Identification of a new curcumin synthase from ginger and construction of a curcuminoid-producing unnatural fusion protein diketide-CoA synthase::curcumin synthase†
Abstract
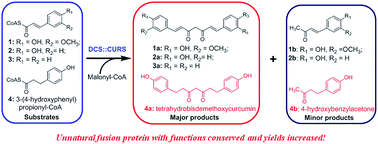
The biosynthesis and metabolic engineering of curcuminoids received considerable attention for their important pharmaceutical properties. In the present study, a new curcumin synthase (ZoCURS) was identified from ginger (Zingiber officinale). Notably, ZoCURS efficiently accepts 3-(4-hydroxyphenyl)propionyl-CoA to produce tetrahydrobisdemethoxycurcumin, which would be meaningful to further understand the biosynthesis of curcuminoids in ginger. In addition, a curcuminoid-producing unnatural fusion protein diketide-CoA synthase::curcumin synthase (DCS::CURS) was constructed. Comparing to ZoCURS, DCS::CURS indicated similar substrate specificities and catalytic potentials to catalyze the formation of various curcuminoids, however, the yield of curcuminoids produced by DCS::CURS was obviously increased, which would be useful for metabolic engineering of pharmaceutically important curcuminoid analogs, particularly including asymmetric dihydrocurcuminoids such as 3-(4-hydroxyphenyl)propionyl-feruloylmethane, 3-(4-hydroxyphenyl)propionyl-p-coumaroylmethane, and 3-(4-hydroxyphenyl)propionyl-cinnamoylmethane.

 Please wait while we load your content...
Please wait while we load your content...