Triazine–benzimidazole conjugates: synthesis, spectroscopic and molecular modelling studies for interaction with calf thymus DNA†
Abstract
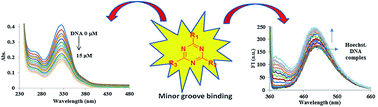
Triazine–benzimidazole analogues with different substitutions of primary and secondary amines as well as aryl groups were synthesized and characterized by 1H, 13C NMR and mass spectrometry. Interactions of these compounds with ct-DNA were explored by spectroscopic and viscometric techniques. These results and molecular modelling studies showed that compounds 7, 9–11, 16 and 21 interacted with ct-DNA through groove binding and not intercalation.

 Please wait while we load your content...
Please wait while we load your content...