index for its future medical potential. Therefore, we treated A549 xenografts nude mice with different dosages of crocin conjugated PEG-SeNPs to examine its in vivo anticancer efficacy. At the end of the experiments, the mice were sacrificed and the tumor weight and tumor volume were measured (Fig. 16). The results show that crocin conjugated PEG-SeNPs significantly inhibited the proliferation of A549 cells in a dose dependent manner, as represented by the decrease in tumor volume (Fig. 16a) and tumor weight (Fig. 16b). Besides, no distinct reduction was observed in the body weight of nude mice, indicating the mineral side effect of PEG-SeNPs after crocin surface decoration (Fig. 16c). These results demonstrate the effective in vivo tumor suppressed capacity of crocin conjugated PEG-SeNPs. Previous studies demonstrated that Tf-SeNPs at similar dosages caused effective in vivo tumor suppression in MCF-7 xenografts nude.36 Further, histological analysis of mice treated with normal saline, crocin conjugated PEG-SeNPs at different concentrations revealed no significant signal of damage from H&E stained organ slices including liver, kidney, and lung (Fig. 16d). Taken together, these findings all indicated that crocin conjugated PEG-SeNPs showed potential therapeutic effect in vivo.4. Conclusion
Our present works provide a design of delivery system by using PEG-SeNPs as a carrier of crocin to achieve anticancer synergism. The studies on in vitro crocin release revealed that faster release of crocin has been observed under the acidic condition, which is exactly what we expect. Crocin thus could principally be distributed around tumor tissues with an acidic microenvironment rather than in the normal section. Therefore, PEG-SeNPs hold a promise as a pH-mediated release delivery vehicle for potential cancer therapy. Crocin conjugated PEG-SeNPs showed perfect hemocompatibility and exhibited enhanced cytotoxicity toward A549 cells (human lung cancer cellines) through induction of apoptosis via mitochondria mediated pathway. Furthermore, crocin conjugated PEG-SeNPs significantly inhibits in vivo tumor growth in nude mice model. Taken together, our results suggest that the strategy to use the PEG-SeNPs as a carrier of crocin could be a highly efficient way to realize synergistic treatment of lung cancer. Furthermore, crocin conjugated PEG-SeNPs may be candidates for further evaluation as a chemotherapeutic agent for other human cancers.Conflict of interest
No conflict of interest was reported by the author of this article.Acknowledgements
This research work was partially supported by UGC – Basic Science for Research (BSR) – RFSMS (Ref. G2/3142/UGC – BSR – RFSMS/2013) and DST-Nano-mission Project, Department of Science and Technology, Nano-mission division, New Delhi (Ref. SR/NM/NS-60/2010 dt. 08-07-2011). We thank the CeNSE Department, Indian Institute of Science (IISc), Bangalore, India, for helping with all the characterizations techniques.References
- R. Prasan and R. Bhandari, Journal of Traditional and Complementary Medicine, 2015, 5, 81–87 CrossRef PubMed.
- L. Wen, L. Xiaoling, W. Yum-Shing, Z. Wenjie, Z. Yibo, C. Wenqiang and C. Tianfeng, ACS Nano, 2012, 6, 6578–6591 CrossRef PubMed.
- W. Yuxia, Z. Xingbo, Z. Jin, X. Songqiang and W. Chaojie, J. Med. Chem., 2012, 55, 3502–3512 CrossRef PubMed.
- Z. Zipeng, T. X Wei, C. X Yen-Jun, T. Trever, Z. Weizhong, L. Xin, N. Gang, L. Gang, W. Lianchun, P. Zhengwei, C. Xiaoyuan and X. Jin, ACS Nano, 2014, 8, 6004–6013 CrossRef PubMed.
- F. Alexis, E. Pridgen, L. K. Molnar and O. C. Farokhzad, Mol. Pharmaceutics, 2008, 5, 505–515 CrossRef CAS PubMed.
- S. K. Sneha and M. R. Theresa, Bioconjugate Chem., 2011, 22, 1879–1903 CrossRef PubMed.
- H. J. Maeda, Controlled-Release Pharm., Symp., 2012, 164, 138–144 CrossRef CAS PubMed.
- L. Pan, Q. He, J. Liu, Y. Chen, M. Ma and L. Zhang, J. Am. Chem. Soc., 2012, 134, 5722–5735 CrossRef CAS PubMed.
- M. Bruchez Jr, M. Moronne, P. Gin, S. Weiss and A. P. Alivisatos, Science, 1998, 281, 2013–2016 CrossRef CAS.
- G. Tkachenko, H. Xie, D. Coleman, W. Glomm, J. Ryan and M. F. Anderson, J. Am. Chem. Soc., 2003, 125, 4700–4711 CrossRef PubMed.
- M. Liong, J. Lu, M. Kovochich, T. Xia, S. G. Ruehm and A. E. Nel, ACS Nano, 2008, 2, 889–896 CrossRef CAS PubMed.
- W. Liu, X. Li, Y. S. Wong, W. Zheng, Y. Zhang and W. Cao, ACS Nano, 2012, 6, 6578–6591 CrossRef CAS PubMed.
- K. K. Vekariya, J. Kaur and K. Tikoo, Nanomedicine, 2012, 8, 1125–1132 CrossRef CAS PubMed.
- L. Kong, Q. Yuan, H. Zhu, Y. Li, Q. Guo and Q. Wang, Biomaterials, 2011, 32, 6515–6522 CrossRef CAS PubMed.
- B. Yu, Y. Zhang, W. Zheng, C. Fan and T. Chen, Inorg. Chem., 2012, 5, 8956–8963 CrossRef PubMed.
- Y. Zhang, X. Li, Z. Huang, W. Zheng, C. Fan and T. Chen, Nanomedicine, 2013, 9, 74–84 CrossRef CAS PubMed.
- S. H. Park, J. Y. Choi, Y. H. Lee, J. T. Park and H. Song, Chem.–Asian J., 2015, 10, 1452–1456 CrossRef CAS PubMed.
- S. Yu, W. Zhang, W. Liu, W. Zhu, R. Guo, Y. Wang, D. Zhang and J. Wang, Nanotechnology, 2015, 26, 145703–145718 CrossRef PubMed.
- J. T. Rotruck, A. L. Pope, H. E. Ganther, A. B. Swanson, D. G. Hafeman and W. G. Hoekstra, Science, 1973, 179, 588–590 CrossRef CAS PubMed.
- J. Zhang, H. Wanh, X. Yan and L. Zhang, Life Sci., 2005, 76, 1099–1109 CrossRef CAS PubMed.
- A. Navas-Acien, J. Bleys and E. Guallar, Curr. Opin. Lipidol., 2008, 19, 43–49 CrossRef CAS PubMed.
- A. L. Ray, R. D. Semba, J. Walston, L. Ferrucci, A. R. Cappola, M. O. Ricks, Q. L. Xue and L. P. Fried, J. Nutr., 2006, 136, 172–176 CrossRef CAS PubMed.
- C. D. Thomson, Eur. J. Clin. Nutr., 2004, 58, 391–402 CrossRef CAS PubMed.
- N. V. Popova, Cancer Lett., 2002, 179, 39–42 CrossRef CAS PubMed.
- C. Redman, J. A. Scott, A. T. Baines, J. L. Basye, L. C. Clark, C. Calley, D. Roe, C. M. Payne and M. A. Nelson, Cancer Lett., 1998, 125, 103–110 CrossRef CAS PubMed.
- B. Prokopczyk, J. G. Rosa, D. Desai, S. Amin, O. S. Sohn, E. S. Fiala and K. El-Bayoumy, Cancer Lett., 2000, 161, 35–46 CrossRef CAS PubMed.
- S. Y. Zheng, X. L. Li, Y. B. Zhang, Q. Xie, Y. S. Wong, Z. W. J. Heng and T. F. Chen, Int. J. Nanomed., 2012, 7, 3939–3949 CAS.
- C. H. Ramamurthy, K. S. Sampath, P. Arunkumar, M. S. Kumar, V. Sujatha, K. Premkumar and C. Thirunavukkarasu, Bioprocess Biosyst. Eng., 2013, 36, 1131–1139 CrossRef CAS PubMed.
- T. F. Chen, Y. S. Wong, W. J. Zheng, Y. Bai and L. Huang, Colloids Surf., B, 2008, 67, 26–31 CrossRef CAS PubMed.
- H. Y. Luo, F. F. Wang, Y. Bai, T. F. Chen and W. J. Zheng, Colloids Surf., B, 2012, 94, 304–308 CrossRef CAS PubMed.
- K. S. Prasad, H. Patel, T. Patel, K. Patel and K. P. Selvaraj, Colloids Surf., B, 2013, 103, 261–266 CrossRef CAS PubMed.
- H. Hu, G. X. Li, L. Wang, J. Watts, G. F. Combs and J. Lu, Cancer Res., 2008, 14, 1150–1158 CAS.
- S. Li, Y. Zhou, R. Wang, H. Zhang, Y. Dong and C. Ip, Mol. Cancer Ther., 2007, 6, 1031–1038 CrossRef CAS PubMed.
- H. Wang, J. Zhang and H. Yu, Free Radical Biol. Med., 2007, 42, 1524–1533 CrossRef CAS PubMed.
- J. Zhang, X. Wang and T. Xu, Toxicol. Sci., 2008, 101, 22–31 CrossRef CAS PubMed.
- P. A. Tran, L. Sarin, R. H. Hurt and T. J. Webster, Int. J. Nanomed., 2010, 5, 351–358 CAS.
- Y. Huang, L. He, W. Liu, C. Fan, W. Zheng, Y. S. Wong and T. Chen, Biomaterials, 2013, 34, 7106–7116 CrossRef CAS PubMed.
- H. H Aung, C. Z. Wang, M. Ni, A. Fishbein, S. R. Mehendale, J. T. Xie, C. Shoyama and C. S. Yuan, Exp. Oncol., 2007, 29, 175–180 Search PubMed.
- C. F. Lv, C. L. Luo, H. Y. Ji and P. Zhao, China J. Chin. Mater. Med., 2008, 33, 1612–1617 Search PubMed.
- H. Hosseinzadeh, H. R. Sadeghnia, T. Ziaee and A. Danaee, J. Pharm. Pharm. Sci., 2005, 8, 387–393 CAS.
- H. Hosseinzadeh and N. B. Noraei, Phytother. Res., 2009, 23, 768–774 CrossRef CAS PubMed.
- N. Pitsikas, A. Boultadakis, G. Georgiadou, P. A. Tarantilis and N. Sakellaridis, Phytomedicine, 2008, 15, 1135–1139 CrossRef CAS PubMed.
- T. Ochiai, H. Shimeno, K. Mishima, K. Iwasaki, M. Fujiwara, H. Tanaka, Y. Shoyama, A. Toda, R. Eyanagi and S. Soeda, Biochim. Biophys. Acta, 2007, 1770, 578–584 CrossRef CAS PubMed.
- H. Hosseinzadeh, M. H. Modaghegh and Z. Saffari, J. Evidence-Based Complementary Altern. Med., 2009, 6, 343–350 CrossRef PubMed.
- H. Hosseinzadeh, A. Abootorabi and H. R. Sadeghnia, DNA Cell Biol., 2008, 27, 657–664 CrossRef CAS PubMed.
- A. Talaei, M. H. Moghadam, S. A. Sajadi Tabassi and S. A. Mohajeri, J. Affective Disord., 2015, 174, 51–56 CrossRef CAS PubMed.
- S. A. Mohajeri, H. Hosseinzadeh, F. Keyhanfar and J. Aghamohammadian, J. Sep. Sci., 2010, 33, 2302–2309 CrossRef CAS PubMed.
- S. Zheng, X. Li, Y. Zhang, Q. Xie, Y. S. Wong, W. Zheng and T. Chen, Int. J. Nanomed., 2012, 7, 3939–3949 CAS.
- W. Liu, X. Li, Y. S. Wong, W. Zheng, Y. Zhang and W. Cao, ACS Nano, 2012, 6, 6578–6591 CrossRef CAS PubMed.
- T. Chen and Y. S. Wong, Biomed. Pharmacother., 2009, 63, 105–113 CrossRef CAS PubMed.
- S. A. Mohajeri, H. Hosseinzadeh, F. Keyhanfar and J. Aghamohammadian, J. Sep. Sci., 2010, 33, 2302–2309 CrossRef CAS PubMed.
- K. S. Prasad, H. Patel, T. Patel, K. Patel and K. Selvaraj, Colloids Surf., B, 2013, 103, 261–266 CrossRef CAS PubMed.
- C. H. Ramamurthy, K. S. Sampath, P. Arunkumar, M. Suresh Kumar, V. Sujatha and K. Premkumar, Bioprocess Biosyst. Eng., 2013, 36, 1131–1139 CrossRef CAS PubMed.
- H. Estevez, J. Carlos Garcia-Lidon, L. Luque-Garcia and C. Camara, Colloids Surf., B, 2014, 122, 184–193 CrossRef CAS PubMed.
- W. Zheng, C. Cao, Y. Liu, Q. Yu, C. Zheng, D. Sun, X. Ren and J. Liu, Acta Biomater., 2015, 11, 368–380 CrossRef CAS PubMed.
- A. Rubio Moraga, O. Ahrazem, J. L. Rambla, A. Granell and L. Gómez Gómez, PLoS One, 2013, 8, 71946 CrossRef PubMed.
- D. C. Litzinger, A. M. J. Buiting, N. Rooijen and L. Huang, Biochim. Biophys. Acta, 1994, 1190, 99–107 CrossRef CAS.
- K. Shanthi, K. Vimala, D. Gopi and S. Kannan, RSC Adv., 2015, 5, 44998–45014 RSC.
- C. Li, Adv. Drug Delivery Rev., 2002, 54, 695–713 CrossRef CAS PubMed.
- C. He, Y. Hu, L. Yin, C. Tang and C. Yin, Biomaterials, 2010, 31, 3657–3666 CrossRef CAS PubMed.
- F. Alexis, E. Pridgen, L. K. Molnar and O. C. Farokhzad, Mol. Pharm., 2008, 5, 505–515 CrossRef CAS PubMed.
- P. Aggarwal, J. B. Hall, C. B. McLeland, M. A. Dobrovolskaia and S. E. McNeil, Adv. Drug Delivery Rev., 2009, 61, 428–437 CrossRef CAS PubMed.
- H. Mu, X. Wang, P. H. Lin, Q. Yao and C. Chen, Atherosclerosis, 2008, 201, 67–75 CrossRef CAS PubMed.
- H. Petersen, P. M. Fechner and A. L. Martin, Bioconjugate Chem., 2002, 13, 845–854 CrossRef CAS PubMed.
- C. Zhang, L. Qi Zhao, Y. Feng Dong, X. Y. Zhang, J. Lin and Z. Chen, Eur. J. Pharm. Biopharm., 2010, 76, 10–16 CrossRef CAS PubMed.
- Z. Liang, M. Yanga, X. Xua, Z. Xiea, J. Huanga, X. Lia and D. Yanga, Sep. Sci. Technol., 2014, 49, 1427–1433 CrossRef CAS.
- K. Manolis, A. Assimiadis, A. T. Petros and M. G. Polissiou, Appl. Spectrosc., 1998, 52, 519–522 CrossRef.
- C. Adrie, M. Bachelet, M. Vayssier-Taussat, F. Russo-Marie, I. Bouchaert, M. Adib-Conquy, J. M. Cavaillon, M. R. Pinsky, J. F. Dhainaut and B. S. Polla, Am. J. Respir. Crit. Care Med., 2001, 164, 389–395 CrossRef CAS PubMed.
- J. Gil, S. Almeida, C. R. Oliveira and A. C. Rego, Free Radical Biol. Med., 2003, 35, 1500–1514 CrossRef CAS PubMed.
- K. Vimala, S. Sundarraj, M. Paulpandi and S. Kannan, Process Biochem., 2014, 49, 160–172 CrossRef CAS.
- R. Vivek, V. Nipun Babu, R. Thangam, K. S. Subramanian and S. Kannan, Colloids Surf., B, 2013, 111, 117–123 CrossRef CAS PubMed.
- M. O. Hengartner, Nature, 2000, 407, 770–776 CrossRef CAS PubMed.
- S. Y. Jeong and D. W. Seol, J. Biochem. Mol. Biol., 2008, 41, 11–22 CAS.
- J. P. Singhal and A. R. Ray, Biomaterials, 2002, 23, 1139–1145 CrossRef CAS PubMed.
- Y. S. Lin and C. L. Haynes, J. Am. Chem. Soc., 2010, 132, 4834–4842 CrossRef PubMed.
Footnote |
| † These authors contributed equally to this manuscript. |
| This journal is © The Royal Society of Chemistry 2016 |