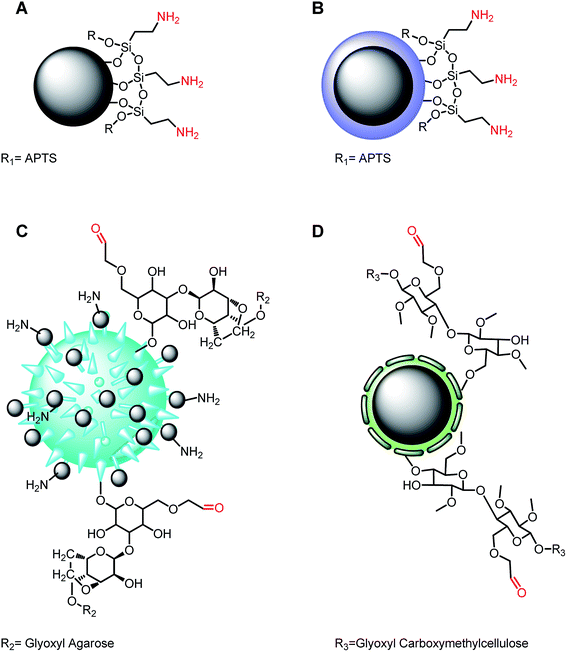
Bovine glutamate dehydrogenase immobilization on magnetic nanoparticles: conformational changes and catalysis†
Caterina G. C.
Marques Netto‡
*a,
Delmárcio G.
da Silva
a,
Sergio H.
Toma
a,
Leandro H.
Andrade
b,
Marcelo
Nakamura
a,
Koiti
Araki
a and
Henrique E.
Toma
a
aSupramolecular NanotechLab, Instituto de Quimica, Universidade de São Paulo, São Paulo-SP, Brazil. E-mail: caterina@ufscar.br
bLaboratory of Fine Chemistry and Biocatalysis, Instituto de Química, Universidade de São Paulo, São Paulo-SP, Brazil
First published on 26th January 2016
Abstract
Glutamate dehydrogenase (GDH) is a well known homohexameric enzyme and its performance in immobilized form was systematically investigated in this work, in order to provide a better understanding of the multimeric enzyme immobilization effects in relation to the monomeric ones. For this purpose, GDH was immobilized on four different magnetic supports and the outcome from such immobilization was characterized in terms of their stabilization and activity. Immobilization procedures involving amine coupling via glutaraldehyde cross-linking yielded the least stable ones, even lower than free GDH, showing no recoverability. However, the immobilization procedures using larger aldehyde cross-linkers presented higher thermal stabilities than free GDH and could be recycled at least 10 times, with a nearly constant activity. Such differences in stability and activity were thoroughly evaluated in terms of the enzymatic structure, which has guided the reasoning behind the intriguing allosteric behavior of the immobilized GDH. The atypical allosteric response exhibited by MagNP–APTS/GDH, MagNP@SiO2–APTS/GDH and MagNP–APTS/glyoxyl-agarose/GDH led us to invoke the intrinsic disorder theory to explain the results. This theory has proved to be an excellent tool to guide research on immobilized enzymes and understand its effects.
Introduction
Enzyme immobilization is a technique used to obtain recoverable, stable and in several cases, generate systems with higher activities than the free protein.1,2 The properties of an immobilized biocatalyst are related to the methodology and supporting material employed, but they also depend on the amino acid composition of the enzyme's surface.3 However, despite of the extensive amount of studies reported in this area,4 the resulting enzyme activity is not easy to predict5 and a previous knowledge of the catalysis is essential for the comprehension of the new system resulting from the protein anchoring to a support.6 Considering that the catalytic properties of an enzyme are intimately dependent on the protein's three-dimensional conformation and the keyhole-lock relationship between enzyme and substrate,7 it is foreseen that any perturbation of the protein's native configuration will impact its activity.3a,8In general, an impact on the enzymatic activity upon immobilization is determined by the induction of microenvironment and conformational changes9 and the extent of impact suffered by a multimeric enzyme after immobilization is expected to be larger than for monomeric ones.10 The greater impact on multimeric enzymes can be related to enzyme subunit dissociation10,11 or to disturbance in the subunits interactions in the quaternary -structures after fixation on a support.12 Therefore, multimeric enzyme immobilization is a rather complex problem since overcoming the issues about enzyme stabilization13,14 includes a supporting material design3b,10,12b,15,16 and study of immobilization procedures.8b,17,18
Another issue faced by multimeric enzymes immobilization is the disturbance of allosteric mechanisms. Some proteins are ordered or disordered prior to ligand binding in other parts of the molecule, regulating a long-range allosteric interaction within the protein.19,20 As a consequence, any modification of these mechanisms through immobilization can alter the precise distribution of states within the native state ensemble21,22 and modulate the allosteric response of the whole system. An example of a useful multimeric enzyme is the 336 kDa homohexameric protein glutamate dehydrogenase (GDH) from bovine liver,23,24 which has its activity tightly controlled by a complex network of allosteric regulators23,25 and its immobilization can induce differences in allosteric response.26 In its immobilized form, glutamate dehydrogenase is normally used as glutamate probes,27–30 employing different supports and methodologies for the attachment on a support.12b,26c,29,31 However, regardless of the extensive amount of study on GDH immobilization, the use of magnetic nanoparticles is still in early stages.32
Therefore, bearing the challenge of obtaining a reusable and stable multimeric biocatalyst12a,33 and the demand for GDH stabilization and activation,24,34 we performed the immobilization of glutamate dehydrogenase on magnetite nanoparticles (MagNP) displaying different characteristics. Aminopropyltriethoxysilane (APTS) was used as a surface modifier35 in two different supports: one with no silica shell and the other one with a silica shell (MagNP–APTS and MagNP@SiO2–APTS), employing glutaraldehyde as a cross-linking agent.36 Differences in enzymatic activities between these two supports would be mainly due to iron exposure to the protein's surface.37 Glyoxyl-carboxymethylcellulose and glyoxyl-agarose38 were the other two cross-linking agents chosen (MagNP@CMC and MagNP–APTS–glyoxyl-agarose), both exhibit a higher loading of aldehyde groups that can help to afford high enzyme loadings, but can also interfere in the catalytic reaction by greater diffusional limitations of the substrates.1,3b,4,5,11a,39 Consequently, beyond the advantage of easy recoverability of magnetic nanoparticles by the use of an external magnetic field,40,41 the difference between these immobilization protocols in stabilization, activation and allosterism should also be investigated. In this work, it became evident that different supports can yield contrasting GDH properties, with higher or lower stability and activity, depending on the enzyme-attachment methodology. The intrinsic disorder mechanism19a was employed for each immobilized system and proved to be an important tool for understanding the effect of different immobilization protocols on the enzyme behavior.
Experimental section
All reagents were purchased from Sigma-Aldrich and were used without previous purification. Glutamate dehydrogenase (type II) from bovine liver exhibited ≥35 units per mg of protein (one unit expresses the reduction of 1 µmol of α-ketoglutarate to L-glutamate per minute in the presence of NADH (2.4 mmol L−1) and ammonium ions at pH 7.3 and 25 °C). Trypsin used in tryptic digestion was TPCK treated from bovine pancreas, exhibiting ≥10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 benzoyl-L-arginine ethyl ester (BAEE) units per mg protein (one BAEE unit will produce a A253 of 0.001 per minute at pH 7.6 at 25 °C using BAEE as a substrate. One BTEE unit = 320 ATEE unit).
000 benzoyl-L-arginine ethyl ester (BAEE) units per mg protein (one BAEE unit will produce a A253 of 0.001 per minute at pH 7.6 at 25 °C using BAEE as a substrate. One BTEE unit = 320 ATEE unit).
Synthesis of MagNP–APTS, MagNP@SiO2–APTS and MagNP@CMC
The synthesis of MagNP–APTS and MagNP@SiO2–APTS has been described elsewhere.37,42 The MagNP@CMC material was generated from freshly prepared magnetic nanoparticles by treating with 0.5% aqueous alkaline solution of carboxymethyl-cellulose (CMC), pH 12, under stirring and reflux for 30 min. The nanoparticles were magnetically concentrated, and washed with deaerated water, yielding stable colloidal solutions displaying average global particle size of 90 nm (based on DLS) and magnetic core of about 25 nm (SEM). MagNP@CMC particles were further dried at 100 °C for 12 hours. Activation of the hydroxyl groups from the CMC shell was performed by suspending 500 mg of MagNP@CMC in 3 mL of deionized water, followed by the addition of 250 µL of 2 mol L−1 NaOH solution (containing 6.7 mg of NaBH4) at 0 °C. Then, 180 µL of glycidol was added dropwise and the reaction proceeded for additional 16 hours at room temperature. The particles were then washed with water (3 × 10 mL) and 15 mg of NaIO4 dissolved in 5 mL of water was added. The reaction proceeded for 3 hours and after that period, the particles were washed with water (10 × 10 mL) and kept in suspension (200 mg mL−1).General procedure of GDH immobilization on MagNP–APTS
MagNP–APTS (0, 1, 2, 4, 8 and 16 mg) suspended in 100 µL of Tris–HCl buffer solution (pH 7.5, 0.1 mol L−1) was mixed with 100 µL of a GDH solution (0.523 mg mL−1, 2 U in 0.1 M Tris–HCl buffer pH 7.5). To this mixture glutaraldehyde (0, 2, 5, 10 and 15 µL of a 0.2% solution in water) was added and the suspension was stirred at 120 rpm for 5, 10, 15 or 20 minutes, at 0 °C. The immobilized enzymes were confined by using an external miniature Nd2Fe14B magnet (1 cm3, 11 kOe from MagTek), and washed 3 times with 0.1 mL of Tris–HCl buffer solution (pH 7.5, 0.1 mol L−1). The best immobilization conditions were obtained by using 2 mg of MagNP–APTS, 2 µL of a glutaraldehyde solution and stirring for 5 minutes, which afforded the system MagNP–APTS/GDH (amount of immobilized GDH: 0.05 mg/2 mg MagNP–APTS).General procedure of GDH immobilization on MagNP@SiO2–APTS
MagNP@SiO2–APTS (0.6, 1.2, 2.4, 4 and 8 mg) suspended in 100 µL of Tris–HCl buffer solution (pH 7.5, 0.1 mol L−1) was mixed with 100 µL of a GDH solution 0.523 mg mL−1 (2 U). Then, glutaraldehyde (0, 2, 5, 10 and 15 µL of a 0.2% solution in water) was added and the resulting mixture was stirred at 120 rpm for 2.5, 5, 10 or 15 minutes, at 0 °C. The immobilized enzymes were confined by using an external miniature Nd2Fe14B magnet, and washed 3 times with 0.1 mL of Tris–HCl buffer solution (pH 7.5, 0.1 mol L−1). The best immobilization conditions were obtained by using 1.2 mg of MagNP@SiO2–APTS and 10 µL of a glutaraldehyde solution and stirring time of 5 minutes, which afforded the system MagNP@SiO2–APTS/GDH (amount of immobilized GDH: 0.05 mg/1.2 mg MagNP@SiO2–APTS).General procedure of GDH immobilization on MagNP–APTS/glyoxyl-agarose
MagNP–APTS (1, 2, 4, 8 and 12 mg) suspended in 100 µL of Tris–HCl buffer solution (pH 7.5, 0.1 mol L−1) was mixed with 100 µL of a GDH solution 0.523 mg mL−1 (2 U). Then a 150 mg mL−1 suspension of glyoxyl-agarose (10, 25, 50, 125 and 150 µL) was added to the enzyme–nanoparticle mixture and the resulting mixture was stirred at 120 rpm at 0 °C for 2, 5, 10 or 20 minutes. The immobilized enzymes were confined by using an external miniature Nd2Fe14B magnet, and washed 3 times with 0.1 mL of Tris–HCl buffer solution (pH 7.5, 0.1 mol L−1). The best immobilization conditions were obtained by using 12 mg of MagNP–APTS, 50 µL of glyoxyl-agarose and stirring time of 5 minutes. The MagNP–APTS/glyoxyl-agarose/GDH material exhibited 0.05 mg of immobilized GDH for 12 mg of MagNP–APTS.General procedure of GDH immobilization on MagNP@CMC
MagNP@CMC (10, 20, 30, 40 and 50 µL) from a 200 mg mL−1 stock suspension was mixed with a certain volume (50, 100, 150, 200 and 150 µL) of GDH solution (0.523 mg mL−1, or 2 U in 0.1 M Tris–HCl buffer pH 7.5). The resulting mixture was stirred at 120 rpm for 2.5, 5, 10 or 20 minutes, at °C. The immobilized enzymes were confined by using an external miniature Nd2Fe14B magnet, and washed 3 times with 0.1 mL of Tris–HCl buffer solution (pH 7.5, 0.1 M). The best immobilization conditions were obtained by using 30 µL of MagNP@CMC suspension, 100 µL of the GDH solution and stirring time of 5 minutes. The MagNP@CMC/GDH material exhibited 0.05 mg of immobilized GDH for 2 mg of MagNP@CMC.Enzyme activity measurements
The as-prepared magnetic GDH were added to an optical quartz cuvette together with 20 µL of a 0.1 mol L−1 glutamate solution, 50 µL of a 2.4 mmol L−1 NAD+ solution and 900 µL of buffer solution. The activity was measured by monitoring the absorbance increase at 340 nm associated with the NADH formation at 25 °C. A fixed time of 10 minutes was maintained for all the reactions and the yields between each system were compared.For the determination of the optimum pH, the experiments were performed at 25 °C and for the optimum temperature, the experiments were performed at pH 7.5 (Tris–HCl buffer solution 0.1 mol L−1).
Thermodynamic measurements
Determination of S0.5 was performed at 25 °C, pH 7.5 with 0.1 mol L−1 Tris–HCl buffer solution, 50 µL of NAD+ (stock solution 0.9 mmol L−1) and glutamate concentration from 0 to 5 mM. Measurements of ΔH≠ and ΔS≠ were performed at 20, 21, 22, 23 and 25 °C and pH 7.5.Trypsin (BRP, European Pharmacopoeia Reference Standard) digestion
The enzyme (free or immobilized) was washed three times with a 0.1 mol L−1 phosphate buffer solution with pH 7 and 0.1 mol L−1 EDTA. From this content, three aliquots were separated:(a) 10 µL of the GDH solution/suspension together with 5 µL of trypsin (1 mg/100 µL).
(b) 10 µL of the GDH solution/suspension together with 0.1 mL of ADP (0.1 mol L−1) and 5 µL of trypsin (1 mg/100 µL).
(c) 10 µL of the GDH solution/suspension together with 0.1 mL of GTP (0.1 mol L−1) and 5 µL of trypsin (1 mg/100 µL).
The tubes were incubated at 25 °C for 12 h. The digestion content was analyzed by MALDI-TOF.
Physical measurements
UV-visible (UV-vis) spectra were recorded on a Hewlett Packard 8453-A diode-array spectrophotometer.For the atomic force microscopy (AFM) measurements the samples were prepared by depositing 5 µL of the nanoparticle solution over mica (Ted Pella Inc.), and allowing it to dry in a clean laminar flow chamber. The AFM images were collected on a PicoSPM I microscope (Molecular Imaging, MI) with PicoScan 2100 (MI) controller coupled with MACMode (MI) unit for intermittent contact AFM, MACMode SFM, and magnetic force microscopy (MFM). Data acquisition was obtained using a PicoScan (MI) device with the scan rate between 0.5 and 1.0 Hz operating from 256 to 512 points per line. For the AFM and MACMode SFM measurements, silicon tips with high aspect ratio from Nanosensors and Agilent (Type II MACLevers, k ∼ 2.8 N m−1; f ∼ 60 kHz) were employed. The MFM images were obtained using silicon tips with magnetic coating of PPP-MFMR model (Nanosensors, k ∼ 2.8 N m−1; f ∼ 60 kHz), and operating with interleave mode enabled with the lift mode active from 25 nm to 100 nm. The magnetic domains were detected by using the phase contrast imaging.
Confocal Raman spectroscopy measurements were recorded on a WITec Alpha 300R microscope equipped with a Nd:YAG laser (λ = 532 nm) and a Nikon objective (20× NA = 0.8).
Results
The supporting materials employed for glutamate dehydrogenase immobilization were composed of magnetite particles covered with: (A) aminopropyl-triethoxysilane (APTS), (B) silica and APTS, (C) glyoxyl-agarose and (D) glyoxyl-β-cellulose, as shown in Fig. 1. Essentially, two groups were derived from these supports: (A), (B) involving the reaction of the amine groups with glutaraldehyde as cross-linking agent, and (C), (D) involving the direct reaction with the available aldehyde groups. In the first group (B) possesses a silica shell coating the magnetic nanoparticles and preventing the iron exposure at the surface, while (A) does not have this feature.37 In the other group, support (C) is made of MagNP–APTS and glyoxyl-agarose, allowing several magnetic nanoparticles to attach to glyoxyl-agarose surface, while support (D) has glyoxyl-β-cellulose coating the magnetic nanoparticles. The reason of using a mixture of MagNP–APTS and glyoxyl-agarose in case (C) was related to the size of cross-linking agent used for the immobilization. Since glutaraldehyde is small, it permits a direct interaction between enzyme and nanoparticle, while a larger cross-linker should prevent it.A complete optimization of the immobilization protocols was performed, changing the amount of magnetic nanoparticles, cross-linking agent and reaction time (ESI†). The best protocols were chosen on the basis of the highest catalytic conversions for each methodology and were characterized according to their thermodynamic properties, pH-dependence, temperature-dependence, atomic force microscopy, recycling, allosteric response, Raman spectroscopy and tryptic digestion behavior. Before enrolling in the characterization, it should be noted that in any immobilization procedure, new interactions between enzyme and the components of the support can result from the immobilization, influencing its final conformation.43,44 This statement was confirmed by the thermodynamic properties shown in Table 1, in which, a significant decrease of ΔS≠ and ΔH≠ (calculated from the Eyring equation45) is observed upon immobilization, in relation to free GDH10,11 (Table 1, entry 1). The diminishment of ΔS≠ can result from the protein–solvent interactions,46,47 e.g. from water molecules structuring around exposed hydrophobic amino acid residues during catalysis48 or either from an internal structure modification of the protein,49 generating a rigid structure.50 It is difficult to describe the exact mechanism responsible for lowering the ΔS≠ in the immobilized GDH, but since immobilization fix the enzyme through several points available on a support, it is plausible to infer that an overall stiffening is occurring. On the other hand, ΔH≠ also decreases for all immobilized systems, as can be seen in Table 1, indicating a decrease in the temperature dependence of the enzyme in catalysis,51 as observed in Fig. 3.
| Entry | System | S 0.5 (µmol L−1) | ΔH≠a (kcal mol−1) | ΔS≠a (cal mol−1) | n | K cat b (s−1) |
|---|---|---|---|---|---|---|
| a Per active site. b Measured at 37 °C and pH 6.8. | ||||||
| 1 | Free GDH | 190 | 34 | 110 | 2.1 | 0.12 |
| 2 | MagNP–APTS/GDH | 350 | 13 | 38 | 3.9 | 0.27 |
| 3 | MagNP@SiO2–APTS/GDH | 75 | 16 | 50 | 3.5 | 0.19 |
| 4 | MagNP–glyoxyl agarose/GDH | 75 | 10 | 28 | 1.6 | 0.89 |
| 5 | MagNP@CMC/GDH | 15 | 8 | 25 | 1.2 | 1.21 |
Likewise, distinct substrate affinity (S0.5) is obtained after the immobilization procedures. For instance, by comparing the free and immobilized GDH, there is a noticeable increase in glutamate affinity for MagNP@SiO2–APTS/GDH, MagNP–APTS/glyoxyl-agarose/GDH and MagNP@CMC/GDH in relation to free GDH as evidenced by a decrease in S0.5 values in Table 1, whereas MagNP–APTS/GDH exhibits lower substrate affinity than free GDH (Table 1, entry 2).
The multimeric enzyme system of GDH also includes subunits cooperativity and the extent of this cooperativity was compared using Hill coefficient (n, Table 1).52 Essentially, what is measured is an estimative of the affinity for other ligand molecules before and after immobilization, with n > 1 indicating a positive cooperativity, while n = 1 indicates no cooperativity.53 Under the conditions of this work, free GDH presented n of 2.1, although after its immobilization on MagNP–APTS and MagNP@SiO2–APTS the n value increased, indicating an enhanced positive cooperativity. However, in the case of MagNP@CMC and MagNP–APTS/glyoxyl-agarose, the enzyme modification seems to produce systems with almost no cooperativity, turning the n value very close to 1. This can be associated with the multi-point binding, promoted by the polymeric coating on the magnetic nanoparticles, preventing the enzyme dissociation and increasing the stability as observed by Garcia-Galan et al.12a These features are also connected to the system thermal stability (Fig. 2), since the systems with smaller n presented higher thermal stability than the free enzyme and glutaraldehyde immobilized GDH (MagNP–APTS/GDH and MagNP@SiO2–APTS/GDH) cases.
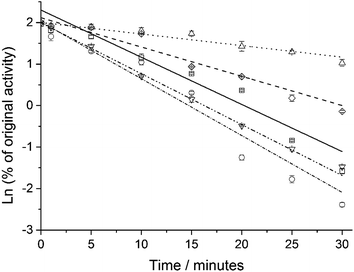 | ||
| Fig. 2 Enzyme inactivation at 50 °C and pH 7.5. Free GDH (solid line, ○), MagNP–APTS/GDH (□), MagNP@SiO2–APTS/GDH (▽), MagNP–APTS/glyoxyl-agarose/GDH (△) and MagNP@CMC/GDH (◊). | ||
In agreement with the ΔH≠ values (Table 1), temperature-dependence catalysis for the immobilized systems presented a more constant dependence of pH vs. activity, as can be seen in Fig. 3. Free GDH (Fig. 3, ▽) reaches 100% of glutamate conversion at 32 °C, while MagNP–APTS/GDH (Fig. 3, □) has its highest conversion (21%) at 25 °C, which is also the optimum temperature for MagNP@SiO2–APTS/GDH (Fig. 3, △), obtaining 30% of glutamate conversion. Immobilization of GDH through glyoxyl-agarose (Fig. 3, ○) presents a maximum conversion of 40% at 32 °C, while MagNP@CMC/GDH (Fig. 3, ◇) has two apparent maximums, one at 25 °C and the other at 47 °C with conversions around 70%, which can be associated with a broad range of working temperature.
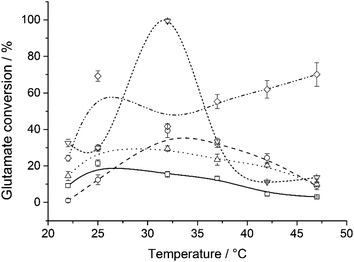 | ||
| Fig. 3 Influence of temperature at pH 7.5 on catalysis for free GDH (▽), MagNP–APTS/GDH (□), MagNP@SiO2–APTS/GDH (△), MagNP–APTS/glyoxyl-agarose/GDH (○) and MagNP@CMC/GDH (◇). | ||
Another feature displaying differences between the immobilized systems and free GDH was the catalytic behavior at different pHs, Fig. 4. An optimum pH at 8.8 was found for free GDH, with glutamate conversion around 30%, while MagNP@SiO2–APTS/GDH and MagNP–APTS/glyoxyl-agarose/GDH had their optimum pHs at 7.5, with conversions of 25% and 12%, respectively. This decrease in optimum pH was also observed by Petach et al.,54 who reasoned that the NH4+ ion does not diffuse away from the bond enzyme, therefore, the local environment around the immobilized enzyme seems more basic. A reduction of 0.8 unit in the optimum pH is observed for MagNP@CMC/GDH (Fig. 4, ◇) and surprisingly presents conversions around 70%, a value 2.5 times higher than free GDH. Remarkably, MagNP–APTS/GDH (Fig. 4, □) presents an increase in its catalytic activity with the increase of pH, reaching 18% of conversion at pH 9.8. An increase in the optimum pH was also observed by Barbotin et al.,26a who also assumed an increase of pH in the NH4+/NH3 equilibrium.
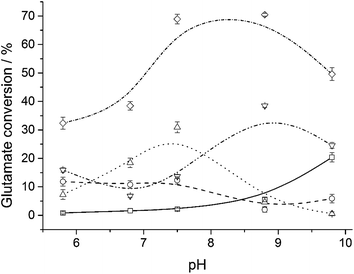 | ||
| Fig. 4 Influence of pH on catalysis at 25 °C for free GDH (▽), MagNP–APTS/GDH (□), MagNP@SiO2–APTS/GDH (△), MagNP–APTS/glyoxyl-agarose/GDH (○) and MagNP@CMC/GDH (◇). | ||
The greatest advantage of immobilized enzymes in contrast to free proteins, is their recyclability. Therefore, a study on the reuse of supported GDH was evaluated at the optimum pH and temperature for each system. MagNP–APTS/GDH and MagNP@SiO2–APTS/GDH were active only during the first reaction cycle as can be seen in Fig. 5, while MagNP@CMC/GDH although exhibiting the best performance in the first reaction cycle, showed a reduced activity after the second cycle and kept an average of 30% conversion up to the 10th cycle. The most stable system among the immobilization methods was MagNP–APTS/glyoxyl-agarose/GDH, demonstrating a nearly constant glutamate conversion activity around 40%, during the 10 monitored cycles.
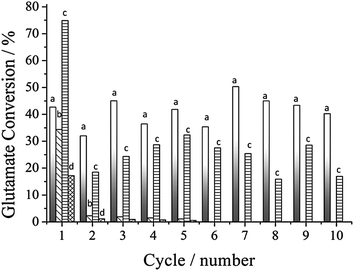 | ||
| Fig. 5 Recyclability of the immobilized systems: MagNP–APTS/glyoxyl-agarose/GDH (a); MagNP@SiO2–APTS/GDH (b); MagNP@CMC/GDH (c) and MagNP–APTS/GDH (d). | ||
For a deeper investigation on the immobilized GDH, topology and phase contrast images were obtained via Atomic Force Microscopy (AFM). In all the systems, after the drying process, a subsequent aggregation was observed with the application on mica, even under highly diluted conditions. This aggregation lead to the formation of clusters of variable sizes as shown in the typical topographic and phase contrast images in Fig. 6. Better information can be taken from the phase contrast images, since mica and the magnetic nanoparticles are hard and anything displaying an opposite contrast can be considered as soft,55 as for example, the enzyme and agarose/cellulose. Therefore, in the phase contrast image for MagNP–APTS/GDH, MagNP–APTS/glyoxyl-agarose/GDH and MagNP@CMC/GDH (Fig. 6A, C and D-II), apparently, magnetic nanoparticles are anchored on organic aggregates. This situation is likely to be reversed in MagNP@SiO2–APTS/GDH system, with the inclusion of magnetic particles in the aggregates, since almost no contrast is observed in Fig. 6B-II.
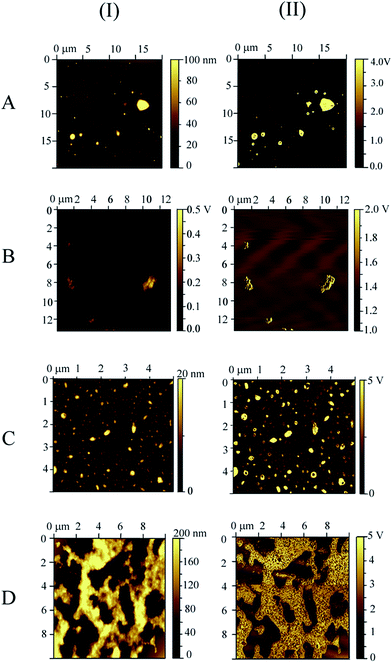 | ||
| Fig. 6 Atomic force microscopy images: (I) topography, (II) phase contrast for: (A) MagNP–APTS/GDH, (B) MagNP@SiO2–APTS/GDH, (C) MagNP–APTS/glyoxyl-agarose/GDH and (D) MagNP@CMC/GDH. | ||
Collectively, all these data indicate that conformational changes took place during the immobilization processes. Suggestion of 3D conformational changes of GDH are explicitly in MagNP–APTS/GDH as the less stable system, in MagNP@CMC/GDH as the most active system and in MagNP–APTS/glyoxyl-agarose/GDH as the most recyclable system. Therefore, other information are needed for a more comprehensive view of these systems.
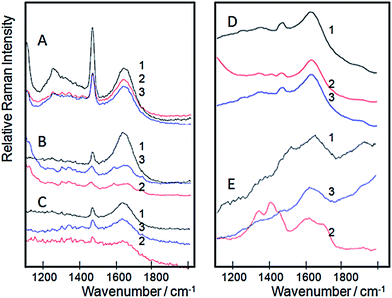
In this regard, Raman spectroscopy is one of the most important tools for structural characterization of proteins.56 Changes in the secondary and/or tertiary structure of proteins during a biochemical transformation57 can be detected by the identification of specific bands associated with α-helix and β-sheet elements.58 However, typically, the interpretation goes in the direction of the shapes of amide I and amide III bands.59 In our case, free and immobilized GDH were submitted to a confocal Raman spectroscopy monitoring before and after glutamate and NAD+ addition (Fig. 7). According to such studies, it was evident that free GDH (Fig. 7A) doesn't present spectroscopic changes with the addition of glutamate (Fig. 7A, red line) and NAD+ (Fig. 7A, blue line), keeping the amide I band centered at 1634 cm−1. The systems that resemble free GDH in this behavior are MagNP@SiO2–APTS/GDH (Fig. 7C) and MagNP–APTS/glyoxyl-agarose/GDH (Fig. 7D) with the amide I band at 1634 and 1629 cm−1, respectively. Also, MagNP@SiO2–APTS/GDH and MagNP–APTS/glyoxyl-agarose/GDH didn't present spectroscopic changes upon the substrates addition.
The initial spectrum of MagNP–APTS/GDH (Fig. 7B) has its amide I band maximum at 1633 cm−1 (black line, Fig. 7B). When glutamate is added to this enzyme its amide I band maximum is blue shifted to 1662 cm−1 (red line, Fig. 7B), with the rise of a small band at 1733 cm−1, which indicates the presence of hydrophobic groups around a carboxyl that changes its position upon glutamate binding.60 With the addition of NAD+ (blue line, Fig. 7B) amide I band red shifts to 1650 cm−1.
The most distinct system is MagNP@CMC/GDH (Fig. 7E) with an amide I band at 1660 cm−1 (black line, Fig. 7E), corresponding to a decrease of intermolecular β structures, however, with the addition of glutamate these structures seem to have an increase in the contribution of intermolecular β structures, presenting a maximum at 1623 cm−1.61 When NAD+ is added, a band at 1733 cm−1 arises, indicating that the carbonyl of an amino acid from the NAD+ binding site is close to a hydrophobic site.60
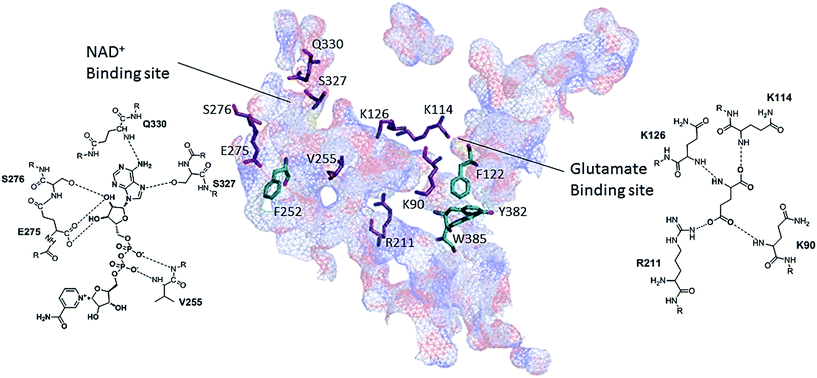
The emergence of the band at 1733 cm−1 can be further inspected, since it indicates an approximation between hydrophobic and carboxylic groups. Therefore, an examination of the 3D structure of bovine liver GDH structure (PDB code, 3MW9) with PyMol program, revealed the residues K90, K126, K114 and R211 as the amino acids responsible for glutamate binding23 and residues V255, E275, S276, Q330 and S327 responsible for NAD+ binding. Close to the carbonyl of E275 in NAD+ binding site there is a phenylalanine residue (F252), as can be seen in Fig. 7, which could be the source of hydrophobic moiety in MagNP@CMC/GDH after NAD+ addition. The system MagNP–APTS/GDH has a similar band which only arises after glutamate addition. In the last case, carbonylic groups from R211 and K90 can approximate to Y382 and F122, respectively, after inclusion of glutamate in its binding site, due to their proximity (Fig. 8) and possible change the active site conformation.
More importantly, unfolding can be observed in confocal Raman spectroscopy, by the sharpening of bands present in amide III region (1230–1300 cm−1).58,62 The extended amide III region is meaningful since the coupling of N–H and Cα–H deformations is very sensitive to geometry and generates informative band structures.63 In our experiments, GDH uncoiling is highly perceptible after glutamate addition in MagNP@CMC/GDH system (Fig. 7E2), with a noticeable increase of the bands in amide III region. This feature can be observed at a smaller extent in MagNP–APTS/glyoxyl-agarose/GDH, while the other systems have inconclusive amide III region. In MagNP@CMC/GDH system, a refolding is also readily observed after NAD+ addition, with the restitution of a similar spectra of the one prior to the substrates addition (Fig. 7E3).
Allegedly, it is expected to observe different allosteric responses in immobilized GDH, since GDH allosterism demand subunits to interact and sense each other,26c and upon immobilization, different 3D conformations were achieved, as were evidenced by Raman spectra (Fig. 6). The antenna from GDH is engaged in the allosterism,64 since the closure of one subunit requires a distortion at the back portion of the antenna.23,65 For example, the inhibition through GTP requires an antenna deformation while GDH closes.23,65 In our experiments, increasing concentrations of the allosteric regulators, ADP, GTP and ATP66 were evaluated by comparison of the initial reaction rates (Tables 2–4, respectively). Increasing concentrations of ADP, activated all the immobilized GDH but MagNP–APTS/GDH, which exhibited a slight inhibition, as evidenced by the decay of Vinitial (Table 2, entry 2).
| Entry | System | Initial reaction rates, Vinitial (mol L−1 s−1) | |||
|---|---|---|---|---|---|
| 0 µM ADP | 10 µM ADP | 50 µM ADP | 100 µM ADP | ||
| 1 | Free GDH | 1.5 × 10−7 (±9 × 10−8) | 2.2 × 10−7 (±6 × 10−8) | 2.2 × 10−7 (±2 × 10−8) | 2.1 × 10−7 (±3 × 10−8) |
| 2 | MagNP–APTS/GDH | 6.9 × 10−8 (±3 × 10−8) | 6.3 × 10−8 (±3 × 10−8) | 5.4 × 10−8 (±1 × 10−8) | 4.3 × 10−8 (±1 × 10−8) |
| 3 | MagNP@SiO2–APTS/GDH | 3.9 × 10−8 (±9 × 10−9) | 4.9 × 10−8 (±2 × 10−8) | 1.5 × 10−7 (±2 × 10−8) | 2.5 × 10−7 (±6 × 10−8) |
| 4 | MagNP–APTS/glyoxyl-agarose/GDH | 1.1 × 10−7 (±5 × 10−8) | 1.0 × 10−7 (±3 × 10−8) | 1.8 × 10−7 (±4 × 10−8) | 2.8 × 10−7 (±2 × 10−7) |
| 5 | MagNP@CMC/GDH | 5.7 × 10−8 (±3 × 10−8) | 3.3 × 10−7 (±2 × 10−7) | 6.8 × 10−7 (±1 × 10−7) | 8.2 × 10−7 (±1 × 10−7) |
| Entry | System | Initial reaction rates (Vinitial (mol L−1 s−1)) | |||
|---|---|---|---|---|---|
| 0 µM GTP | 10 µM GTP | 50 µM GTP | 100 µM GTP | ||
| 1 | Free GDH | 6.4 × 10−7 (±2 × 10−8) | 4.2 × 10−7 (±5 × 10−8) | 9.2 × 10−8 (±7 × 10−8) | 4.4 × 10−8 (±1 × 10−8) |
| 2 | MagNP–APTS/GDH | 8.3 × 10−9 (±1 × 10−9) | 3.8 × 10−8 (±2 × 10−9) | 5.34 × 10−8 (±1 × 10−9) | 8.7 × 10−8 (±7 × 10−9) |
| 3 | MagNP@SiO2–APTS/GDH | 1.8 × 10−7 (±4 × 10−8) | 1.3 × 10−7 (±2 × 10−8) | 1.6 × 10−7 (±2 × 10−8) | 1.2 × 10−7 (±4 × 10−8) |
| 4 | MagNP–APTS/glyoxyl-agarose/GDH | 6.0 × 10−8 (±2 × 10−8) | 5.26 × 10−8 (±9 × 10−9) | 9.2 × 10−8 (±2 × 10−8) | 7.3 × 10−8 (±6 × 10−9) |
| 5 | MagNP@CMC/GDH | 4.7 × 10−7 (±1 × 10−7) | 2.4 × 10−7 (±9 × 10−8) | 1.17 × 10−7 (±1 × 10−8) | 1.8 × 10−7 (±2 × 10−8) |
| Entry | System | Initial reaction rates (Vinitial (mol L−1 s−1)) | ||||
|---|---|---|---|---|---|---|
| 0 µM ATP | 100 µM ATP | 1 mM ATP | 10 mM ATP | 100 mM ATP | ||
| 1 | Free GDH | 9.8 × 10−8 (±2 × 10−8) | 1.1 × 10−7 (±1 × 10−8) | 1.6 × 10−7 (±2 × 10−8) | 9.0 × 10−8 (±2 × 10−8) | 1.6 × 10−8 (±2 × 10−8) |
| 2 | MagNP–APTS/GDH | 2.8 × 10−8 (±1 × 10−8) | 3.4 × 10−8 (±1 × 10−8) | 1.2 × 10−8 (±1 × 10−8) | 2.4 × 10−8 (±1 × 10−8) | 2.8 × 10−8 (±2 × 10−8) |
| 3 | MagNP@SiO2–APTS/GDH | 4.4 × 10−8 (±1 × 10−8) | 5.7 × 10−8 (±8 × 10−9) | 6.0 × 10−8 (±2 × 10−9) | 7.9 × 10−10 (±7 × 10−10) | 3.5 × 10−8 (±2 × 10−8) |
| 4 | MagNP–APTS/glyoxyl-agarose/GDH | 1.6 × 10−8 (±2 × 10−8) | 2.4 × 10−8 (±7 × 10−9) | 2.9 × 10−8 (±3 × 10−9) | 7.9 × 10−9 (±7 × 10−9) | 3.8 × 10−9 (±3 × 10−9) |
| 5 | MagNP@CMC/GDH | 3.16 × 10−8 (±1 × 10−8) | 1.0 × 10−7 (±9 × 10−8) | 1.3 × 10−7 (±8 × 10−8) | 2.4 × 10−9 (±1 × 10−9) | 3.1 × 10−9 (±2 × 10−8) |
Glutamate dehydrogenase natural inhibitor, GTP,67 only inhibited free GDH and MagNP@CMC/GDH (Table 3, entries 1 and 5). The systems MagNP–APTS/glyoxyl-agarose/GDH and MagNP@SiO2–APTS/GDH essentially weren't affected by increasing concentrations of GTP (Table 3, entries 3 and 4). Curiously, MagNP–APTS/GDH exhibited activation upon increasing GTP concentrations (Table 3, entry 2).
Due to the difference in allosteric response to ADP and GTP, and lack of information whether adenine or triphosphate moiety were the responsible moieties for the distinct responses, increasing concentrations of ATP were also evaluated. The ATP-based allosterism is more complex than ADP and GTP ones,68 since it inhibits glutamate dehydrogenase at low (1–100 µmol L−1) and high (1–10 mmol L−1) concentrations due to binding at the GTP site, but at intermediate concentrations (100 µmol L−1 to 1 mmol L−1) it becomes an activating agent as it binds to the ADP site.64,69 Consequently, it would be expected that systems with ADP-activation behavior, to be activated when intermediate concentrations (100 µmol L−1 to 1 mmol L−1) of ATP and vice versa. From the immobilized GDH systems, consistently, MagNP–APTS/GDH shows the most unusual behavior, being inhibited at 100 µM of ATP, while at higher concentrations of ATP, it did not exhibit allosterism (Table 4, entry 2). The other systems, in higher or minor magnitudes, exhibited a similar behavior to free GDH.
In support to the allosteric kinetics, enzyme digestion studies using trypsin were carried out and the fragments were analyzed by MALDI-TOF. A careful analysis of the fragments was performed in accordance to the amino acid sequence of GDH in a similar manner to Wacker et al.70 aiming the evaluation of sites blockage and any possible conformational change of GDH upon immobilization to magnetic supports. The enzymatic digestion was carried out in the presence and absence of the allosteric molecules.
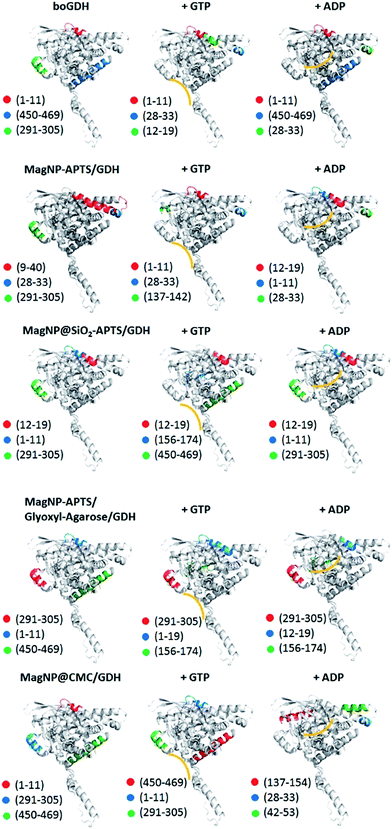
The most intense peaks observed in these digestion experiments are highlighted in Fig. 9. Essentially, 10 main fragments are obtained: 1–11, 450–469, 291–305, 12–19, 28–33, 9–40, 156–174, 137–154 and 476–501, which showed to be characteristic of the enzyme motion after addition of ADP or GTP. For instance, in the case of free GDH in the absence of ADP and GTP the main fragment is composed by the amino acids sequence 1–11 (m/z 1353), followed by the fragment 450–469 (m/z 2311) and 291–305 (m/z 1714). In the presence of ADP and GTP, the fragment 1–11 remains the major one, however in smaller extent. Exposure of the fragments 12–19 (m/z 100) and 28–33 (m/z 715) is detected when GDH is in presence of GTP. Both fragments (12–19 and 28–33) are directly opposite to the GTP site (Fig. 8, yellow line), indicating the enzyme conformation change upon GTP binding. On the other hand, ADP increases the preferential digestion of the fragment 450–469, which is opposite to its binding site.
It is expected therefore, to observe similar patterns of digestion when inhibition or activation is observed, independently of the regulator used. This assumption was proved to be correct, since MagNP–APTS/GDH when in presence of its inhibitor, ADP, presented the fragment 12–19, which is characteristic of GTP binding to free GDH. This fragment is also observed in MagNP@SiO2–APTS/GDH without allosteric regulators and in MagNP–APTS–glyoxyl-agarose/GDH in the presence of both GTP and ADP. Another important fragment to be highlighted is fragment 156–174, which is only observed for MagNP@SiO2–APTS/GDH and MagNP–APTS–glyoxyl-agarose/GDH when in presence of GTP, which suggests an antenna torsion and possible blockage of GTP site. The fragment 476–501, seen in MagNP@SiO2–APTS/GDH, is associated with an internal conformation of free GDH and occurs both in presence of ADP and GTP, thus, possibly, a partial protein unfolding occurs to expose internal structures of GDH after the addition of allosteric regulators for MagNP@SiO2–APTS/GDH.
MagNP@CMC/GDH is perhaps the most similar system to free GDH, since fragments 1–11, 291–305 and 450–469 are favored in the digestion experiments. The addition of ADP to this system, permits a preferential fragmentation of 137–154, which would indicate an exposure of the internal parts that connects the subunits.
Discussion
The obtained results corroborate to a conclusion that each magnetic support is influencing to a certain extent the final 3D structure from glutamate dehydrogenase. For instance, glutaraldehyde-based immobilization gives less stable enzymatic systems (Fig. 1, supports (A) and (B)), while larger aldehyde cross-coupling results in higher stability of GDH (Fig. 1, supports (C) and (D)), in agreement to other reported studies.71–74 This singularity is also pronounced in the allosteric-response experiments, with ADP activation of all immobilized systems but MagNP–APTS/GDH. In addition, GDH inhibition occurs in free GDH and MagNP@CMC/GDH, though in MagNP–APTS/GDH, GTP actually acts as an activator, and in the other systems, MagNP–APTS/glyoxyl-agarose/GDH and MagNP@SiO2–APTS/GDH, GTP doesn't have a marked influence in their catalysis. Even more surprisingly are the results with ATP, because MagNP–APTS/GDH is inhibited at activating concentrations, while the other systems had similar activity to free GDH.A classical argument on allosterism is around the influence of conformational changes between two well-defined structures, with protein flexibility playing an essential role.75 In glutamate dehydrogenase for example, it is said that GTP preferentially binds via the phosphates to Iso216 and Ser217 to the open form of GDH,76 while ADP binds via adenosine to Arg463 in an hydrophobic site of both forms of GDH, apo and holo.76 However, allosteric behavior is not exclusive of highly ordered enzymes, since even disordered proteins75 and proteins with no conformational change77 display this behavior, meaning that there is an entropic contribution to allosterism.75 In account to entropic contributions, are the works describing environmental conditions that change the cooperativity between subunits which transform an activator into a repressor, or vice versa. This theory is called intrinsic disorder mechanism.19a The first description of this theory was reported by Cooper and Dryden,77 who guide to the dynamics of proteins and consider that intramolecular motions (hinge-bending, breathing, local unfolding, among others) influence the overall allosterism. For instance, local unfolding has been shown to give thermodynamic stability due to the reduction of a protein's domain flexibility and result in allosteric response.78 In addition, it is known that increase in binding affinity can be due to local unfolding, enhancing protein's conformational flexibility.79,80
As a consequence from these relationships between intramolecular motions and allosterism we might develop a theory behind the anomaly in our GDH immobilized systems. In this study, the lowest S0.5 is observed in MagNP@CMC/GDH (Table 1), implying the highest substrate affinity among all GDH used and consequently, a higher protein flexibility. The elasticity of GDH on MagNP@CMC leads to a larger globally extended conformation since it should accompany the reduction in binding affinity. As could be observed in Raman spectroscopy (Fig. 7E), GDH supported on MagNP@CMC uncoiled after glutamate addition and this local unfolding was less evident in MagNP–APTS/glyoxyl-agarose/GDH and MagNP@SiO2–APTS/GDH (Fig. 7C and D), which present the second lowest S0.5 (Table 1). Without changing a protein's conformation, Petit et al.81 observed a reduced binding affinity when a α-helix was removed from a protein. In our case, it is believed that MagNP–APTS is bond on the GDH's antenna (allosteric response experiments: Tables 2–4 and trypsin digestion), and precisely MagNP–APTS/GDH, is the system with a diminished substrate binding affinity. Comparing MagNP–APTS/GDH behavior to Petit et al.81 study, the almost 2 fold increase in S0.5 for MagNP–APTS/GDH can be attributed to an entropic nature of glutamate binding which can be also related to its peculiar allosteric response.82
In another study, tethering a protein through a α-helix produced a more thermal-stable protein than when the attachment was through a flexible loop.83 From this finding it is expected that MagNP–APTS/GDH to be tethered through a loop since it is the least thermal stable system. A careful analysis of GDH antenna indicates that 27% of the more exposed loops are concentrated in the antenna region, therefore, due to the fact that MagNP–APTS/GDH is the least thermal stable system and has the most abnormal allosteric response, it would indicate a higher probability of GDH immobilization on MagNP–APTS via the loop of the antenna. Following this reasoning, MagNP@SiO2–APTS/GDH should also be immobilizing GDH through a loop, since it is the second least thermal-stable system. The tryptic digestion and allosteric-response experiments suggest that GTP binding site is possibly blocked, suggesting that MagNP@SiO2–APTS is most probably bond to GTP site. From the 3D structure from GDH, there is an indication that 36% of the exposed loops are next to GTP binding site, which corroborates to our assumption of immobilization next to the GTP binding site.
However, the conjecture of possible conformational states that are favorably immobilized should accompany the other behaviors of the immobilized systems, such as for example, the allosteric response experiments. To answer whether GDH allosterism is largely dependent on entropy issues more than on enthalpy ones, the relative stability of all the possible states should be addressed.75,84 Here, to determine the most probable states present in the immobilized systems, a comparison between fragments from free GDH and immobilized GDH was performed. After the determination of states with higher probability of occurring, the local folding/unfolding of these states was assumed according to Hilser et al.19a method (ESI†).
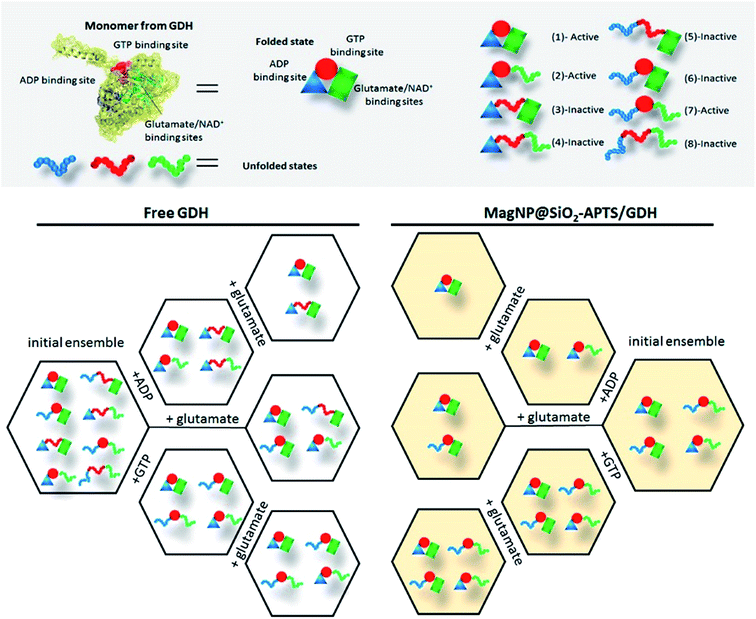
First of all, free GDH is assumed to bear 8 possible states for each monomer as shown in Fig. 10. Hence, the homohexameric structure would have 36 possible states, but for sake of simplification, we are assuming just one monomer. The next step is to determine whether a molecule is activating or inactivating, which would mean a positive or negative ΔGint (difference of the free energy of unfolding each domain and the energy of breaking the interactions between domains), for activator and inhibitor molecules, respectively. Therefore, upon ADP addition, the domain responsible for ADP binding will be preferentially stabilized and states 1, 2, 3 and 4 (Fig. 10) will possess a higher probability in the ensemble. Since ADP is an activator of GDH, ΔGint has a positive value, meaning that domains from ADP and glutamate binding are positively coupled. The positive coupling indicates that breaking the interaction between ADP and glutamate sites will be unfavorable, consequently, after addition of glutamate and NAD+, a stabilization of glutamate binding site occurs, increasing the probability of finding states 1 and 3 in the ensemble (as shown in Fig. 10). For GTP interaction, an opposite effect is observed, since ΔGint has a negative value (interaction between GTP and glutamate binding sites) and the unfolded states dominate the ensemble probabilities (states 1, 2, 6 and 7). Comparing the addition of allosteric regulators with an ensemble without regulators (preferential states 1, 2, 5 and 6) it is clear that ADP addition increases the probability of active GDH in solution, while GTP decreases this probability, demonstrating this effect in the total enzyme activity.
To illustrate this methodology in an immobilized GDH, we have chosen MagNP@SiO2–APTS/GDH system, since its visualization is straightforward. Fragmentation of this system indicate the existence of fragment 12–19 in the presence and absence of the allosteric regulators ADP and GTP. This fragment suggest the major probability of conformations where GTP site is folded, as shown in Fig. 10 (states 1, 2, 6 and 7). ADP addition increases the probability of states 1 and 2, while GTP addition has no influence on the ensemble probability as can be seen in Fig. 10. This result corroborate to the experimental data obtained, with GTP not acting as an inhibitor for MagNP@SiO2–APTS/GDH system.
The same reasoning can be used for the other 3 immobilized systems, as can be seen in the ESI.† Briefly, MagNP–APTS/GDH possibly presents as major conformations in the initial ensemble the unfolded glutamate and NAD+ binding site, which would also explain the higher S0.5 for this system. This statement can be reasoned by the trypsin digestion experiments and by Raman spectroscopy, in which, the later indicates a folding of the glutamate binding site after addition of glutamate (amide I band shift from 1633 to 1662 cm−1). The increase in optimum pH can be also reasoned by the conformational change, since pKa of K126 increases in 1 unit of pH (from 8.8 to 9.8), shifting to a pKa of an exposed lysine (≈10.3), which agrees with the higher percentage of unfolded states in the initial ensemble. More importantly, one can highlight the difference on immobilization and obtained dynamics for systems MagNP–APTS/GDH and MagNP@SiO2–APTS/GDH, since their difference is only on the iron exposure to the surface. MagNP–APTS/GDH have as an initial ensemble, a higher probability of unfolded states (high S0.5), which may be due to iron coordination and interaction to the enzyme residues, while MagNP@SiO2–APTS/GDH would have a silica shell preventing this interaction, but the immobilized preferential conformation using the silica-shell magnetite is in the inactive form, which explains the lower Kcat for MagNP@SiO2–APTS/GDH (0.19 s−1) than for MagNP–APTS/GDH (0.27 s−1).
The pKa of K126 in MagNP@SiO2–APTS/GDH and MagNP–APTS/glyoxyl-agarose/GDH is also modified (pKa ≈ 7.5). However, their Raman spectra is very similar to free GDH's one and the argument behind the different catalysis are based on the tryptic digestion and the initial ensemble states. Differently from MagNP@SiO2–APTS/GDH, system MagNP–APTS/glyoxyl-agarose/GDH exhibits as most probable states the completely folded one and the ADP unfolded one. The first state is active and the second one is inactive and since GDH has the glutamate-site most probably immobilized in the folded conformation, the higher recoverability of this system could be explained as the favoring of active and folded conformational states. Another system that presents a folded preference of glutamate binding site is MagNP@CMC/GDH, but in that case, 75% of the states are active, therefore, the higher activity (2.5 time higher than free GDH) exhibited for this system is explained as the immobilization of preferential active states.
As a conclusion of this theory, the immobilization of GDH on different magnetic supports resulted in distinct enzymatic activities majorly due to the stabilization of different dynamic-conformational states, which is a result of the system's entropy. The only system that actually behaves abnormally is MagNP–APTS/GDH, and this deviation is possibly due to a negative ΔGint energy between ADP and glutamate binding sites, while a positive ΔGint is observed between GTP and glutamate binding sites, resulting in ADP/ATP ([ATP] = 100 µmol L−1 to 1 mmol L−1) inhibition and GTP/ATP ([ATP] = 1–100 µmol L−1 and 1–10 mmol L−1) activation.
Conclusion
Upon immobilization on four different magnetic supports, glutamate dehydrogenase presented unusual catalytic behavior. The highly active MagNP@CMC/GDH was shown to be not as stable as MagNP–APTS/glyoxyl-agarose/GDH, while MagNP–APTS/GDH presented opposite allosteric responses. The intrinsic disorder mechanism was used to understand these behaviors, bringing explanations on the probability of each system to be active or inactive with ADP and GTP additions. MagNP@SiO2–APTS/GDH was majorly immobilized on its inactive conformation, therefore, clarifying the reason behind the null effect observed in MagNP@SiO2–APTS/GDH catalysis after GTP addition. On the other hand, the highest activity of system MagNP@CMC/GDH can be explained by the existence of active major conformations, than in the free enzyme. In conclusion, the intrinsic disorder mechanism proved to be an important tool for understanding immobilized enzymes and the influence of the support on the protein total ensemble.Acknowledgements
The support from FAPESP Grant 2008/10177-7 (Fundação de Amparo à Pesquisa do Estado de São Paulo) and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) are gratefully acknowledged.References
- C. Mateo, J. M. Palomo, G. Fernandez-Lorente, J. M. Guisan and R. Fernandez-Lafuente, Improvement of enzyme activity, stability and selectivity via immobilization techniques, Enzyme Microb. Technol., 2007, 40(6), 1451–1463 CrossRef CAS.
- R. A. Sheldon, Enzyme Immobilization: The Quest for Optimum Performance, Adv. Synth. Catal., 2007, 349, 1289 CrossRef CAS.
- (a) F. Secundo, Conformational changes of enzymes upon immobilisation, Chem. Soc. Rev., 2013, 46, 6250 RSC; (b) J. C. S. D. Santos, O. Barbosa, C. Ortiz, A. Berenguer-Murcia, R. C. Rodrigues and R. Fernandez-Lafuente, Importance of the support properties for immobilization or purification of enzymes, ChemCatChem, 2015, 7(16), 2413 CrossRef.
- C. G. C. M. Netto, H. E. Toma and L. H. Andrade, Superparamagnetic nanoparticles as versatile carriers and supporting materials for enzymes, J. Mol. Catal. B: Enzym., 2013, 85–86, 71–92 CrossRef CAS.
- B. M. Brena and F. Batista-Viera, Immobilization of Enzymes, in Immobilization of Enzymes and Cells, ed. Guisán J. M., Humana Press Inc., Totowa, NJ, 2nd edn, 2006 Search PubMed.
- B. Trewyn, Heterogeneous Catalysis for Today's Challenges: Synthesis, Characterization and Applications, The Royal Society of Chemistry, Cambridge, UK, 2015 Search PubMed.
- L. J. Kingsley and M. A. Lill, Substrate tunnels in enzymes: structure–function relationships and computational methodology, Proteins: Struct., Funct., Bioinf., 2015, 83(4), 599 CrossRef CAS PubMed.
- (a) R. C. Rodrigues, C. Ortiz, Á. Berenguer-Murcia, R. Torres and R. Fernández-Lafuente, Modifying enzyme activity and selectivity by immobilization, Chem. Soc. Rev., 2013, 42, 6290 RSC; (b) U. Hanefeld, L. Cao and E. Magner, Enzyme immobilization: fundamentals and application, Chem. Soc. Rev., 2013, 42(15), 6211 RSC; (c) D. S. Clark, Can immobilization be exploited to modify enzyme activity?, TIBTECH, 1994, vol. 12, p. 439 Search PubMed.
- R. K. Singh, M. K. Tiwari, R. Singh and J.-K. Lee, From Protein Engineering to Immobilization: Promising Strategies for the Upgrade of Industrial Enzymes, Int. J. Mol. Sci., 2013, 14, 1232 CrossRef CAS PubMed.
- R. Fernandez-Lafuente, Stabilization of multimeric enzymes: strategies to prevent subunit dissociation, Enzyme Microb. Technol., 2009, 45(6–7), 405 CrossRef CAS.
- (a) J. Pedroche, M. d. M. Yust, C. Mateo, R. Fernández-Lafuente, J. Girón-Calle, M. Alaiz, J. Vioque, J. M. Guisán and F. Millána, Effect of the support and experimental conditions in the intensity of the multipoint covalent attachment of proteins on glyoxyl-agarose supports: correlation between enzyme–support linkages and thermal stability, Enzyme Microb. Technol., 2007, 40(5), 1160 CrossRef CAS; (b) C. S. Bezerra, C. M. G. d. F. Lemos, M. d. Sousa and L. R. B. Gonçalves, Enzyme immobilization onto renewable polymeric matrixes: past, present, and future trends, J. Appl. Polym. Sci., 2015, 132(26), 42125 CrossRef.
- (a) C. Garcia-Galan, O. Barbosa and R. Fernandez-Lafuente, Stabilization of the hexameric glutamate dehydrogenase from Escherichia coli by cations and polyethyleneimine, Enzyme Microb. Technol., 2013, 52, 211–217 CrossRef CAS PubMed; (b) J. M. Bolivar, C. Mateo, V. Grazu, A. V. Carrascosa, B. C. Pessela and J. M. Guisan, Heterofunctional supports for the one-step purification, immobilization and stabilization of large multimeric enzymes: amino-glyoxyl versus amino-epoxy supports, Process Biochem., 2010, 45, 1692–1698 CrossRef CAS.
- S. S. Wong and L.-J. C. Wong, Chemical crosslinking and the stabilization of proteins and enzymes, Enzyme Microb. Technol., 1992, 14, 866 CrossRef CAS PubMed.
- (a) R. W. Lencki, J. Arul and R. J. Neufeld, Effect of Subunit Dissociation, Denaturation, Aggregation, Coagulation, and Decomposition on Enzyme Inactivation Kinetics: II. Biphasic and Grace Period Behavior, Biotechnol. Bioeng., 1992, 40, 1427 CrossRef CAS PubMed; (b) O. M. Poltorak, E. S. Chukhray and I. Y. Torshin, Dissociative thermal inactivation, stability, and activity of oligomeric enzymes, Biochemistry, 1998, 63(3), 303 CAS.
- F. Jia, B. Narasimhan and S. Mallapragada, Materials-based strategies for multi-enzyme immobilization and co-localization: a review, Biotechnol. Bioeng., 2014, 111(2), 209 CrossRef CAS PubMed.
- J. P. d. Costa, R. Oliveira-Silva, A. L. Daniel-da-Silva and R. Vitorino, Bionanoconjugation for Proteomics applications—An overview, Biotechnol. Adv., 2014, 32(5), 952 CrossRef PubMed.
- S. Ding, A. A. Cargill, I. L. Medintz and J. C. Claussen, Increasing the activity of immobilized enzymes with nanoparticle conjugation, Curr. Opin. Biotechnol., 2015, 34, 242 CrossRef CAS PubMed.
- J. M. Palomo and M. Filice, New emerging bio-catalysts design in biotransformations, Biotechnol. Adv., 2015, 33(5), 605 CrossRef CAS PubMed.
- (a) V. J. Hilser and E. B. Thompson, Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(20), 8311 CrossRef CAS PubMed; (b) J. Habchi, P. Tompa, S. Longhi and V. N. Uversky, Introducing Protein Intrinsic Disorder, Chem. Rev., 2014, 114, 6561 CrossRef CAS PubMed.
- (a) B. J. Reed, M. N. Locke and R. G. Gardner, A Conserved Deubiquitinating Enzyme Uses Intrinsically Disordered Regions to Scaffold Multiple Protein Interaction Sites, J. Biol. Chem., 2015, 290, 20601 CrossRef CAS PubMed; (b) J. Ramírez, R. Recht, S. Charbonnier, E. Ennifar, R. A. Atkinson, G. Travé, Y. Nominé and B. Kieffer, Disorder-To-Order Transition of MAGI-1 PDZ1 C-Terminal Extension upon Peptide Binding: Thermodynamic and Dynamic Insights, Biochemistry, 2015, 54(6), 1327 CrossRef PubMed; (c) V. N. Uversky, The multifaceted roles of intrinsic disorder in protein complexes, FEBS Lett., 2015, 589(19), 2498 CrossRef CAS PubMed.
- H. N. Motlagh and V. J. Hilser, Agonism/antagonism switching in allosteric ensembles, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(11), 4134 CrossRef CAS PubMed.
- M.-H. Seo, J. Park, E. Kim, S. Hohng and H.-S. Kim, Protein conformational dynamics dictate the binding affinity for a ligand, Nat. Commun., 2014, 5, 3724 CAS.
- P. E. Peterson and T. J. Smith, The structure of bovine glutamate dehydrogenase provides insights into the mechanism of allostery, Structure, 1999, 7, 769–782 CrossRef CAS PubMed.
- S. A. Wacker, M. J. Bradley, J. Marion and E. Bell, Ligand-induced changes in the conformational stability and flexibility of glutamate dehydrogenase and their role in catalysis and regulation, Protein Sci., 2010, 19(10), 1820 CrossRef CAS PubMed.
- J. B. Carrigan and P. C. Engel, The structural basis of proteolytic activation of bovine glutamate dehydrogenase, Protein Sci., 2008, 17, 1346 CrossRef CAS PubMed.
- (a) J. N. Barbotin and M. Breuil, Immobilization of Glutamate Dehydrogenase into proteic films: Stability and Kinetic Modulation effectors, Biochim. Biophys. Acta, 1978, 525, 18–27 CrossRef CAS PubMed; (b) J.-N. Barbotin and B. Thomasset, Immobilization of L-glutamate dehydrogenase into soluble cross-linked polymers, Biochim. Biophys. Acta, 1979, 570, 11–21 CrossRef CAS PubMed; (c) S. Bigdeli, A. H. Talasaz, P. Stahl, H. H. J. Persson, M. Ronaghi, R. W. Davis and M. Nemat-Gorgani, Conformational Flexibility of a Model Protein Upon Immobilization on Self-Assembled Monolayers, Biotechnol. Bioeng., 2008, 100(1), 19–27 CrossRef CAS PubMed.
- J. Cordek, X. Wang and W. Tan, Direct Immobilization of Glutamate Dehydrogenase on Optical Fiber Probes for Ultrasensitive Glutamate Detection, Anal. Chem., 1999, 71, 1529–1533 CrossRef CAS.
- R.-a. Doong and H.-m. Shih, Glutamate optical biosensor based on the immobilization of glutamate dehydrogenase in titanium dioxide sol–gel matrix, Biosens. Bioelectron., 2006, 22, 185–191 CrossRef CAS PubMed.
- L. Blasi, L. Longo, G. Vasapollo, R. Cingolani, R. Rinaldi, T. Rizzello, R. Acierno and M. Maffia, Characterization of glutamate dehydrogenase immobilization on silica surface by atomic force microscopy and kinetic analyses, Enzyme Microb. Technol., 2005, 36, 818–823 CrossRef CAS.
- L. Blasi, L. Longo, P. P. Pompa, L. Manna, G. Ciccarella, G. Vasapollo, R. Cingolani, R. Rinaldi, A. Rizzello, R. Acierno, C. Storelli and M. Maffia, Formation and characterization of glutamate dehydrogenase monolayers on silicon supports, Biosens. Bioelectron., 2005, 21, 30–40 CrossRef CAS PubMed.
- (a) C. D. Stalikas, M. I. Karaynnis and S. Tzouwara-Karayan, Immobilization of glutamate dehydrogenase on glass derivatives: a method for the assay of glutamates in real samples with simplex optimized automated FIA-system, Talanta, 1994, 41(9), 1561–1567 CrossRef CAS PubMed; (b) H. H. Petach and J. Driscoll, Transparent Chitosan Derivatives for the Immobilization of Glutamate Dehydrogenase, Biotechnol. Bioeng., 1994, 44, 1018–1022 CrossRef CAS PubMed; (c) M.-L. Hourdou and F. Besson, Enzymatic Activities and Infrared Studies of Glutamate Dehydrogenase Immobilized on Langmuir–Blodgett Films, Biotechnol. Tech., 1995, 9(9), 643–648 CrossRef CAS; (d) C. Jeffries, N. Pasco, K. Baronian and L. Gorton, Evaluation of a thermophile enzyme for a carbon paste amperometric biosensor: L-glutamate dehydrogenase, Biosens. Bioelectron., 1997, 12(3), 225–232 CrossRef CAS; (e) A. P. Girard-Egrot, R. M. Morélis and P. R. Coulet, Bioactive nanostructure with glutamate dehydrogenase associated with LB films: protecting role of the enzyme molecules on the structural lipidic organization, Thin Solid Films, 1997, 292, 282–289 CrossRef CAS; (f) A. Gambhir, M. Gerard, A. K. Mulchandani and B. D. Malhotra, Coimmobilization of Urease and Glutamate Dehydrogenase in Electrochemically Prepared Polypyrrole–Polyvinyl Sulfonate Films, Appl. Biochem. Biotechnol., 2001, 96, 249 CrossRef CAS PubMed; (g) P. P. Pompa, L. Blasi, L. Longo, R. Cingolani, G. Ciccarella, G. Vasapollo and R. Rinaldi, Optical characterization of glutamate dehydrogenase monolayers chemisorbed on SiO2, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 2003, 67, 041902 CrossRef CAS PubMed; (h) L. Longo, G. Vasapollo, M. R. Guascito and C. Malitesta, New insights from X-ray photoelectron spectroscopy into the chemistry of covalent enzyme immobilization, with glutamate dehydrogenase (GDH) on silicon dioxide as an example, Anal. Bioanal. Chem., 2006, 385, 146–152 CrossRef CAS PubMed; (i) J. M. Bolivar, F. Cava, C. Mateo, J. Rocha-Martín, J. M. Guisán, J. Berenguer and R. Fernandez-Lafuente, Immobilization–stabilization of a new recombinant glutamate dehydrogenase from Thermus thermophilus, Appl. Microbiol. Biotechnol., 2008, 80, 49–58 CrossRef CAS PubMed; (j) J. M. Bolivar, J. Rocha-Martin, C. Godoy, R. C. Rodrigues and J. M. Guisan, Complete reactivation of immobilized derivatives of a trimeric glutamate dehydrogenase from Thermus thermophilus, Process Biochemistry, 2010, 45, 107–113 CrossRef CAS.
- (a) B. El-Zahab, D. Donnelly and P. Wang, Particle-tethered NADH for production of methanol from CO2 catalysed by coimmobilized enzymes, Biotechnol. Bioeng., 2008, 99(3), 508 CrossRef CAS PubMed; (b) M. Zheng, S. Zhang, G. Ma and P. Wang, Effect of molecular mobility on coupled enzymatic reactions involving cofactor regeneration using nanoparticle-attached enzymes, J. Biotechnol., 2011, 154, 274–280 CrossRef CAS PubMed; (c) M. Zheng, Z. Su, X. Ji, G. Ma, P. Wang and S. Zhan, Magnetic field intensified bi-enzyme system with in situ cofactor regeneration supported by magnetic nanoparticles, J. Biotechnol., 2013, 168(2), 212–217 CrossRef CAS PubMed.
- J. M. Bolivar, C. Mateo, J. Rocha-Martin, F. Cava, J. Berenguer, R. Fernandez-Lafuente and J. M. Guisan, The adsorption of multimeric enzymes on very lowly activated supports involves more enzyme subunits: stabilization of a glutamate dehydrogenase from Thermus thermophilus by immobilization on heterofunctional supports, Enzyme Microb. Technol., 2009, 44, 139–144 CrossRef CAS.
- E. Reisler, J. Pouyet and H. Eisenberg, Molecular Weights, Association, and Frictional Resistance of Bovine Liver Glutamate Dehydrogenase at Low Concentrations. Equilibrium and Velocity Sedimentation, Light-Scattering Studies, and Settling Experiments with Macroscopic Models of the Enzyme Oligomer, Biochemistry, 1970, 9(15), 3095 CrossRef CAS PubMed.
- P. Zucca and E. Sanjust, Inorganic Materials as Supports for Covalent Enzyme Immobilization: Methods and Mechanisms, Molecules, 2014, 19, 14139 CrossRef PubMed.
- (a) I. Migneault, C. Dartiguenave, M. J. Bertrand and K. C. Waldron, Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking, BioTechniques, 2004, 37(5), 790 CAS; (b) O. Barbosa, C. Ortiz, Á. Berenguer-Murcia, R. Torres, R. C. Rodrigues and R. Fernandez-Lafuente, Glutaraldehyde in bio-catalysts design: a useful crosslinker and a versatile tool in enzyme immobilization, RSC Adv., 2014, 4, 1583 RSC.
- C. G. C. M. Netto, E. H. Nakamatsu, L. E. S. Netto, M. A. Novak, A. Zuin, M. Nakamura, K. Araki and H. E. Toma, Catalytic properties of thioredoxin immobilized on superparamagnetic nanoparticles, J. Inorg. Biochem., 2011, 105(5), 738–744 CrossRef CAS PubMed.
- G. S. Chauhan, Evaluation of nanogels as supports for enzyme immobilization, Polym. Int., 2014, 63(11), 1889 CrossRef CAS.
- (a) B. K. Hamilton, L. J. Stockmeyer and C. K. Colton, Comments on Diffusive and Electrostatic Effects with Immobilized Enzymes, J. Theor. Biol., 1973, 41, 547 CrossRef CAS PubMed; (b) W. Tischer and F. Wedekind, Immobilized Enzymes: Methods and Applications, in Biocatalysis – From Discovery to Application, ed. Fessner W.-D., ArchelasA., DemirjianD. C., FurstossR., GrienglH., JaegerK.-E., Morís-VarasE., ÖhrleinR., ReetzM. T., ReymondJ.-L., SchmidtM., ServiS., ShahP. C., TischerW. and hideF. W., 1999, vol. 200, pp. 95–126 Search PubMed.
- J. Xu, J. Sun, Y. Wang, J. Sheng, F. Wang and M. Sun, Application of Iron Magnetic Nanoparticles in Protein Immobilization, Molecules, 2014, 19, 11465 CrossRef PubMed.
- J. Govan and Y. K. Gun'ko, Recent Advances in the Application of Magnetic Nanoparticles as a Support for Homogeneous Catalysts, Nanomaterials, 2014, 4(2), 222 CrossRef CAS.
- C. G. C. Marques Netto, L. H. Andrade and H. E. Toma, Enantioselective transesterification catalysis by Candida antarctica lipase, Tetrahedron: Asymmetry, 2009, 20, 2299–2304 CrossRef.
- C. Garcia-Galan, Á. Berenguer-Murcia, R. Fernandez-Lafuente and R. C. Rodrigues, Potential of different enzyme immobilization strategies to improve enzyme performance, Adv. Synth. Catal., 2011, 353, 2885 CrossRef CAS.
- B. J. Johnson, W. R. Algar, A. P. Malanoski, M. G. Ancona and I. L. Medintz, Understanding enzymatic acceleration at nanoparticle interfaces: approaches and challenges, Nano Today, 2014, 9, 102 CrossRef CAS.
- R. G. Mortimer and H. Eyring, Elementary transition state theory of the Soret and Dufour effects, Proc. Natl. Acad. Sci. U. S. A., 1980, 77(4), 1728–1731 CrossRef CAS.
- (a) S. N. Timasheff, Protein–Solvent Interactions and Protein Conformation, Acc. Chem. Res., 1970, 3(2), 62 CrossRef CAS; (b) F. Franks, Solvation interactions of proteins in solution, Philos. Trans. R. Soc. London, Ser. B, 1977, 278, 89 CrossRef CAS PubMed.
- P. Sashi, U. M. Yasin, H. Balasubramanian, M. U. Sree, D. Ramakrishna and a. A. K. Bhuyan, Preferential Water Exclusion in Protein Unfolding, J. Phys. Chem. B, 2014, 118, 717 CrossRef CAS PubMed.
- E. v. Dijk, A. Hoogeveen and S. Abeln, The Hydrophobic Temperature Dependence of Amino Acids Directly Calculated from Protein Structures, PLoS Comput. Biol., 2015, 11(5), e1004277 Search PubMed.
- G. Castronuovo, V. Elia, C. Postiglione and F. Velleca, Interactions of amino acids in concentrated aqueous solutions of urea or ethanol. Implications for the mechanism of protein denaturation, Thermochim. Acta, 1999, 339, 11 CrossRef CAS.
- P. S. Low, J. L. Bada and G. N. Somero, Temperature Adaptation of Enzymes: Roles of the Free Energy, the Enthalpy and the Entropy of Activation, Proc. Natl. Acad. Sci. U. S. A., 1973, 70(2), 430–432 CrossRef CAS.
- K. S. Siddiqui, T. Thomas and V. N. Uversky, Protein Adaptation in Extremophiles, Nova Science Publishers, New York, USA, 2008 Search PubMed.
- J. N. Weiss, The Hill equation revisited: uses and misuses, FASEB J., 1997, 11(11), 835 CAS.
- V. I. Krupyanko, An additional Possibility of Using the Hill Coefficients, Eur. Chem. Bull., 2015, 4(7), 340 Search PubMed.
- H. H. Petach and J. Driscoll, Transparent Chitosan Derivatives for the Immobilization of Glutamate Dehydrogenase, Biotechnol. Bioeng., 1994, 44, 1018–1022 CrossRef CAS PubMed.
- J. Tamayo and R. García, Deformation, Contact Time, and Phase Contrast in Tapping Mode Scanning Force Microscopy, Langmuir, 1996, 12, 4430 CrossRef CAS.
- A. Y. Chikisev, C. Otto, N. N. Brandt, V. V. Molodozhenya, I. K. Sakodynskaya, J. Greve and N. I. Koroteev, Function-related conformational changes of protein molecules revealed by Raman spectroscopy, in Spectroscopy of Biological Molecules: New Directions, ed. GreveJ., PuppelsG. J. and OttoC., Kluver Academic Publishers, Netherlands, 1999 Search PubMed.
- (a) S. E. Babbitt, B. S. Francisco, D. L. Mendez, G. S. Lukat-Rodgers, K. R. Rodgers, E. C. Bretsnyder and R. G. Kranz, Mechanisms of Mitochondrial Holocytochrome c Synthase and the Key Roles Played by Cysteines and Histidine of the Heme Attachment Site, Cys-XX-Cys-His, J. Biol. Chem., 2014, 289(42), 28795–28807 CrossRef CAS PubMed; (b) J. Zhao, J. Moretto, P. Le and S. Franzen, Measurement of Internal Substrate Binding in Dehaloperoxidase–Hemoglobin by Competition with the Heme–Fluoride Binding Equilibrium, J. Phys. Chem. B, 2015, 119, 2827–2838 CrossRef CAS PubMed.
- N. C. Maiti, M. M. Apetri, M. G. Zagorski, P. R. Carey and V. E. Anderson, Raman Spectroscopic Characterization of Secondary Structure in Natively Unfolded Proteins: a-Synuclein, J. Am. Chem. Soc., 2004, 126, 2399–2408 CrossRef CAS PubMed.
- A. A. Bunaciu, H. Y. Aboul-Enein and V. D. Hoang, Raman Spectroscopy for Protein Analysis, Appl. Spectrosc. Rev., 2015, 50, 377 CrossRef CAS.
- P. R. Carey and A. C. Storer, Characterization of transient enzyme–substrate bonds by resonance Raman spectroscopy, Annu. Rev. Biophys. Bioeng., 1984, 13, 25–49 CrossRef CAS PubMed.
- W. Dzwolak and V. Smirnovas, A conformational a-helix to h-sheet transition accompanies racemic self-assembly of polylysine: an FT-IR spectroscopic study, Biophys. Chem., 2005, 115, 49 CrossRef CAS PubMed.
- Z. Chi and S. A. Ashe, UV Resonance Raman Determination of Protein Acid Denaturation: Selective Unfolding of Helical Segments of Horse Myoglobin, Biochemistry, 1998, 37, 2865–2872 CrossRef CAS PubMed.
- D. L. Barron, E. W. Blanch and L. Hecht, Advances in Protein Chemistry, in Unfolded Proteins, Academic Press, San Diego, California, 2002, vol. 62 Search PubMed.
- M. Li, C. Li, A. Allen, C. A. Stanley and T. J. Smith, The structure and allosteric regulation of glutamate dehydrogenase, Neurochem. Int., 2011, 59(4), 445 CrossRef CAS PubMed.
- H. F. Fisher and J. Tally, Isoergonic Cooperativity in Glutamate Dehydrogenase Complexes: A New Form of Allostery, Biochemistry, 1997, 36, 10807–10810 CrossRef CAS PubMed.
- E. L. Smith, M. Landon, D. Piszkiewicz, W. J. Brattin, T. J. Langley and M. D. Melamed, Bovine Liver Glutamate Dehydrogenase: Tentative Amino Acid Sequence; Identification of a Reactive Lysine; Nitration of a Specific Tyrosine and Loss of Allosteric Inhibition by Guanosine Triphosphate, Proc. Natl. Acad. Sci. U. S. A., 1970, 67(2), 724–730 CrossRef CAS.
- C. A. Stanley, Regulation of glutamate metabolism and insulin secretion by glutamate dehydrogenase in hypoglycemic children, Am. J. Clin. Nutr., 2009, 90(3), 8625 CrossRef PubMed.
- T. J. Smith and C. A. Stanley, Untangling the glutamate dehydrogenase allosteric nightmare, Trends Biochem. Sci., 2008, 33(11), 557 CrossRef CAS PubMed.
- S. Banerjee, T. Schmidt, J. Fang, C. A. Stanley and T. J. Smith, Structural Studies on ADP Activation of Mammalian Glutamate Dehydrogenase and the Evolution of Regulation, Biochemistry, 2003, 42, 3446–3456 CrossRef CAS PubMed.
- S. A. Wacker, M. J. Bradley, J. Marion and E. Bell, Ligand-induced changes in the conformational stability and flexibility of glutamate dehydrogenase and their role in catalysis and regulation, Protein Sci., 2010, 19, 1820 CrossRef CAS PubMed.
- J. M. Guisán, G. Alvaro, R. Fernandez-Lafuente, C. M. Rosell, J. L. Garcia and A. Tagliani, Stabilization of heterodimeric enzyme by multipoint covalent immobilization: penicillin G acylase from Kluyvera citrophila, Biotechnol. Bioeng., 1993, 42(4), 455 CrossRef PubMed.
- C. Mateo, O. Abian, R. Fernandez–Lafuente and J. M. Guisan, Increase in conformational stability of enzymes immobilized on epoxy-activated supports by favoring additional multipoint covalent attachment, Enzyme Microb. Technol., 2000, 26(7), 509 CrossRef CAS PubMed.
- V. Grazú, O. Abian, C. Mateo, F. Batista-Viera, R. Fernández-Lafuente and J. M. Guisán, Stabilization of enzymes by multipoint immobilization of thiolated proteins on new epoxy–thiol supports, Biotechnol. Bioeng., 2005, 90(5), 597 CrossRef PubMed.
- A. Manrich, A. Komesu, W. S. Adriano, P. W. Tardioli and R. L. C. Giordano, Immobilization and stabilization of xylanase by multipoint covalent attachment on agarose and on chitosan supports, Appl. Biochem. Biotechnol., 2010, 161, 455 CrossRef CAS PubMed.
- H. N. Motlagh, J. O. Wrabl, J. Li and V. J. Hilser, The ensemble nature of allostery, Nature, 2014, 508, 331 CrossRef CAS PubMed.
- T. J. Smith, T. Schmidt, J. Fang, J. Wu, G. Siuzdak and C. A. Stanley, The Structure of Apo Human Glutamate Dehydrogenase Details Subunit Communication and Allostery, J. Mol. Biol., 2002, 318, 765–777 CrossRef CAS PubMed.
- A. Cooper and D. T. E. Dryden, Allostery without conformational change, Eur. Biophys. J., 1984, 11, 103 CrossRef CAS PubMed.
- S. E. Reichheld, Z. Yu and A. R. Davidson, The induction of folding cooperativity by ligand binding drives the allosteric response of tetracycline repressor, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(52), 22263 CrossRef CAS PubMed.
- I. Luque and E. Freire, Structural stability of binding sites: consequences for binding affinity and allosteric effects, Proteins: Struct., Funct., Bioinf., 2000, 4, 63 CrossRef.
- H. J. Dyson and P. E. Wright, Coupling of folding and binding for unstructured proteins, Curr. Opin. Struct. Biol., 2002, 12, 54 CrossRef CAS PubMed.
- C. M. Petit, J. Zhang, P. J. Sapienza, E. J. Fuentes and A. L. Lee, Hidden dynamic allostery in a PDZ domain, Proc. Natl. Acad. Sci. U. S. A., 2009, 206(43), 18249 CrossRef PubMed.
- K. K. Frederick, M. S. Marlow, K. G. Valentine and A. J. Wand, Conformational entropy in molecular recognition by proteins, Nature, 2007, 448, 325 CrossRef CAS PubMed.
- T. L. Ogorzalek, S. Wei, Y. Liu, Q. Wang, C. L. Brooks, Z. Chen and E. N. G. Marsh, Molecular-Level Insights into Orientation-Dependent Changes in the Thermal Stability of Enzymes Covalently Immobilized on Surfaces, Langmuir, 2015, 31, 6145 CrossRef CAS PubMed.
- P. Cioni and G. B. Strambini, Dynamical Structure of Glutamate Dehydrogenase as Monitored by Tryptophan Phosphorescence: Signal Transmission Following Binding of Allosteric Effecters, J. Mol. Biol., 1989, 207, 237 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra24637g |
| ‡ Present address: Metalloenzymes and Mimetics Laboratory, UFSCar, São Carlos-SP, Brazil. |
| This journal is © The Royal Society of Chemistry 2016 |