Effects of the i-motif DNA loop on the fluorescence of silver nanoclusters†
Taotao Lia,
Nongyue Heab,
Jiuhai Wanga,
Song Lib,
Yan Dengb and
Zunliang Wang*a
aState Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University, Si Pai Lou 2, Nanjing 210096, China. E-mail: zunliang.wang@gmail.com; Tel: +86 25 83792245
bEconomical Forest Cultivation and Utilization of 2011 Collaborative Innovation Center in Hunan Province, Hunan Key Laboratory of Green Packaging and Biological Nanotechnology, Hunan University of Technology, Zhuzhou 412007, China
First published on 22nd February 2016
Abstract
Cytosine-rich DNA strands, due to their distinct i-motif structure, are often used as stabilization templates for the formation of silver nanoclusters (Ag NCs). DNA i-motif conformation and stability are largely determined by the DNA loop region. Therefore, the optimal loop sequence contributes to the DNA template, forming a stable i-motif structure, thus promoting the formation of Ag NCs. In the present work, we prepared Ag NCs stabilized by a set of DNA sequences CCCC-X1X2X3X4-CCCC (C denotes cytosine, X denotes adenine or thymine) and investigated the loop sequence dependence of the fluorescence quantum yields (QYs) of Ag NCs. In all of the template DNA, the sequence 5′-CCCC-ATAT-CCCC-3′ was screened to be the most optimal template which stabilized clusters and could emit the brightest red fluorescence with a quantum yield (QY) of 0.78. However, for the reversed template sequence 5′-CCCC-TATA-CCCC-3′, the QY of the Ag NCs is lowered to 0.15. Such a significant difference in QYs indicates that even for the same base sequence, a different base alignment direction (starting from the 5′ or 3′ end) can have an important effect on the fluorescence of nanoclusters. Additionally, to optimize the preparation conditions of Ag NCs, the effects of reductants and pH environment on the clusters stabilized by the template sequence 5′-CCCC-ATAT-CCCC-3′ were further examined by using CD, UV and fluorescence spectroscopy, respectively.
Introduction
DNA-templated silver nanoclusters (Ag NCs), as a new type of fluorescent probe, have been successfully applied in highly sensitive biological detection due to their good water solubility, small sizes, high brightness and photochemical stability and biocompatibility.1–6 Previous reports have been shown that the sequences of DNA templates had a very critical impact on the fluorescence properties of Ag NCs, and the fluorescent emissions of Ag NCs could be adjusted by changing the sequences of the DNA templates.7–9 The binding interaction between the DNA sequence and silver clusters is mainly dependent on the different binding affinities of silver ions to four bases in the DNA sequences.10–12 Since silver ions prefer Lewis acid–base interactions with electron-rich nitrogen and oxygen atoms of nucleobases, cytosine (C) and guanine (G) can interact with Ag+ strongly.13 In addition, the secondary structure of DNA templates is also considered to be important for the stability of Ag NCs.14In recent years, C-rich DNA sequences, due to their potential to form distinct i-motif structures, have been widely used as the effective templates for the preparation of Ag NCs.7–12 It has been shown that the stability of the DNA i-motif structure was related to the loop sequence of i-motifs.15–18 Meanwhile, many studies demonstrated that the fluorescence property of silver clusters is associated with the loop sequence of i-motif DNA templates.19–22 However, due to the variety of the bases and arrangements of loop sequences in DNA templates, there is little discussion on how the loop sequences affects the fluorescence properties of Ag NCs. Moreover, the reason why the loop sequences affect silver clusters' fluorescence properties still remains unclear. To address these issues, a systematic understanding of the correlation between loop sequences and Ag clusters' fluorescence properties is required.
In this work, we have prepared the Ag NCs by using a serial of sequences C4-X4-C4 (here, C denotes cytosine, X denotes adenine or thymine) as the templates. By comparisons of the quantum yields (QYs) of the prepared Ag clusters, it was found that difference in fluorescence QYs was correlated with the variation of the loop sequences and the sequence 5′-C4-ATAT-C4-3′ was screened out to be optimal template that could give the clusters with the maximal QYs of 0.78. Interestingly, we found this correlation is also related to the base arrangement direction of the loop sequences in DNA templates. When reversing the DNA loop sequence with TATA, the prepared Ag clusters exhibited significantly lowered QY of 0.15. Such observations offer vital evidence for the effect of i-motif loop sequences on the fluorescence of Ag NCs. To optimize the preparation conditions, the effects of pH, the phosphate buffer (PB, a mixed solution of Na2HPO4 and NaH2PO4) concentration, and the molar ratio of DNA/AgNO3/NaBH4 on the clusters' fluorescence emission were also investigated, respectively.
Materials and methods
Materials
In this work, the silver nitrate (99.999%) and sodium borohydride were purchased from Sigma-Aldrich Co. All PAGE-purified oligonucleotides (purity > 95%) were purchased from Sangon Biotech (Shanghai) Co., Ltd. The DNA sequences are summarized in Table 1.| Template | Em/Ex (nm) | QY | Template | Em/Ex (nm) | QY |
|---|---|---|---|---|---|
| a Note: “—” means no signal detected. | |||||
| C4-AAAA-C4 | 630/580 | 0.24 | C4-TTTT-C4 | — | — |
| C4-TAAA-C4 | 620/570 | 0.29 | C4-ATTT-C4 | 615/570 | 0.12 |
| C4-ATAA-C4 | 630/580 | 0.23 | C4-TATT-C4 | — | — |
| C4-AATA-C4 | 615/570 | 0.19 | C4-TTAT-C4 | 625/580 | 0.06 |
| C4-AAAT-C4 | 630/590 | 0.48 | C4-TTTA-C4 | 615/570 | 0.26 |
| C4-TAAT-C4 | 630/580 | 0.09 | C4-ATTA-C4 | 615/570 | 0.22 |
| C4-ATAT-C4 | 635/590 | 0.78 | C4-TATA-C4 | 620/570 | 0.15 |
| C4-AATT-C4 | 630/580 | 0.32 | C4-TTAA-C4 | 615/570 | 0.21 |
Preparation method for Ag NCs
In this work, the DNA–Ag+ conjugated complex were firstly prepared by adding AgNO3 to the aqueous solution containing the DNA templates. After that the silver clusters were prepared by reduction of NaBH4. The DNA concentration in the buffer solution is 10 μM and the molar ratio of DNA/Ag+/BH4− is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. To enhance the fluorescence signals from the clusters, we made an investigation into the changes in the fluorescence intensities of the Ag NCs at different molar ratios of DNA/Ag+ ranged from 1
1. To enhance the fluorescence signals from the clusters, we made an investigation into the changes in the fluorescence intensities of the Ag NCs at different molar ratios of DNA/Ag+ ranged from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 to 1
6 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 27 and different molar ratios of Ag+/BH4− in the range of 6
27 and different molar ratios of Ag+/BH4− in the range of 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5–6
0.5–6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6, respectively. We also measured the fluorescence intensities of the Ag clusters over a wide range of pH values from 3.6 to 10.6 by CD spectroscopy. During the preparation of the Ag clusters, the DNA–Ag+ mixtures were firstly incubated for 10 min at 80 °C and then naturally cooled to room temperature for about 25 min. After the addition of NaBH4 to the buffer solution, the DNA–Ag+ mixtures were then reduced and the reaction was kept for 12 h at room temperature.
6, respectively. We also measured the fluorescence intensities of the Ag clusters over a wide range of pH values from 3.6 to 10.6 by CD spectroscopy. During the preparation of the Ag clusters, the DNA–Ag+ mixtures were firstly incubated for 10 min at 80 °C and then naturally cooled to room temperature for about 25 min. After the addition of NaBH4 to the buffer solution, the DNA–Ag+ mixtures were then reduced and the reaction was kept for 12 h at room temperature.
Circular dichroism
To characterize the secondary structure of the DNA, DNA–Ag+ and DNA–Ag cluster complex, circular dichroism (CD) spectroscopy experiments were carried out by using a J-810 spectropolarimeter. All of the samples in this work were scanned within a wavelength ranged from 200 to 340 nm under the protection of N2.Fluorescence emission spectroscopy
The measurements of fluorescence emissions were performed with an F-7000 fluorescence spectrophotometer (Hitachi) at 25 °C. The emission/excitation spectra were obtained at an interval of 5 nm excitation wavelength.Transmission electron microscopy
In this work, the size distribution of the Ag NCs can be calculated by using the transmission electron microscopy (TEM) images of the Ag clusters, which were captured by a JEM-2100 transmission electron microscope (JEOL). The samples of the Ag clusters were dropped on the carbon-coated copper grids and dried at 25 °C.Calculation of fluorescence QYs
The fluorescence QYs are defined as the ratio of the number of the photons emitted vs. the number of photons absorbed, which can be obtained by comparing the integrated fluorescence intensity and absorption of clusters vs. a reference fluorophore dye with a known fluorescence QY.23 Then, the fluorescence QYs of clusters can be calculated with the equation below:where φdye is the QYs of reference sample (rhodamine B with the QY of 0.31), mNC and mdye denote the fitted slopes to fluorescence vs. absorption for the cluster and reference fluorophore, respectively. In this work, for all the cluster samples, we prepared a serial solution to ensure that the absorbance value at the peak excitation wavelength was approximately 0.08, 0.06, 0.04, 0.02, and 0.01, respectively. All the fluorescence intensities were integrated over the entire emission spectral range.
Results and discussion
Effect of the DNA loop sequence on Ag NCs
To investigate the effects of the loop sequences in i-motif DNA templates on Ag NCs, a set of sequences C4-X1X2X3X4-C4 were used as the DNA templates for stabilizing Ag NCs (Table 1). It is well known that CD spectroscopy is currently the most effective method for characterizing DNA secondary structure including i-motif structure.18,24 All of the DNA i-motif structures had been confirmed by appearance of the two CD spectral bands with a positive helicity at 290 nm and a negative helicity at 255 nm (Fig. S1, ESI†). As illustrated in Table 1, for the template sequence C4-AAAA-C4, the Ag NCs exhibited an emission peak at the λem = 630 nm and their average QY is 0.24. When the template sequence is C4-TTTT-C4, no fluorescence can be measured. We further evaluated the effect of the position of thymine in the loop sequence on the fluorescence of clusters by calculating the QYs of the Ag NCs (Table 1). Compared with the sequence C4-AAAA-C4, the sequence C4-TAAA-C4 or C4-AAAT-C4 can be used to prepare the silver clusters with high QYs. However, when the X2 or X4 in the loop sequence were replaced by thymine (e.t -ATAA-, or -AATA-), the fluorescence QYs were obviously lowered, indicating that the QYs of clusters are strongly dependent on the loop base composition. Furthermore, this result shows that changing the position of thymine can modulate the fluorescence QY of Ag NCs. The interaction of the central four bases with Ag+ ions may influence the loop structural flexibility and thus further affect the overall stability of i-motif structure. This result suggests that the alignment of single thymine adjacent to the cytosine (e.t C4-TAAA-C4, or C4-AAAT-C4) is favorable for high QYs clusters. Meanwhile, as illustrated in Table 1, Ag NCs showed significantly difference in the QYs for the loop “AAAT” (QY = 0.48) and for the loop “TAAA” (QY = 0.29).By comparing the QYs of the prepared Ag clusters, the sequence C4-ATAT-C4 is the best template for the preparation of the bright fluorescent Ag NCs with a high QY of 0.78. However, when using the reversed sequence C4-TATA-C4 as the template, the Ag NCs exhibited a much lowered QY of 0.15. This distinction in the fluorescence QYs further shows that the base arrangement direction of loop sequence is well correlated with the fluorescence emission of Ag NCs. To better understanding this correlation, we compared the CD spectra of the two sequences and their complex binding with silver ions. Both of the sequences (C4-TATA-C4, C4-ATAT-C4) exhibited almost the same CD spectra, suggesting the formation of the same i-motif structure (Fig. S2A, ESI†). However, after adding Ag+ ions, the CD spectra of the two DNA–Ag+ complexes showed the significant changes in their respective ellipticities at 220 nm (herein, the ellipticity is −14 for the sequence C4-ATAT-C4 and -23 for the sequence C4-TATA-C4, respectively) (Fig. S2B, ESI†). This difference in the relative ellipticities indicates that the binding affinities of the two sequences to silver ions are different, thus leading to the formation of the silver clusters with different sizes.25
A possible reason for the effect of the loop sequences on the Ag NCs is that the variation of base sequences could lead to the different microenvironments required for the preparation and stabilization of Ag NCs, thus causing their different fluorescence properties.
Effect of the length of DNA loop sequence on Ag NCs
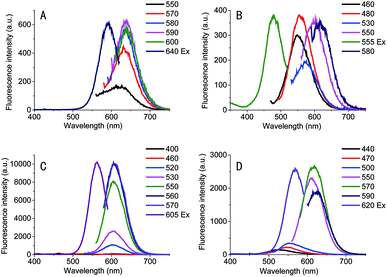
To further examine the length of loop sequences on the fluorescence of Ag NCs, we measured the fluorescence spectra of the Ag NCs stabilized by the sequence C4-An-C4 (herein, n = 1, 2, 3, and 4), as illustrated in Fig. 1. It was found that the Ag NCs stabilized by the sequence C4-A-C4 showed a sharp spectral peak at the λem = 640 nm under various excitation wavelengths, suggesting that single type of clusters was formed. When an additional base A is added into the loop region (e.t. C4-A2-C4), some different emission peaks appear at series of excitation wavelength, suggesting the formation of various types of silver species (Fig. 1). When using sequence C4-A3-C4 as the template, the Ag NCs showed a spectral peak at the λex = 605 nm under various excitations and fluorescent intensity is much larger than that of other Ag NCs using the template sequence C4-A-C4 or C4-A2-C4. However, in the case of C4-A4-C4, the fluorescent intensity was lower obviously than that of Ag clusters stabilized by the sequence C4-A3-C4. These variations in fluorescence QYs may be associated with the loop structural change and is dependent on the binding affinity of silver ions and loop bases. With the increase in the number of base A at loop region, the loop-Ag+ binding affinity may be enhanced accordingly, thus likely leading to a more tightly loop structure. Our result indicates that the length of loop sequences in i-motif DNA templates is important for the stability of i-motif structures.Characterization of Ag NCs
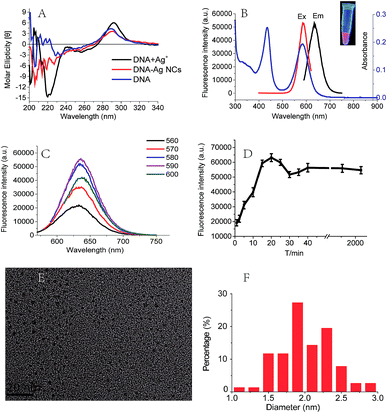
Since the DNA–Ag+ binding interactions is important for the formation of Ag NCs,12,13,26 we further investigated the interactions between the sequence C4-ATAT-C4 and Ag+ using CD and UV spectroscopy.14,27 As illustrated in Fig. 2A, the CD spectra for the DNA (C4-ATAT-C4) and Ag+ mixture showed the obvious and changes at 220 and 260 nm, respectively, which suggest the binding of DNA with silver ions. The Ag NCs can emit a prominent red fluorescence at the λem = 635 nm (Fig. 2B). In the range of λex = 560–600 nm, an emission peak can be found at the λem = 635 nm and its maximum peak occurred at the λex = 590 nm (Fig. 2C). In addition, the UV absorption spectrum of DNA–cluster complex showed an obvious peak at 588 nm (Fig. 2B, blue line), which is close to the maximum λex = 590 nm, further suggesting the aggregated silver ions had been reduced into the silver clusters. It should be noted that the UV absorption peak at 440 nm (Fig. 2B, blue line) indicates the formation of larger Ag nanoparticles, which agrees well with the previous work.28 As shown in Fig. 2D, the fluorescence intensity of DNA–cluster complexes increased to 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 after one minute upon the addition of NaBH4 and then reached up to its maximum peak within 20 minutes. Finally, the fluorescence intensity maintained 55
000 after one minute upon the addition of NaBH4 and then reached up to its maximum peak within 20 minutes. Finally, the fluorescence intensity maintained 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 for more than 48 hours. The red fluorescence of the Ag NCs can be maintained for more than 5 days at room temperature and for more than 6 weeks at 4 °C.
000 for more than 48 hours. The red fluorescence of the Ag NCs can be maintained for more than 5 days at room temperature and for more than 6 weeks at 4 °C.
The morphologies and sizes of Ag NCs were determined by TEM. The TEM image (Fig. 2E) showed the Ag NCs stabilized by the sequence C4-ATAT-C4 were of a spherical shape and well dispersed. The size distribution histogram (Fig. 2F) showed the observed diameters of the Ag NCs ranged from 1.0 to 3.0 nm and the average diameter was 2.0 nm. For the Ag NCs stabilized by the sequence C4-TATA-C4, the observed diameters of the Ag NCs ranged from 2.3 to 4.7 nm and the average diameter was 3.3 nm (Fig. S3A and B, ESI†). It has been known that the fluorescence emissions of the Ag clusters are closely related to the sizes of Ag NCs.23 Such differences in the size distributions and diameters of the Ag NCs indicate that the two loop sequences with the opposite directions have a different impact on the formation of the Ag NCs thus leading to the silver clusters' completely different QYs, which further confirms the above CD spectral results.
Effect of the reductants and Ag+ concentration on Ag NCs
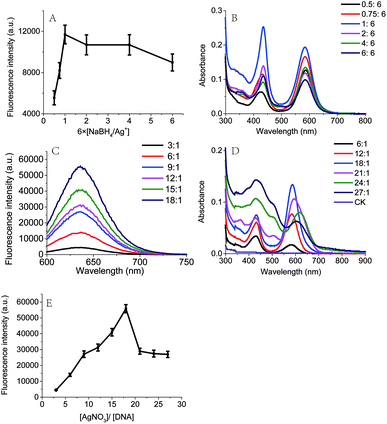
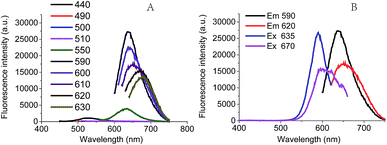
In this work, the red fluorescence emitted from Ag NCs was found to be enhanced with the increase in molar ratio [NaBH4/Ag+], and reached its maximal value upon the [NaBH4/Ag+] = 6 (Fig. 3A), at which the clusters exhibited the maximal UV absorption peak (Fig. 3B). We found that the fluorescence intensity of Ag NCs at low concentration of NaBH4 was much higher than that at high concentration of NaBH4. This result shows that the fluorescence character of Ag NCs can be affected by the molar ratio [NaBH4/Ag+], which is in good agreement with the previous report.29We further examined the effect of the molar ratio [Ag+/DNA] on Ag NCs by increasing the Ag+ concentration. As shown in Fig. 3C, the fluorescence intensity can be increased with the concentration of silver ions, and the maximal fluorescence emission peak occurred when the molar ratio [Ag+/DNA] is 18. This result indicates that the formation of Ag NCs is closely correlated with the concentration of silver ions. When the molar ratio [Ag+/DNA] is higher than 21, the fluorescence intensity became lowered and the red shift can be obviously observed in Fig. 3D. When the molar ratio [Ag+/DNA] was 24, the UV absorption peak occurred at 620 nm (Fig. 3D, green), suggesting that the most optimal excitation wavelength should be 620 nm. However, (under the same condition), as illustrated in Fig. 4A, the maximum fluorescence emission peak occurred at the λex = 590 nm, and the red shift occurred as the excitation wavelength increased. Moreover, when the excitation wavelength is 620 nm, the fluorescence emission peak occurred at 670 nm (Fig. 4B). This red shift indicates the formation of the larger silver nanoparticles at high concentration of Ag+ ions (the molar concentration ratio [Ag+/DNA] = 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). This result indicates that the formation of silver clusters is also closely correlated with Ag+ concentrations.
1). This result indicates that the formation of silver clusters is also closely correlated with Ag+ concentrations.
Effect of the pH on Ag NCs
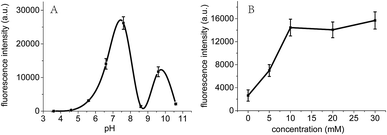
It is known that C-rich DNA template sequences are easy to form i-motif structures due to the hemi-protonated C–C pairing interactions in weakly acidic or neutral solution.30,31 Therefore, the preparation and stabilization of Ag NCs could be influenced by pH of solvents. Fig. 5 showed a significant increase (∼100 times) in the fluorescence intensity of Ag NCs as the pH increased from 4.6 to 7.6. The Ag NCs showed a maximum fluorescence intensity at pH 7.6 at which the atom N3 of cytosine (pKa = 4.6) and thymine (pKa = 9.6) are primarily in deprotonated and protonated states, respectively. This distinction between the pKa's and the optimal pH for fluorescence indicates that the degree of protonation of the individual bases could not influence cluster formation, which is in good agreement with previous findings.27,30,32 It is likely that the coordination interactions between multiple loop bases and Ag clusters at pH 7.6 could offer the most preferred microenvironments for the formation of homogenous Ag NCs, thus leading to their maximum fluorescence QYs. To further examine the effects of pH on i-motif structures, we further measured the CD spectra of the sequence C4-ATAT-C4 over a wide pH range from 4.6 to 9.6 (Fig. S4, ESI†). It was shown that almost all of the spectra (except for that at pH 9.6) showed similar spectral band variations, which is efficient to confirm that a specific i-motif structure can be formed under various pH conditions. Meanwhile, it should be noted that the relative ellipticities had gradually lowered as pH increased, suggesting the gradually weakened i-motif structure above pH 4.6.Since PB is associated with the degree of the protonation of cytosine bases,33 we measured the fluorescence intensity of the prepared Ag clusters as a function of the PB concentration to examine the effect of PB concentrations on Ag clusters. As shown in Fig. 5B, the emitting fluorescence intensity of the Ag NCs gradually increased to 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 as the PB concentration increased to 10 mM, then maintained at ∼14
000 as the PB concentration increased to 10 mM, then maintained at ∼14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. This result indicates that the optimization of PB concentrations can enhance the binding interactions of cytosine bases with silver ions, thus promoting the formation of Ag NCs, which is in good agreement with the previous report.34
000. This result indicates that the optimization of PB concentrations can enhance the binding interactions of cytosine bases with silver ions, thus promoting the formation of Ag NCs, which is in good agreement with the previous report.34
To further investigate the effect of temperature on the fluorescence of Ag NCs, we have measured the UV absorption spectra and fluorescence intensity of Ag NCs as a function of temperature, respectively (Fig. S5, ESI†). Our results showed that the room temperature (25–30 °C) was the most favorable for the formation of the high-QYs Ag NCs in this study.
Conclusion
In this work, by comparisons of the fluorescence QYs of the prepared Ag clusters, we have uncovered the correlation between loop sequences and Ag clusters' fluorescence properties. Our results shows that loop base compositions, loop sequence arrangement directions and loop lengths in i-motif DNA templates can have important effects on the fluorescence properties of Ag NCs. From a serial of DNA sequences C4-X1X2X3X4-C4, we found that best DNA template is the sequence C4-ATAT-C4 which can be used to prepare Ag NCs with a high QY of 0.78. Such high-QYs silver clusters in combination with DNA aptamer probes are expected to be used for high-resolution imaging and screening of tumor cells. Additionally, we have examined the effects of reductants, Ag+ concentrations, pH, and PB concentrations on Ag NCs' fluorescence, respectively. Due to the variety of base compositions and arrangements of loop sequences in i-motif DNA templates, the synthesis condition of the Ag NCs optimized for the loop ATAT may not be directly applied for the preparation of Ag NCs stabilized by other DNA sequences. Nevertheless, we believe our research will aid to a better understanding of the dependence of fluorescent Ag clusters on the loop sequences in i-motif DNA templates. Future work will be devoted to exploring the exact reason why the loop sequences can affect the fluorescence of Ag NCs.Acknowledgements
This research was financially supported by the National Key Program for Developing Basic Research (2014CB744501), the Chinese National Key Project of Science and Technology (2013ZX10004103-002), the NSFC (61271056, 61471168, 61201100 and 61527806), and the Economical Forest Cultivation and Utilization of 2011 Collaborative Innovation Center in Hunan Province [(2013) 448].References
- J. T. Petty, S. P. Story, J. Hsiang and R. M. Dickson, J. Phys. Chem. Lett., 2013, 4, 1148–1155 CrossRef CAS PubMed.
- Y. Shiang, C. Huang, W. Chen, P. Chen and H. Chang, J. Mater. Chem., 2012, 22, 12972–12982 RSC.
- Z. Yuan, Y. Chen, H. Li and H. Chang, Chem. Commun., 2014, 50, 9800–9815 RSC.
- W. Guo, J. Yuan, Q. Dong and E. Wang, J. Am. Chem. Soc., 2010, 132, 932–934 CrossRef CAS PubMed.
- T. Zhao, Q. Chen, P. Wang and Z. Chen, RSC Adv., 2014, 4, 10390–10394 RSC.
- X. Hou, W. Guo, F. Xia, F. Nie, H. Dong, Y. Tian, L. Wen, L. Wang, L. Cao, Y. Yang, J. Xue, Y. Song, Y. Wang, D. Liu and L. Jiang, J. Am. Chem. Soc., 2009, 131, 7800–7805 CrossRef CAS PubMed.
- L. Feng, Z. Huang, J. Ren and X. Qu, Nucleic Acids Res., 2012, 40, e122 CrossRef CAS PubMed.
- G. Lan, W. Chen and H. Chang, RSC Adv., 2011, 1, 802–807 RSC.
- D. Schultz and E. Gwinn, Chem. Commun., 2011, 47, 4715–4717 RSC.
- G. H. Clever and M. Shionoya, Coord. Chem. Rev., 2010, 254, 2391–2402 CrossRef CAS.
- A. Ono, S. Cao, H. Togashi, M. Tashiro, T. Fujimoto, T. Machinami, S. Oda, Y. Miyake, I. Okamoto and Y. Tanaka, Chem. Commun., 2008, 4825–4827 RSC.
- H. Urata, E. Yamaguchi, Y. Nakamura and S.-i. Wada, Chem. Commun., 2011, 47, 941–943 RSC.
- A. Ono, H. Torigoe, Y. Tanaka and I. Okamoto, Chem. Soc. Rev., 2011, 40, 5855–5866 RSC.
- J. Sharma, R. C. Rocha, M. L. Phipps, H. Yeh, K. A. Balatsky, D. M. Vu, A. P. Shreve, J. H. Werner and J. S. Martinez, Nanoscale, 2012, 4, 4107–4110 RSC.
- N. Zhang, A. Gorin, A. Majumdar, A. Kettani, N. Chernichenko, E. Skripkin and D. J. Patel, J. Mol. Biol., 2001, 312, 1073–1088 CrossRef CAS PubMed.
- Y. Yang, Y. Sun, Y. Yang, Y. Xing, T. Zhang, Z. Wang, Z. Yang and D. Liu, Macromolecules, 2012, 45, 2643–2647 CrossRef CAS.
- N. Escaja, E. Pedroso, M. Rico and C. González, J. Am. Chem. Soc., 2000, 122, 12732–12742 CrossRef CAS.
- S. Kendrick, Y. Akiyama, S. M. Hecht and L. H. Hurley, J. Am. Chem. Soc., 2009, 131, 17667–17676 CrossRef CAS PubMed.
- C. I. Richards, S. Choi, J. Hsiang, Y. Antoku, T. Vosch, A. Bongiorno, Y. Tzeng and R. M. Dickson, J. Am. Chem. Soc., 2008, 130, 5038–5039 CrossRef CAS PubMed.
- G.-Y. Lan, C.-C. Huang and H.-T. Chang, Chem. Commun., 2010, 46, 1257–1259 RSC.
- Y. Fu, J. Zhang, X. Chen, T. Huang, X. Duan, W. Li and J. Wang, J. Phys. Chem. C, 2011, 115, 10370–10379 CrossRef CAS.
- S. A. Patel, C. I. Richards, J.-C. Hsiang and R. M. Dickson, J. Am. Chem. Soc., 2008, 130, 11602–11603 CrossRef CAS PubMed.
- D. Schultz, K. Gardner, S. S. R. Oemrawsingh, N. Markešević, K. Olsson, M. Debord, D. Bouwmeester and E. Gwinn, Adv. Mater., 2013, 25, 2797–2803 CrossRef CAS PubMed.
- N. r. Escaja, J. l. Viladoms, M. Garavís, A. Villasante, E. Pedroso and C. González, Nucleic Acids Res., 2012, 40, 11737–11747 CrossRef CAS PubMed.
- G.-Y. Lan, C.-C. Huang and H.-T. Chang, Chem. Commun., 2010, 46, 1257–1259 RSC.
- S. Johannsen, N. Megger, D. BÖhme, R. K. O. Sigel and J. Müller, Nat. Chem., 2010, 2, 229–234 CrossRef CAS PubMed.
- C. M. Ritchie, K. R. Johnsen, J. R. Kiser, Y. Antoku, R. M. Dickson and J. T. Petty, J. Phys. Chem. C, 2007, 111, 175–181 CrossRef CAS PubMed.
- S. H. Yau, N. Abeyasinghe, M. Orr, L. Upton, O. Varnavski, J. H. Werner, H. Yeh, J. Sharma, A. P. Shreve, J. S. Martinez and T. Goodson, Nanoscale, 2012, 4, 4247–4254 RSC.
- P. R. O'Neill, L. R. Velazquez, D. G. Dunn, E. G. Gwinn and D. K. Fygenson, J. Phys. Chem. C, 2009, 113, 4229–4233 CrossRef.
- B. Sengupta, K. Springer, J. G. Buckman, S. P. Story, O. H. Abe, Z. W. Hasan, Z. D. Prudowsky, S. E. Rudisill, N. N. Degtyareva and J. T. Petty, J. Phys. Chem. C, 2009, 113, 19518–19524 CrossRef CAS.
- J. M. Dettler, R. Buscaglia, J. Cui, D. Cashman, M. Blynn and E. A. Lewis, Biophys. J., 2010, 99, 561–567 CrossRef CAS PubMed.
- B. Sengupta, C. M. Ritchie, J. G. Buckman, K. R. Johnsen, P. M. Goodwin and J. T. Petty, J. Phys. Chem. C, 2008, 112, 18776–18782 CrossRef CAS.
- J. Smiatek and A. Heuer, RSC Adv., 2014, 4, 17110–17113 RSC.
- P. Shah, S. K. Cho, P. W. Thulstrup, Y.-J. Bhang, J. C. Ahn, S. W. Choi, A. Rørvig-Lund and S. W. Yang, Nanotechnology, 2014, 25, 111–120 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra22489f |
| This journal is © The Royal Society of Chemistry 2016 |