Intrinsically disordered proteins in PubMed: what can the tip of the iceberg tell us about what lies below?†
Abstract
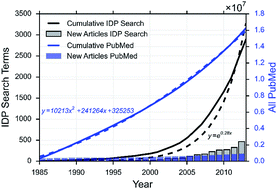
Intrinsically disordered proteins (IDPs) have a troubled history in the literature. Historically, a wide variety of terminology has been used to describe these strange proteins that do not adopt a stable three-dimensional structure. We provide here a survey of the current status of both IDPs and IDP terminology in PubMed. We have performed an extensive search of the literature from 1978 through 2014 and compiled a list of 1127 proteins and protein domains and the corresponding citations that refer to these proteins using IDP terminology. We show that papers that use IDP terminology are only the tip of the iceberg in terms of the larger body of literature referring to this group of proteins. Furthermore, our analysis suggests that this is likely due to a lack of perceived relevance rather than a lack of awareness. Finally, we have analyzed the language provided by author keywords, MeSH terms, and abstracts as well as the journals that are currently publishing IDP articles. Our results demonstrate a convergence on a common set of terminology and a rise in the number of papers using this terminology. However, our results also demonstrate that we have not reached the point where IDP terminology is fully accepted and embraced in the literature.

 Please wait while we load your content...
Please wait while we load your content...