Design, synthesis, biological evaluation and molecular docking of amide and sulfamide derivatives as Escherichia coli pyruvate dehydrogenase complex E1 inhibitors†
Abstract
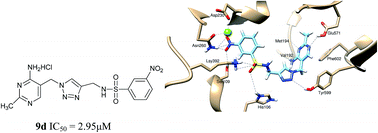
In this study, a series of novel amide derivatives and sulfamide derivatives as potential E. coli PDHc E1 inhibitors were designed and synthesized by optimizing the linker between triazole and benzene ring moieties based on the structure of lead compound I as thiamin diphosphate (ThDP) analogs. Their inhibitory activity against E. coli PDHc E1 were examined in vitro and their inhibitory activity against microbial diseases were further evaluated. Most of these compounds exhibit good inhibitory activity against E. coli PHDc E1 (IC50 1.99 to 25.66 μM) and obvious antibacterial activity. 5a, 5c and 9i showed 90–100% antibacterial activity against Xanthomonas oryzae pv. oryzae (Xoo), Acidovorax avenae subsp. avenae (Aaa) and cyanobacteria. Sulfamide derivatives 9 showed more potent inhibitory activity against E. coli PDHc E1 (IC50 < 14 μM) than that of amide derivatives 5 or lead compound I. Especially 9d (IC50 = 2.95 μM) and 9k (IC50 = 1.99 μM) exhibited not only the most powerful inhibitory potency against E. coli PDHc E1, but also 9k showed 99% antibacterial activity against Aaa at 500 μg mL−1 and almost the best inhibition of 97% against cyanobacteria at 20 μg mL−1. Furthermore, the binding mode of 5d and 9d to E. coli PDHc E1 was analyzed by a molecular docking method. The possible interactions of 9d with the important residues of E. coli PDHc E1 were further verified via site-directed mutagenesis enzymatic assays, and fluorescence spectral analysis. Both theoretical and experimental results revealed that 9d could display a more powerful interaction than that of 5d or I by forming a hydrogen bond between a sulfamide linkage and residues Lsy392, Tyr599 and His106 at active site of E. coli PDHc E1. 9k, 9d and 9i with both potent enzyme inhibition and significant antibacterial activity, could be used as novel lead compounds for further optimization. These results proved that a series of compounds with potential antibacterial activity could be obtained by the biorational design of E. coli PDHc E1 inhibitors.
- This article is part of the themed collection: A Decade of Progress in Click Reactions Based on CuAAC

 Please wait while we load your content...
Please wait while we load your content...