Open Access Article
Open Access ArticleCross-species biosynthesis of maytansine in Maytenus serrata†‡
Parijat Kusaria,
Souvik Kusari *b,
Dennis Eckelmannb,
Sebastian Zühlkeb,
Oliver Kaysera and
Michael Spiteller*b
*b,
Dennis Eckelmannb,
Sebastian Zühlkeb,
Oliver Kaysera and
Michael Spiteller*b
aDepartment of Biochemical and Chemical Engineering, Chair of Technical Biochemistry, TU Dortmund, Emil-Figge-Str. 66, 44227 Dortmund, Germany
bInstitute of Environmental Research (INFU), Department of Chemistry and Chemical Biology, Chair of Environmental Chemistry and Analytical Chemistry, TU Dortmund, Otto-Hahn-Str. 6, 44221 Dortmund, Germany. E-mail: souvik.kusari@infu.tu-dortmund.de; m.spiteller@infu.tu-dortmund.de; Fax: +49-231-755-4084; Fax: +49-231-755-4085; Tel: +49-231-755-4086 Tel: +49-231-755-4080
First published on 18th January 2016
Abstract
Endophytic bacterial communities harboring different tissues of Maytenus serrata originating from Cameroon were investigated using targeted genome mining techniques coupled to bioanalytical approaches to elucidate the source of maytansine biosynthesis. It was revealed that the host plant, along with its cryptic endophytic microflora, produces the biosynthetically unique core structural moiety 3-amino-5-hydroxybenzoic acid (AHBA) that serves as the unique starter unit for maytansine biosynthesis. However, the biosynthetic step of halogenase-mediated incorporation of chlorine, which is missing in the host plant, is accomplished by the culturable stem endophytic bacterial community. Our results provide new insights into plant-endophyte communication for the biosynthesis of maytansine.
Endophytic microorganisms are ubiquitous in plants in the tissues and apoplastic compartments, both above- and below-ground, where they establish complex communities that can substantially contribute to the overall fitness of the host plants.1–5 Microorganisms rarely exist in “axenic forms” in nature and endophytes are no exception.6 In microbial communities within plants, the interplay between endophytes and plants, and the production of secondary metabolites results in a variety of functional traits of both agricultural and ecological importance.6–11 Thus far, it has been firmly established that suitable plant-endophyte and endophyte–endophyte interactions (fungi–fungi, fungi–bacteria, and/or bacteria–bacteria) play a major role in the production of secondary metabolites.
In the last decades, studies on interspecies and multispecies crosstalk of endophytes have led to the elucidation of functional traits of endophytes concomitant to associated organisms.12 Their coexistence within complex and mixed communities within their specific niches lead to their coevolution such that they can “withstand ecological forces” and function in ways that mostly cannot be accomplished by single organisms.13 For example, it was recently shown that a root-associated bacterial community of the tobacco plant, Nicotiana attenuata, functions as a consortium to provide protection to the host plant against sudden-wilt disease.14 Notably, this important ecological function could not be accomplished by single organisms of the root-associated bacterial community.14 Several recent studies have further lent evidence to the fact that certain so-called plant metabolites are actually biosynthesized by associated microbial community. For instance, we have shown that the original source organisms responsible for the biosynthesis of the important anticancer and cytotoxic compound maytansine (Fig. 1a) is the endophytic bacterial community harbored specifically within the roots of Putterlickia verrucosa and Putterlickia retrospinosa plants (Celastraceae).15 Chemical and molecular characterization of the root endophytic bacterial community revealed that maytansine is actually a biosynthetic product of Putterlickia root-associated endophytic bacterial community.
Given the fact that maytansine was discovered in the 1970s not only in Putterlickia species but also in other Celastraceae plants of the genus Maytenus,16,17 along with our aforementioned results on Putterlickia endophytes, we aimed to investigate the endophytes harbored in Maytenus serrata plants. The main difference, however, was that maytansine was found to be localized in the above-ground tissues of Maytenus plants compared to its accumulation in the roots of Putterlickia plants.16,17 Given these facts, in the present work, we answer the following open questions:
(A) Are endophytic communities responsible for maytansine biosynthesis in Maytenus serrata plants, similar to that of Putterlickia species?
(B) If so, where are these endophytes localized and what is their phylogenetic relationship to those harbored in Putterlickia species?
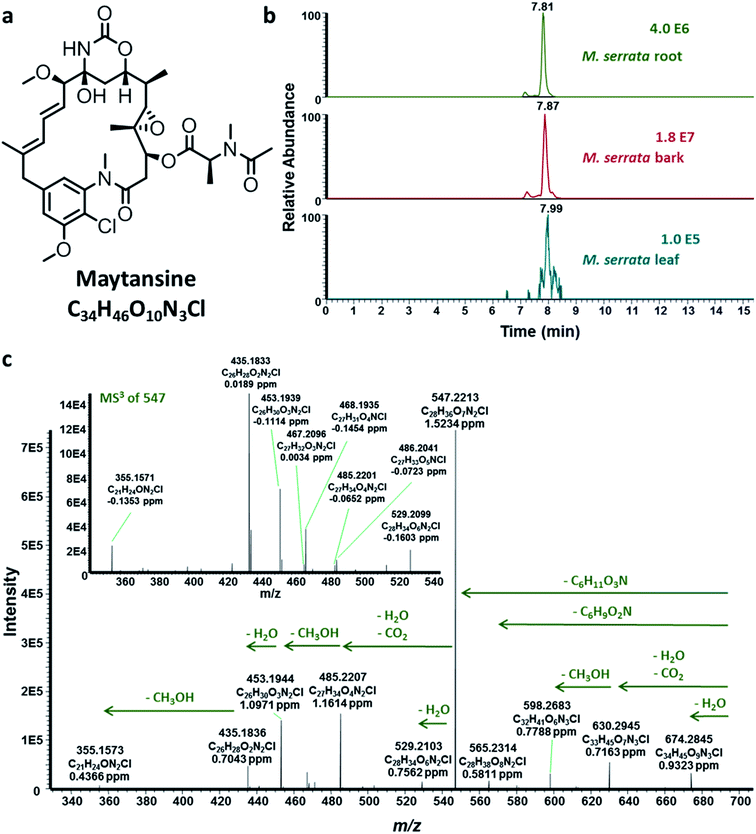
In order to shed light on the aforementioned questions, we collected M. serrata plants from two different locations encompassing very different ecological niches; the first location was the rainforest at Bafoussam in Cameroon, whereas the second sampling was done at the Asakraka-Asikam district in Eastern Ghana. The above- and below-ground plant tissues were extracted and analyzed using HPLC-ESI-HRMSn (see ESI‡), which revealed that maytansine was present in the tissues of M. serrata sampled only from Cameroon (Fig. 1b and c and ESI, Fig. S1‡), but not from Ghana. The striking results reinforced the notion that biosynthesis of maytansine in M. serrata plants is performed by the endophytes and not by the plants themselves,5 similar to what was demonstrated earlier for Putterlickia plants.15
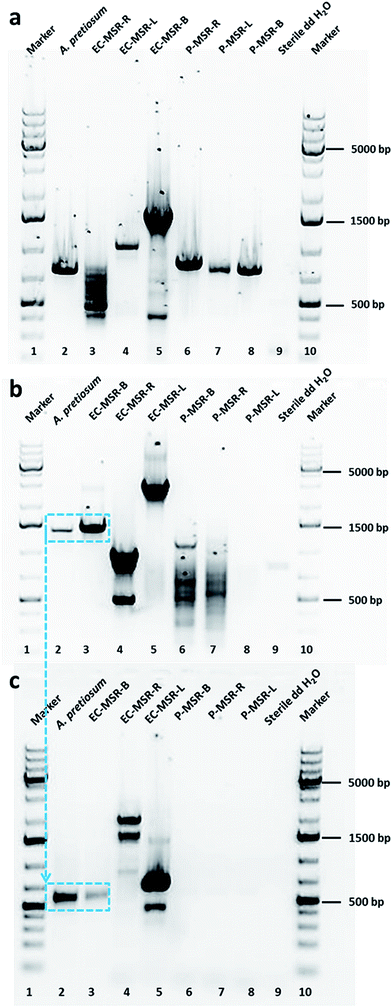
Taking cues from our preliminary results, we examined the biosynthesis of maytansine in the Cameroonian M. serrata and the possible contribution of endophytic bacterial community harbored in different tissues (root, bark and leaf) of the plant. Firstly, we evaluated the different tissues of Maytenus plant and the endophytic bacterial community for the presence of genes biosynthesizing the important starter enzyme, AHBA synthase (3-amino-5-hydroxybenzoic acid synthase).15 This enzyme comprises of a unique mC7N structural moiety which forms the common precursor of different ansamycins.18 Thus, a pair of degenerate primers (primer pair P3; ESI, Table S1‡) were designed amplifying the conserved amino acid regions of different genes namely asnF, asm24, mitA, napF and rifK.15 As a positive control, the genomic DNA of Actinosynnema pretiosum subsp. auranticum (DSM 44131) was amplified resulting in an approx. 755 bp fragment. Further, the root, leaf and bark tissues and the respective endophytic bacterial communities were subjected to the optimized PCR conditions using the same degenerate primer pair. All the plant tissues amplified the expected fragment of AHBA synthase gene (Fig. 2a). Interestingly, none of the endophytic bacterial communities produced the desired amplification product. The positive PCR amplified products were sequenced, aligned, identified, and further translated into protein sequences. The positive control showed 100% homology to 3-amino-5-hydroxybenzoic acid synthase (AHBA synthase; EMBL AAC13997.1 and UniProt Identifier Q44131). Similarly, the amplified products from the different plant tissues also showed sequence homology to 3-amino-5-hydroxybenzoic acid synthase (ESI, Table S2‡). The sequences of the amplified PCR products have been submitted to the EMBL-Bank. We also evaluated the aerial and non-aerial tissues of the Ghanaian M. serrata, which does not contain maytansine, for the presence of AHBA synthase genes. Interestingly, AHBA synthase genes were found to be absent in this plant (data not shown). Thus far, we could conclude that only the M. serrata plant growing in the rainforest of Cameroon contains the genetic blueprint of the starter enzyme (AHBA synthase) for the biosynthesis of maytansine.
Taking ansamitocin biosynthetic pathway into consideration, we decided to further investigate the post-polyketide synthase modification of the ansamitocin pathway. One of the key steps in this pathway is the conversion of proansamitocin to 19-chloroproansamitocin that introduces chlorine into the structure. Chlorination is an important FADH2-dependent halogenation catalyzed by different halogenase enzymes. In the ansamitocin biosynthetic pathway of positive control A. pretiosum, this step is achieved by the gene asm12, which is a FADH2-dependent halogenase.19 We, therefore, aimed to evaluate the plant tissues and the endophytic communities for the presence of halogenase genes in order to elucidate which organism (host plant or endophytic bacterial community) is responsible for incorporation of chlorine towards completing the biosynthesis of maytansine. We employed two strategies to answer this question. In our first approach, we evaluated for the presence of halogenase gene asm12, which catalyzes the first step post-polyketide synthase modification responsible for the chlorination step in ansamitocin biosynthesis. For the detection of asm12 gene, the genomic DNA of plant tissues and endophytic community were subjected to PCR amplification using primer pairs P1 (ESI, Table S1‡). The P1 primers were designed based on the sequence of asm12 gene from A. pretiosum (DSM 44131). None of the plant tissues (leaf, stem, or root) amplified the desired product (Fig. 2b). Furthermore, the leaf and root endophytic bacterial community also did not amplify the desired product (Fig. 2b). Interestingly, only the bark endophytic bacterial community (EC-MSR-B) could produce the desired fragment (Fig. 2b). As a positive control, the genomic DNA of A. pretiosum was amplified (spanning around 1300 bp). The positive fragments were sequenced, aligned, identified, and further translated into protein sequences. The positive control showed 99% homology to asm12 halogenase gene (EMBL AAM54090.1) and 100% homology to UniProt Identifier Q8KUG0. Although the bark endophytic community amplified the desired fragment, the homology with phylogenetically-related reference halogenase sequence (i.e., asm12 of A. pretiosum) was relatively low. In order to avoid any false positives, in our second approach we further analyzed the presence of any FADH2-dependent halogenase irrespective of asm12 gene in particular. Therefore, we performed a second PCR analysis with degenerate primers deduced from highly conserved regions of six different FADH2-dependent halogenases (ansamitocin being only one of them) having different enzyme specificities to produce different halogenated metabolites.20 We re-evaluated the plant tissue and endophytic bacterial communities for the presence of halogenase genes by subjecting them to PCR amplification with primer pair P2 (ESI, Table S1‡). For the positive control, we used the PCR purified 1300 bp asm12 gene of A. pretiosum as template for primer walking with P2. Simultaneously, the 1300 bp positive fragment of bark endophytic bacterial community was also subjected to second PCR using primer pair P2. As expected, we observed an approximately 550 bp amplified fragment for positive control and only bark endophytic bacterial community (Fig. 2c). None of the plant tissues or other endophytic bacterial communities amplified this fragment. The positive fragments were evaluated as detailed above. The positive control naturally showed 99% homology EMBL AF453501.1 and 97.2% homology to UniProt Identifier Q8KUG0. Interestingly, this time the bark endophytic bacterial community showed 100% homology to halogenase (EMBL CP001630.1) and 100% homology to UniProt Identifier C6W874 (tryptophan halogenase).
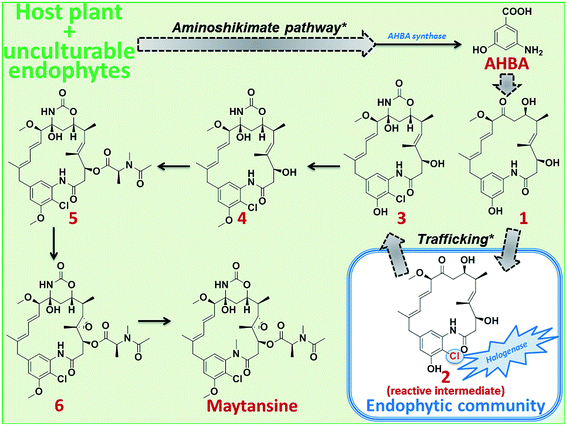
As expected, none of the isolated endophytes and endophytic bacterial communities could produce maytansine when fermented under laboratory conditions (outside the host plant). Furthermore, HRMS/MS allowed us to complement the genetic data; we could detect the major precursors (1–6) of maytansine in the plant tissues, particularly in the bark (ESI, Fig. S2 and Table S3‡). Thus far, using the combination of molecular and analytical data, we propose a plant-endophyte cross-species biosynthetic pathway for production of maytansine that occurs only in some M. serrata plants (such as the one sampled from Cameroonian rainforest) (Scheme 1). The host plant along with its unculturable or cryptic endophytes produces AHBA, the unique starter moiety of maytansine biosynthesis, by means of an aminoshikimate pathway21,22 that is absent in the cultivable bark endophytic bacterial community. AHBA is further converted to precursor 1, which is taken up by the bark endophytic bacterial community to introduce chlorine (precursor 3 via reactive intermediate 2) using the halogenase enzyme that is absent in the host plant and its cryptic endophytes (ESI, Fig. S2 and Table S3‡). Further biosynthetic steps through intermediates 4–6 to the production of maytansine is achieved by the host plant (Scheme 1).
Finally, we aimed to elucidate the phylogenetic relationship between the different endophytic bacterial communities isolated from the leaves, stems, and roots of the maytansine-containing M. serrata plant and those harbored in Putterlickia species.15 The 16S rRNA amplification of the endophytic bacterial communities (Fig. 3a) followed by phylogenetic evaluation using T-Coffee (ESI, Fig. S3‡) was done according to Kusari et al. (2014).15 The phenogram (Fig. 3b) revealed a clear clustering of leaf, bark and root endophytic bacterial communities of M. serrata to that of primary root endophytic bacterial communities of Putterlickia plants. Interestingly, the bark endophytic bacterial community containing the halogenase gene (EC-MSR-B) clustered very close to the root endophytic bacterial communities of Putterlickia plants that were found to produce maytansine (PRR1 and PVR1).15 This shows plausible species-level relatedness between endophytic bacterial communities that contain the genes involved in the biosynthetic pathway of maytansine. Interestingly, the endophytic bacterial communities of M. serrata under our study also showed evolutionary relationships to bacteria harboring diverse hosts (niches) ranging from termites to earthworms (ESI, Fig. S4‡).
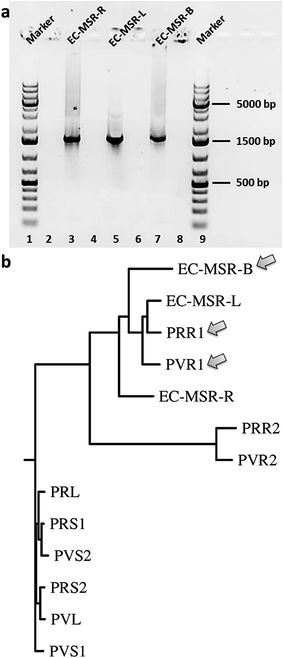 | ||
| Fig. 3 (a) Stained agarose gels of purified PCR products of 16S rRNA analyses of endophytic bacterial communities of Maytenus serrata (spanning around 1500 bp). (b) The phenogram-like rooted phylogenetic tree diagram of endophytic bacterial communities of the present study (EC-MSR-B, EC-MSR-L, EC-MSR-R), compared to ones reported earlier from Putterlickia plants,15 using Drawgram in PHYLIP 3.66 based on their 16S rRNA regions. | ||
Taken together, our study reveals a unique aspect of plant-endophyte communication where only the bark endophytic bacterial community of a selected M. serrata plant had coevolved a means to produce maytansine jointly with the host plant, including trafficking of the precursors from and to the host plant. In other M. serrata plants where this endophytic community is absent, there is no production of maytansine, as exemplified not only by the Ghanaian M. serrata plant but also other Maytenus plants of different origin presently under our investigation (data not shown). Our results provide a scientific handle to delve into further details of plant-endophyte communication not only on the transcriptome level but also the metabolome level to pinpoint regulation of host-endophyte communication. Our study further opens avenues to determine qualitative and quantitative pattern of localization and trafficking of endophytes with regard to shared biosynthesis of maytansine. Further studies along these lines are currently underway.
Acknowledgements
This work was funded by the “Welcome to Africa” initiative of the German Academic Exchange Service (DAAD) and the German Federal Ministry of Education and Research (BMBF). The Ministry of Innovation, Science, Research and Technology of the State of North Rhine-Westphalia, Germany, and the German Research Foundation (DFG) are thankfully acknowledged for granting a high-resolution mass spectrometer. We thankfully acknowledge Ms K. Louven (TU Dortmund) for technical assistance during the isolation of the endophytic communities and fermentation. We appreciate the assistance of Prof. S. F. Kouam (University of Yaoundé I, Cameroon) and Ms E. A. Mireku (INFU, TU Dortmund) in the collection of plants from Cameroon and Ghana, respectively.Notes and references
- T. Hosni, C. Moretti, G. Devescovi, Z. R. Suarez-Moreno, M. B. Fatmi and C. Guarnaccia, et al., ISME J., 2011, 5, 1857 CrossRef CAS PubMed.
- D. G. Meldau, S. Meldau, L. H. Hoang, S. Underberg, H. Wünsche and I. T. Baldwin, Plant Cell, 2013, 25, 2731 CrossRef CAS PubMed.
- S. Kusari, S. Singh and C. Jayabaskaran, Trends Biotechnol., 2014, 32, 304 CrossRef CAS PubMed.
- P. Kusari, S. Kusari, M. Lamshöft, S. Sezgin, M. Spiteller and O. Kayser, Appl. Microbiol. Biotechnol., 2014, 98, 7173 CrossRef CAS PubMed.
- D. J. Newman and G. M. Cragg, Front. Chem., 2015, 3, 34 Search PubMed.
- S. Kusari, C. Hertweck and M. Spiteller, Chem. Biol., 2012, 19, 792 CrossRef CAS PubMed.
- V. Schroeckh, K. Scherlach, H. W. Nützmann, E. Shelest, W. Schmidt-Heck and J. Schuemann, et al., Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 14558 CrossRef CAS PubMed.
- B. Reinhold-Hurek and T. Hurek, Curr. Opin. Plant Biol., 2011, 14, 435 CrossRef PubMed.
- A. Debbab, A. H. Aly and P. Proksch, Fungal Divers., 2013, 61, 1 CrossRef.
- A. L. Navarro-Meléndez and M. Heil, J. Chem. Ecol., 2014, 40, 816 CrossRef PubMed.
- L. Tyvaert, S. C. França, J. Debode and M. Höfte, J. Appl. Microbiol., 2014, 116, 1563 CrossRef CAS PubMed.
- S. Kusari, S. P. Pandey and M. Spiteller, Phytochemistry, 2013, 91, 81 CrossRef CAS PubMed.
- J. de Vrieze, Science, 2015, 349, 680 CrossRef CAS PubMed.
- R. Santhanam, V. T. Luu, A. Weinhold, J. Goldberg, Y. Oh and I. T. Baldwin, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, e5013 CrossRef CAS PubMed.
- S. Kusari, M. Lamshöft, P. Kusari, S. Gottfried, S. Zühlke and K. Louven, et al., J. Nat. Prod., 2014, 77, 2577 CrossRef CAS PubMed.
- S. M. Kupchan, Y. Komoda, W. A. Court, G. J. Thomas, R. M. Smith and A. Karim, et al., J. Am. Chem. Soc., 1972, 94, 1354 CrossRef CAS PubMed.
- S. M. Kupchan, Y. Komoda, A. R. Branfman, A. T. Sneden, W. A. Court and G. J. Thomas, et al., J. Org. Chem., 1977, 42, 2349 CrossRef CAS PubMed.
- Q. Kang, Y. Shen and L. Bai, Nat. Prod. Rep., 2012, 29, 243 RSC.
- Y. Wu, Q. Kang, Y. Shen, W. Su and L. Bai, Mol. BioSyst., 2011, 7, 2459 RSC.
- A. Hornung, M. Bertazzo, A. Dziarnowski, K. Schneider, K. Welzel and S. E. Wohlert, et al., ChemBioChem, 2007, 8, 757 CrossRef CAS PubMed.
- H. G. Floss, Nat. Prod. Rep., 1997, 14, 433 RSC.
- H. Maeda and N. Dudareva, Annu. Rev. Plant Biol., 2012, 63, 73 CrossRef CAS PubMed.
Footnotes |
| † Accession numbers: All the sequences and products verified/discovered in the present study have been deposited at the EMBL-Bank under the accession numbers LN874065 to LN874068. |
| ‡ Electronic supplementary information (ESI) available: Experimental section, Fig. S1–S4 and Tables S1–S3. See DOI: 10.1039/c5ra25042k |
| This journal is © The Royal Society of Chemistry 2016 |