Multiplex lateral flow immunoassay for five antibiotics detection based on gold nanoparticle aggregations†
Juan Penga,
Yongwei Wangb,
Liqiang Liu a,
Hua Kuanga,
Aike Lib and
Chuanlai Xu*a
a,
Hua Kuanga,
Aike Lib and
Chuanlai Xu*a
aState Key Lab of Food Science and Technology, School of Food Science and Technology, Jiangnan University, Wuxi, JiangSu 214122, People's Republic of China. E-mail: xcl@jiangnan.edu.cn; lak@chinagrain.org; Tel: +86-510-85329076
bCereals & Oils Nutrition Research Group, Academy of Science & Technology of State Administration of Grain, Beijing100037, People's Republic of China
First published on 11th January 2016
Abstract
A new immunochromatographic assay (ICA) was developed for the simultaneous screening of five antibiotics that can coexist in milk, namely lincomycin (LIN), gentamicin (GEN), kanamycin (KAN), streptomycin (STR), and neomycin (NEO), using five corresponding monoclonal antibodies (mAbs). The five mAbs were conjugated to colloidal gold nanoparticles (AuNPs), forming AuNP-labeled antibodies, which were placed in a microtiter well after freeze-drying; the five antigens were immobilized separately on five test lines to align with their corresponding AuNP-labeled antibodies. Using this method, the cutoff values for the strip test in milk were 25 ng mL−1 for LIN, 25 ng mL−1 for GEN, 50 ng mL−1 for KAN, 50 ng mL−1 for STR, and 100 ng mL−1 for NEO, which are below the maximum residue levels set by the European Union. Based on the strip reader, the multiplex strip could detect the concentrations of LIN, GEN, KAN, STR, and NEO as low as 2.5 ng mL−1, 2.5 ng mL−1, 2.5 ng mL−1, 2.5 ng mL−1, and 5 ng mL−1 in milk, respectively. The accuracy and reproducibility of the assay were acceptable when tested on food samples. In conclusion, our ICA strip is useful for the rapid and high-throughout screening of antibiotics in food and environments on-site.
1. Introduction
Antibiotics are widely used in veterinary treatment. However, their use can have numerous negative effects, such as enhanced drug resistance and contamination of the environment, and are a threat to public health.1–3 Lincomycin (LIN) belongs to the lincosamide class of antibiotics, and was derived from the bacterium, Streptomyces lincolnensis. This antibiotic is often injected into dairy cattle via the intramuscular or intramammary routes to protect against Gram-positive bacteria.4 Genomic damage can be induced after a long-term or high-level exposure of LIN, and can be enhanced by concomitant use of other antibiotics.5,6 Gentamicin (GEN), kanamycin (KAN), streptomycin (STR) and neomycin (NEO) are often found as antibiotic residues in milk, and are grouped in a subclass of aminoglycosides (AMGs).7 GEN is used to treat both Gram-positive and -negative bacteria by parenteral or oral injection in cattle.8 KAN, produced by Streptomyces kanamyceticus, can inhibit the growth of both Gram-positive and -negative bacteria,9,10 while STR is used for treating Gram-negative bacteria, and works by blocking the synthesis of protein.11 Neomycin (NEO), a broad-spectrum antibiotic, is injected orally to treat gastrointestinal infections and via the intramammary route to treat mastitis in cattle.12 Excessive and long-term usage of AMGs may lead to ototoxicity and nephrotoxicity.12 The presence of residues in milk is easily detected if the suggested withdrawal time is not respected, and is particularly obvious after intramammary or intrauterine injections rather than intravenous or intramuscular injections.13,14 Therefore, the following maximum residue levels (MRLs) in milk were established by the European Union (EU): 150 μg kg−1 for LIN, 100 μg kg−1 for GEN, 150 μg kg−1 for KAN, 200 μg kg−1 for STR, and 1500 μg kg−1 for NEO.15Many analytical methods have been developed for monitoring the contamination levels of these five antibiotics in milk, including microbiological assays, instrument methods, and immunoassay methods.16 Microbiological assays generally lack sensitivity and specificity,17 while instrument methods, including high-performance liquid chromatography, liquid chromatography-mass spectrometry, and liquid chromatography-tandem mass spectrometry,7,18,19 are costly, time consuming, require complicated sample preparation, and are unsuitable for field applications, despite the advantages of higher sensitivity and multiplex detectability.20 Previous studies have reported the use of immunoassay methods such as ELISA and biosensor methods for monitoring antibiotic residues.4,8,21–26 Among these methods, antibody-based multicomponent strip sensors are a particularly good method for field use due to their time efficiency and simple preparation compared with the complex steps required for ELISA such as washing and incubation.27–29 For this purpose, an immunochromatographic assay (ICA) has been proposed for detecting these antibiotic residues in milk. Several studies have been published on the detection of these five antibiotic residues using ICA methods.21,30–33 However, to our knowledge, there have been no reports of ICA for the simultaneous detection of these five analytes using different monoclonal antibodies (mAbs). In this paper, we aimed to develop a one-step ICA for the rapid and simultaneous screening of LIN, GEN, KAN, STR, and NEO, and to assess its application in spiked milk.
2. Materials and methods
2.1. Reagents and apparatus
LIN, GEN, KAN, STR, and NEO were purchased from J&K Scientific (Shanghai, China). Goat anti-mouse immunoglobulin (IgG) antibody was supplied by Jackson ImmunoResearch Laboratories (West Grove, PA, USA). All buffer solutions were prepared with ultrapure water from a Milli-Q Ultrapure System (Millipore Corporation, Bedford, MA, USA). All other reagents and chemicals were obtained from the National Pharmaceutical Group Chemical Reagent Co., Ltd. (Shanghai, China). The anti-LIN mAb, anti-GEN mAb, anti-KAN mAb, anti-STR mAb, and anti-NEO mAb were produced in our laboratory.34The polyvinylchloride (PVC) backing material, the sample pad (glass fiber membrane, GL-b01), the absorbance pad (H5079), and the conjugate pad (Ahlstrom 8964) were obtained from JieYi Biotechnology Co., Ltd. (Shanghai, China). The nitrocellulose (NC) membrane was purchased from Millipore Corporation. The CM4000 Guillotine Cutting Module and the Dispensing Platform were obtained from Shanghai Kinbio Tech Co., Ltd. (China). The BioDot TSR3000 membrane strip reader was supplied by Gene Company Limited (Shanghai, China).
2.2. Preparation of the hapten–protein conjugates
The hapten–bovine serum albumin (BSA) conjugates (NEO–BSA, KAN–BSA, STR–BSA, and GEN–BSA) were synthesized using the glutaraldehyde method, as described previously.35 Briefly, 5.37 μL glutaraldehyde solution (25%) diluted 10 times with 0.01 M phosphate buffered saline (PBS, pH 7.4) was added dropwise into 13.4 μmol AMGs (GEN, KAN, STR, and NEO) dissolved in 600 μL 0.01 M PBS. After stirring for 10 min at room temperature, the solution mixture was slowly added to 10 mg BSA dissolved in 3.4 mL 0.01 M PBS. After reacting for 40 min at room temperature, 16 mg sodium borohydride was added. The mixture was allowed to react for 2 h at 4 °C, and then dialyzed against 0.01 M PBS for 3 days.The LIN conjugate (LIN–BSA) was prepared using the following method. LIN succinate (LIN-HS) was conjugated to BSA by the 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC)/N-hydroxysuccinimide (NHS) method.4 Briefly, 3.3 mg EDC (17.2 μmol) and 1.5 mg NHS (13 μmol) were added to LIN-HS (8.9 μmol) dissolved in 400 μL N,N-dimethylformamide (DMF). The mixture was reacted for 4 h at room temperature and then slowly added to 10 mg BSA dissolved in 5 mL 0.01 M borate buffer (BB, pH 8.4). The mixture was maintained at room temperature overnight, followed by dialysis against 0.01 M PBS for 3 days.
2.3. Preparation of the immunochromatographic strip and its characterization
The AuNP solution was prepared as previously reported,36 with some modifications. Briefly, 10 mL of 1% trisodium citrate solution was rapidly added to 500 mL of 0.01% HAuCl4 solution, and then boiled with constant stirring. The reaction solution changed to the color of red wine within around 1 min, and then we continued boiling the mixture for another 5 min. The solution was then cooled to room temperature and stored at 4 °C for following experiment.The anti-AMGs/LIN mAbs produced in our laboratory were purified by the caprylic acid–ammonium method, and then evaluated by indirect competitive ELISA and our single strip method. The indirect competitive ELISA was performed as previously described.37 Briefly, 100 μL of AMGs/LIN–BSA diluted with 0.05 M carbonate buffer (CB) were coated onto 96-microwell plates for 2 h at 37 °C. Then, the plates were washed three times with 0.01 M PBS with 0.05% Tween 20 (PBST) and blocked with 0.05 M CB with 0.2% w/v gelatin for 2 h at 37 °C. After being washed thrice again, 50 μL of different concentrations of the analytes in PBS and 50 μL mAb dissolved in antibody diluent (0.01 M PBS containing 0.05% v/v Tween 20 and 0.1% w/v gelatin) were added to each well. After incubation for 30 min at 37 °C, the plates were washed with PBST, and 100 μL of peroxidase-labeled goat anti-mouse IgG (diluted 3000 times with the antibody diluent) were added to each well and incubated for 30 min at 37 °C. After being washed with PBST, 100 μL of 3,3′,5,5′-tetramethylbenzidine substrate was added, and the reaction was stopped by the addition of 50 μL of 2 M H2SO4, followed by incubation at 37 °C for 15 min. The optical density was then read by the microplate reader at 450 nm.
The AuNP-labeled antibodies were prepared using the following method.38 First, the pH of the AuNP solution (1 nM) was adjusted to 8 with K2CO3. Then, the purified mAb, which was dissolved in ultrapure water, was added dropwise to 10 mL of the above-mentioned AuNP solution. After gentle stirring for 30 min, 0.5 mL of 10% (w/v) BSA was added to block the unreacted sites. After 1 h, the mixture was centrifuged twice at 8000g for 20 min. Finally, the precipitate was resuspended in 1 mL of boric acid buffer solution (0.002 M boric acid, 0.2% BSA, and 0.02% NaN3).
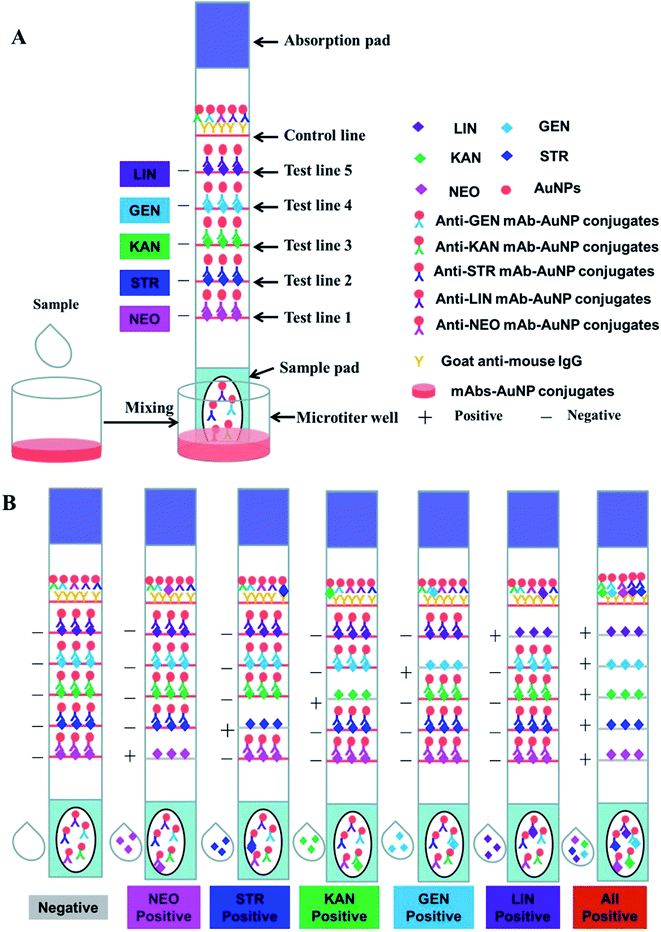
The immunochromatographic strip was assembled as shown in Fig. 1A. The NC membrane was glued to the middle of the PVC support plate, and the sample pad and absorbent pad were glued to the bottom and upper regions, respectively. The goat anti-mouse IgG antibody and antigens were sprayed onto the NC membrane part of the strip as the control and test lines (C line and T lines, respectively) at a speed of 1 μL cm−1 using the rapid test dispenser platform (HM3035). The distance between the T lines and C line was 5 mm. After drying at 37 °C for 2 h, the strips were cut to the specification of 3 mm × 60 mm using a guillotine cutter, and then stored in a sealed bag at room temperature together with the mAb–AuNP conjugates, which were freeze-dried using a vacuum freeze dryer. The antigens specific to the five different kinds of antibiotics (NEO, STR, KAN, GEN, and LIN) were evenly distributed on the NC membrane in the tomographic flow direction to form five separate T lines for simultaneous detection. In this study, a 200 μL sample solution containing all five analytes (LIN, GEN, KAN, STR, and NEO) at different concentrations was added to the mAb–AuNP conjugates in the microtiter wells. The optimal concentrations of the antigens synthesized with the standard bioconjugation techniques were 0.5 mg mL−1 for LIN–BSA, GEN–BSA, KAN–BSA, and STR–BSA, and 1 mg mL−1 for NEO–BSA on the test lines. After reacting for 5 min at room temperature, strips were then added to the mixtures. Under these conditions, the antigen–antibody complex was formed. The amount of complex was proportional to the amount of analyte in the original sample. These complex, in solution, were contacted with the strip sample pad and migrates into the NC membrane. The five coating reagents and the goat anti-mouse IgG was sprayed on the NC membrane and formed the five test lines and control line before. If there is no analyte in the sample solution, the GNP-labeled antibodies bind to the immobile coating antigens on the T lines (TL-1, -2, -3, -4, and -5) and the goat anti-mouse IgG on the C line separately. This result was considered negative. If a certain amount of analytes exist in the sample solution, free analytes (in the sample solution) would combine with the corresponding mAb–AuNP conjugates. Less mAb–AuNP conjugates would combine with the immobile coating antigens and the intensity of the T lines will decrease. If all the five analytes concentration is high enough, the five T lines will all disappear. Excess reagents that cross the NC membrane become entrapped in an absorbent pad.
2.4. Sensitivity of the multiple strips
Five kinds of mixed standards were prepared for detecting the visual detection limit of the multiple strips. LIN, STR, KAN, and GEN were diluted to 0, 2.5, 5, 10, 25, and 50 ng mL−1, and NEO was diluted to 0, 5, 10, 25, 50, and 100 ng mL−1. PBS (100 μL) with and without the five analytes was added to the microtiter wells with the mAb–AuNP conjugate mixtures. After 5 min reaction time between the analytes and the mAb–AuNP conjugates, a strip was put into each microporous well. The solution moved along the NC membrane with the help of capillary action and aggregated on the test and control lines due to the antigen–antibody reaction. After 10 min, the results were evaluated with the naked eye. The anti-AMGs/LIN mAb–AuNP conjugates freely migrated into the NC membrane and combined with the immobilized AMGs/LIN–BSA antigen on the test zones in the absence of the five analytes in the sample solution. Therefore, five visible red lines were formed, as showed in Fig. 1A. In contrast, if the concentrations of the five analytes reached certain values, the mAb–AuNP conjugates would first bind to the corresponding free analytes in the sample; thus, few or no mAb–AuNP conjugates would migrate and combine with the immobilized AMGs/LIN–BSA antigen. Thereafter, a weakly colored line or no red line was then observed in the corresponding zone as showed in Fig. 1B. All the uncombined mAb–AuNPs conjugates gathered on the control line, forming a deep red band after a complete reaction between the mAbs–AuNP conjugates and the goat–mouse IgG antibody.2.5. Assay of milk samples with the multiplex strip
Milk samples were used to estimate the practicability of the multiplex strip. Different concentrations of LIN, GEN, KAN, STR, and NEO were spiked in negative milk samples previously confirmed by ELISA. The analyses were repeated 20 times for each concentration (n = 20).3. Results and discussion
3.1. Preparation of antibody–AuNP conjugates
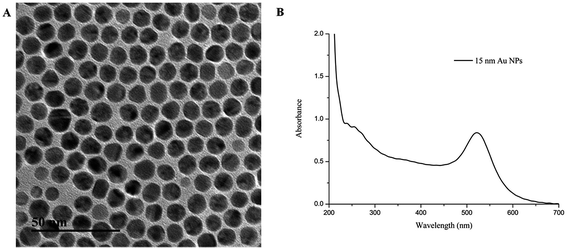
AuNPs (15 ± 1 nm) contributes to the plasmon peak absorption at 520 nm (Fig. 2). The antibody–AuNP conjugates formed due to the high electron density of the gold particles and the fact that their surfaces can bind to biological macromolecules such as antibodies.39,40 The pH of the AuNP solution and the amount of the mAbs were optimized during the preparation of the antibody–AuNP conjugates. Briefly, different volume of 0.1 M K2CO3 (2 μL, 4 μL, 8 μL, 10 μL) was added to tubes containing 1 mL AuNP solutions to determine the optimal pH value. 20 μL of 1 mg mL−1 mAb solution was then added and incubated for 10 min at room temperature. Then, 100 μL of 10% NaCl solution was added dropwise to each tube. Based on the results of our experiment, the color of the mAb–AuNP conjugates were showed no color change through the UV spectrum and the naked eye observation when 4 μL of 0.1 M K2CO3 was added to 1 mL AuNP solution to adjust the pH (pH = 7.2). 2 μL K2CO3 will result in low pH (pH = 6.8) and unstable mAb–AuNP conjugates. The color of conjugates were commonly bluish violet and the AuNP will aggregate after 10% NaCl were added or after centrifugation. 8 μL K2CO3 (pH = 7.8) or 10 μL K2CO3 (pH = 8.2) could be used but not recommended because the antibody labelling efficiency will decrease as the charge of antibody will be more negative at pH above its isoelectric point (Table S1†). Therefore, the addition of 4 μL of 0.1 M K2CO3 to each 1 mL AuNP solution optimized the pH value. To determine the optimal amount of mAb to conjugate to the gold nanoparticles, a gradient concentration of each mAb solution was added into 1 mL AuNP solution adjusted with 4 μL of 0.1 M K2CO3. After 10 min, 100 μL of 10% NaCl solution was added dropwise and incubated for 10 min. The optimal amount of mAb to conjugate to the gold nanoparticles was in excess of 10% of the lowest amount of mAb, which was effective for preventing aggregation of the colloidal solution. The results showed that the optimal concentrations for the five mAb–AuNP conjugates were 8 μg mL−1 for anti-LIN and anti-KAN, 10 μg mL−1 for anti-GEN and anti-STR, and 15 μg mL−1 for anti-NEO mAbs. To reduce nonspecific reactions, 50 μL of 10% BSA was added to 1 mL of each mAb–AuNP conjugate with 1 h of slight shaking to block any unconjugated sites.3.2. Optimization of the immunochromatographic strip
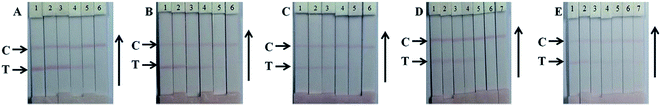
High sensitivity and specificity of the antibodies and the corresponding antigens play key roles in the performance of this multiplex strip. In our study, five highly sensitive antibodies were prepared and the half maximal inhibitory concentration values were confirmed with indirect ELISAs and the single strips (Table S2†); Fig. 3 shows the single test immunochromatographic strips for LIN, GEN, KAN, STR, and NEO in milk. On the control lines, the optimal concentration of goat anti-mouse IgG antibody was determined to be 0.5 mg mL−1. Under these optimized conditions, the developed ICA had clear bands, with high sensitivity.3.3. Evaluation of the multiplex strip
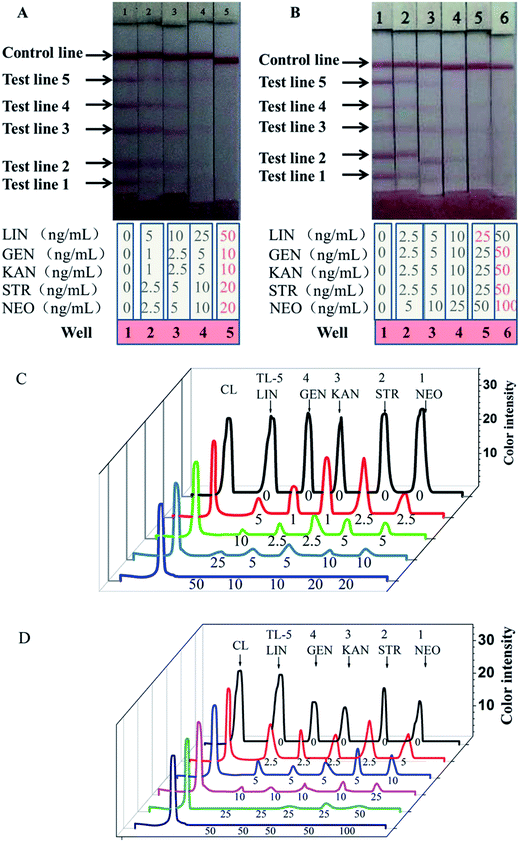
Five analytes at different concentrations were mixed in PBS to generate standards for evaluating the multiplex strip. For the assay, the end of the test-strip sample pad was placed into the microtiter wells and 100 μL of extracts with and without standards was added to the wells with the five freeze-dried antibody–AuNP conjugates. The 100 μL extracts in different wells contained different levels of LIN, GEN, KAN, STR, or NEO as shown in Fig. 4. The standards were first bonded with the antibody–AuNP conjugates in the solution in the wells, and then, driven by capillary forces, migrated from the sample pad to the absorbent pad by way of the hapten–protein conjugates on the test lines and goat anti-mouse IgG antibody on the control line. The whole process took 5 min. As shown in Fig. 4A, the intensity of the color of the test lines decreases as the concentration of the analytes increases. Based on visual inspection, the cutoff levels for this method in PBS were 50 ng mL−1 for LIN, 10 ng mL−1 for GEN, 10 ng mL−1 for KAN, 20 ng mL−1 for STR, and 20 ng mL−1 for NEO. The optical density profiles of the test lines (TL-1, -2, -3, -4, and -5) and the control line (CL) were recorded by strip reader and showed in Fig. 4C. The weakly positive results (the test line is clearly lighter than the negative result but has not disappeared) indicate that the multiplex strip works when the concentrations of LIN, GEN, KAN, STR, and NEO are 5 ng mL−1, 1 ng mL−1, 1 ng mL−1, 2.5 ng mL−1, and 2.5 ng mL−1, respectively.Similarly, milk samples spiked with different concentrations of the five standards were tested using our multiplex strip. As shown in Fig. 4B, the cutoff levels for this method in milk were 25 ng mL−1 for LIN, 25 ng mL−1 for GEN, 50 ng mL−1 for KAN, 50 ng mL−1 for STR, and 100 ng mL−1 for NEO, which are lower than the MRLs in milk set by the EU. The optical density profiles of the test lines (TL-1, -2, -3, -4, and -5) and the control line (CL) were recorded by strip reader and showed in Fig. 4D. The weakly positive results indicate that the multiplex strip works when the concentrations of LIN, GEN, KAN, STR, and NEO are 2.5 ng mL−1, 2.5 ng mL−1, 2.5 ng mL−1, 2.5 ng mL−1, and 5 ng mL−1, respectively. In the milk matrix, antibody could interact with LIN more sensitively and the detection limit was decreased 2-fold. The detection limits for GEN, KAN, STR, and NEO was unchanged or increased.
Table 1 shows no cross-reactivity of these five analytes by ELISA. This can be attributed to the different structures of the analytes shown in Table S2† and the specific antibodies we prepared.
| Analytes | Analogues | Cross-reactivity (%) |
|---|---|---|
| LIN | GEN | <0.01 |
| KAN | <0.01 | |
| STR | <0.01 | |
| NEO | <0.01 | |
| GEN | LIN | <0.01 |
| KAN | <0.01 | |
| STR | <0.01 | |
| NEO | <0.01 | |
| KAN | LIN | <0.01 |
| GEN | <0.01 | |
| STR | <0.01 | |
| NEO | <0.01 | |
| NEO | LIN | <0.01 |
| GEN | <0.01 | |
| KAN | <0.01 | |
| STR | <0.01 |
The accuracy and reproducibility of the developed ICA strips were further evaluated by spiking four different concentrations of the analytes into negative milk samples. The semi-quantitative results in Table S3† indicate that the ICA results are consistent with the spiked analyte concentrations based on visual inspection. The quantitative results in Table S4† indicate the ICA good stability and application with the recoveries ranging from 86.2% to 121.7%. Concordant results were obtained for 20 replicates of the strip test.
Thus, our ICA strip method could be a useful tool for the multiple screening of LIN, GEN, KAN, STR, and NEO residues in milk.
4. Conclusions
In the present study, a newly developed ICA was used to screen five antibiotics that can coexist in milk using five corresponding mAbs. Highly sensitive and specific antibody–antigen systems ensure the sensitivity and specificity of the developed ICA. The lack of complicated sample pretreatment, naked eye analysis within 10 min, and ease of use, makes the strip suitable for on-site analysis.Acknowledgements
This work is financially supported by the Key Programs from MOST (2012BAD29B05, 2013AA065501, 2012BAK08B01).References
- T. P. van Boeckel, C. Brower, M. Gilbert, B. T. Grenfell, S. A. Levin, T. P. Robinson, A. Teillant and R. Laxminarayan, Global trends in antimicrobial use in food animals, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 5649–5654 CrossRef CAS.
- H. Wang, B. Wang, Q. Zhao, Y. Zhao, C. Fu, X. Feng, N. Wang, M. Su, C. Tang and F. Jiang, Antibiotic Body Burden of Chinese School Children: A Multisite Biomonitoring-Based Study, Environ. Sci. Technol., 2015, 49, 5070–5079 CrossRef CAS.
- Q. Q. Zhang, G. G. Ying, C. G. Pan, Y. S. Liu and J. L. Zhao, A comprehensive evaluation of antibiotics emission and fate in the river basins of China: source analysis, multimedia modelling, and linkage to bacterial resistance, Environ. Sci. Technol., 2015, 49, 6772–6782 CrossRef CAS.
- M. A. Burkin and I. A. Galvidis, Development of a Competitive Indirect ELISA for the Determination of Lincomycin in Milk, Eggs, and Honey, J. Agric. Food Chem., 2010, 58, 9893–9898 CrossRef CAS.
- I. M. Hagenbuch and J. L. Pinckney, Toxic effect of the combined antibiotics ciprofloxacin, lincomycin, and tylosin on two species of marine diatoms, Water Res., 2012, 46, 5028–5036 CrossRef CAS.
- L. Rocco, C. Peluso and V. Stingo, Micronucleus test and comet assay for the evaluation of zebrafish genomic damage induced by erythromycin and lincomycin, Environ. Toxicol., 2012, 27, 598–604 CrossRef CAS.
- Y. Tao, D. Chen, H. Yu, L. Huang, Z. Liu, X. Cao, C. Yan, Y. Pan, Z. Liu and Z. Yuan, Simultaneous determination of 15 aminoglycoside(s) residues in animal derived foods by automated solid-phase extraction and liquid chromatography-tandem mass spectrometry, Food Chem., 2012, 135, 676–683 CrossRef CAS.
- Y. Chen, Y. Shang, X. Li, X. Wu and X. Xiao, Development of an enzyme-linked immunoassay for the detection of gentamicin in swine tissues, Food Chem., 2008a, 108, 304–309 Search PubMed.
- J. W. Park, S. R. Park, K. K. Nepal, A. R. Han, Y. H. Ban, Y. J. Yoo, E. J. Kim, E. M. Kim, D. Kim, J. K. Sohng and Y. J. Yoon, Discovery of parallel pathways of kanamycin biosynthesis allows antibiotic manipulation, Nat. Chem. Biol., 2011, 7, 843–852 CrossRef CAS.
- M. A. Zaunbrecher, R. D. Sikes, B. Metchock, T. M. Shinnick and J. E. Posey, Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 20004–20009 CrossRef CAS.
- K. Mølbak, D. L. Baggesen, F. M. Aarestrup, J. M. Ebbesen, J. Engberg, K. Frydendahl, P. Gerner-Smidt, A. M. Petersen and H. C. Wegener, An outbreak of multidrug-resistant, quinolone-resistant Salmonella enterica serotype Typhimurium DT104, N. Engl. J. Med., 1999, 341, 1420–1425 CrossRef.
- V. Gaudin, N. Cadieu and P. Sanders, Results of a European proficiency test for the detection of streptomycin/dihydrostreptomycin, gentamicin and neomycin in milk by ELISA and biosensor methods, Anal. Chim. Acta, 2005, 529, 273–283 CrossRef CAS.
- T. A. McGlinchey, P. A. Rafter, F. Regan and G. P. McMahon, A review of analytical methods for the determination of aminoglycoside and macrolide residues in food matrices, Anal. Chim. Acta, 2008, 624, 1–15 CrossRef CAS.
- A. Smyth, K. H. Tan, P. Hyman-Taylor, M. Mulheran, S. Lewis, D. Stableforth, A. Knox and T. S. Group, Once versus three-times daily regimens of tobramycin treatment for pulmonary exacerbations of cystic fibrosis—the TOPIC study: a randomised controlled trial, Lancet, 2005, 365, 573–578 CrossRef CAS.
- C. Regulation, No 470/2009 of the European Parliament and of the Council of 6 May 2009 laying down a community procedures for the establishment of residue limits of pharmacologically active substances in foodstuffs of animal origin, repealing Council Regulation (EEC) No 2377/90 and amending Directive 2001/82, Off. J. Eur. Communities: Legis., 2009, 152, 11–22 Search PubMed.
- D. A. Stead, Current methodologies for the analysis of aminoglycosides, J. Chromatogr. B: Biomed. Sci. Appl., 2000, 747, 69–93 CrossRef CAS.
- E. Adams, L. Liu, K. Dierick, S. Guyomard, P. Nabet, S. Rico, P. Louis, E. Roets and J. Hoogmartens, Neomycin: microbiological assay or liquid chromatography?, J. Pharm. Biomed. Anal., 1998, 17, 757–766 CrossRef CAS.
- R. Han, N. Zheng, Z. Yu, J. Wang, X. Xu, X. Qu, S. Li, Y. Zhang and J. Wang, Simultaneous determination of 38 veterinary antibiotic residues in raw milk by UPLC–MS/MS, Food Chem., 2015, 181, 119–126 CrossRef CAS.
- S. B. Turnipseed, W. C. Andersen, C. M. Karbiwnyk, M. R. Madson and K. E. Miller, Multi-class, multi-residue liquid chromatography/tandem mass spectrometry screening and confirmation methods for drug residues in milk, Rapid Commun. Mass Spectrom., 2008, 22, 1467–1480 CrossRef CAS.
- S. K. Grebe and R. J. Singh, LC-MS/MS in the clinical laboratory – where to from here?, Clin. Biochem. Rev., 2011, 32, 5 Search PubMed.
- Y. Chen, Z. Wang, Z. Wang, S. Tang, Y. Zhu and X. Xiaos, Rapid enzyme-linked immunosorbent assay and colloidal gold immunoassay for kanamycin and tobramacin in swine tissues, J. Agric. Food Chem., 2008, 56, 2944–2952 CrossRef CAS.
- Y. P. Chen, M. Q. Zou, C. Qi, M. X. Xie, D. N. Wang, Y. F. Wang, Q. Xue, J. F. Li and Y. Chen, Immunosensor based on magnetic relaxation switch and biotin–streptavidin system for the detection of kanamycin in milk, Biosens. Bioelectron., 2013, 39, 112–117 CrossRef CAS.
- L. R. Schoukroun-Barnes, S. Wagan and R. J. White, Enhancing the analytical performance of electrochemical RNA aptamer-based sensors for sensitive detection of aminoglycoside antibiotics, Anal. Chem., 2014, 86, 1131–1137 CrossRef CAS.
- N. A. Taranova, A. N. Berlina, A. V. Zherdev and B. B. Dzantiev, ‘Traffic light’ immunochromatographic test based on multicolor quantum dots for the simultaneous detection of several antibiotics in milk, Biosens. Bioelectron., 2015, 63, 255–261 CrossRef CAS.
- J. Zhou, K. Zhu, F. Xu, W. Wang, H. Jiang, Z. Wang and S. Ding, Development of a Microsphere-Based Fluorescence Immunochromatographic Assay for Monitoring Lincomycin in Milk, Honey, Beef, and Swine Urine, J. Agric. Food Chem., 2014, 62, 12061–12066 CrossRef CAS.
- B. Liu, B. Zhang, Y. Cui, H. Chen, Z. Gao and D. Tang, Multifunctional Gold-Silica Nanostructures for Ultrasensitive Electrochemical Immunoassay of Streptomycin Residues, ACS Appl. Mater. Interfaces, 2011, 3, 4668–4676 CAS.
- Y. Guo, B. Ngom, T. Le, X. Jin, L. Wang, D. Shi, X. Wang and D. Bi, Utilizing three monoclonal antibodies in the development of an immunochromatographic assay for simultaneous detection of sulfamethazine, sulfadiazine, and sulfaquinoxaline residues in egg and chicken muscle, Anal. Chem., 2010, 82, 7550–7555 CrossRef CAS.
- C. Xing, L. Liu, S. Song, M. Feng, H. Kuang and C. Xu, Ultrasensitive immunochromatographic assay for the simultaneous detection of five chemicals in drinking water, Biosens. Bioelectron., 2015, 66, 445–453 CrossRef CAS.
- M. Ren, H. Xu, X. Huang, M. Kuang, Y. Xiong, H. Xu, Y. Xu, H. Chen and A. Wang, Immunochromatographic Assay for Ultrasensitive Detection of Aflatoxin B-1 in Maize by Highly Luminescent Quantum Dot Beads, ACS Appl. Mater. Interfaces, 2014, 6, 14215–14222 CAS.
- N. A. Byzova, E. A. Zvereva, A. V. Zherdev, S. A. Eremin, P. G. Sveshnikov and B. B. Dzantiev, Pretreatment-free immunochromatographic assay for the detection of streptomycin and its application to the control of milk and dairy products, Anal. Chim. Acta, 2011, 701, 209–217 CrossRef CAS.
- S. Cao, S. Song, L. Liu, N. Kong, H. Kuang and C. Xu, Comparison of an Enzyme-Linked Immunosorbent Assay with an Immunochromatographic Assay for Detection of Lincomycin in Milk and Honey, Immunol. Invest., 2015, 44, 438–450 CrossRef CAS.
- Y. Jin, J. W. Jang, C. H. Han and M. H. Lee, Development of ELISA and immunochromatographic assay for the detection of gentamicin, J. Agric. Food Chem., 2005, 53, 7639–7643 CrossRef CAS.
- Y. Jin, J. W. Jang, M. H. Lee and C. H. Han, Development of ELISA and immunochromatographic assay for the detection of neomycin, Clin. Chim. Acta, 2006, 364, 260–266 CrossRef CAS.
- N. Xu, C. Qu, W. Ma, L. Xu, L. Liu, H. Kuang and C. Xu, Development and application of one-step ELISA for the detection of neomycin in milk, Food Agric. Immunol., 2011, 22, 259–269 CrossRef CAS.
- F. Xu, W. Jiang, J. Zhou, K. Wen, Z. Wang, H. Jiang and S. Ding, Production of Monoclonal Antibody and Development of a New Immunoassay for Apramycin in Food, J. Agric. Food Chem., 2014, 62, 3108–3113 CrossRef CAS.
- L. Xu, H. Kuang, C. Xu, W. Ma, L. Wang and N. A. Kotov, Regiospecific Plasmonic Assemblies for in situ Raman Spectroscopy in Live Cells, J. Am. Chem. Soc., 2012, 134, 1699–1709 CrossRef CAS.
- J. Peng, D. Kong, L. Liu, S. Song, H. Kuang and C. Xu, Determination of quinoxaline antibiotics in fish feed by enzyme-linked immunosorbent assay using a monoclonal antibody, Anal. Methods, 2015, 7, 5204–5209 RSC.
- L. Liu, L. Luo, S. Suryoprabowo, J. Peng, H. Kuang and C. Xu, Development of an immunochromatographic strip test for rapid detection of ciprofloxacin in milk samples, Sensors, 2014, 14, 16785–16798 CrossRef.
- Y. Zhao, L. Liu, D. Kong, H. Kuang, L. Wang and C. Xu, Dual Amplified Electrochemical Immunosensor for Highly Sensitive Detection of Pantoea stewartii subsp. stewartii, ACS Appl. Mater. Interfaces, 2014, 6, 21178–21183 CAS.
- S. Nara, V. Tripathi, H. Singh and T. G. Shrivastav, Colloidal gold probe based rapid immunochromatographic strip assay for cortisol, Anal. Chim. Acta, 2010, 682(1), 66–71 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra22583c |
| This journal is © The Royal Society of Chemistry 2016 |