Probing molecular basis of single-walled carbon nanotube degradation and nondegradation by enzymes based on manganese peroxidase and lignin peroxidase†
Ming Chenabc,
Xiaosheng Qin*c,
Jian Lic and
Guangming Zengab
aCollege of Environmental Science and Engineering, Hunan University, Changsha 410082, China
bKey Laboratory of Environmental Biology and Pollution Control (Hunan University), Ministry of Education, Changsha 410082, China
cSchool of Civil and Environmental Engineering, Nanyang Technological University, 639798, Singapore. E-mail: xsqin@ntu.edu.sg; Fax: +65-67921650; Tel: +65-67905288
First published on 18th December 2015
Abstract
Increasing evidence has shown that carbon nanotubes (CNTs) present adverse effects on the environment and human health, which stresses the importance of exploring CNT biodegradation. In this study, we describe the molecular basis of single-walled carbon nanotube (SWCNT) biodegradation using a CNT-degrading enzyme (manganese peroxidase, MnP) and a CNT-nondegrading enzyme (lignin peroxidase, LiP, from Phanerochaete chrysosporium) with similar catalytic cycles. Our results showed that SWCNT impeded the native conformational changes in free LiP by anchoring its loop regions to avoid the degradation. In contrast, SWCNT did not limit conformational transitions in MnP and might induce larger conformational fluctuations than in free MnP by interacting with its helical and loop regions, providing the molecular basis of SWCNT degradation. SWCNT slightly affected the secondary structures and the mean smallest distances between residue pairs in LiP and MnP. These findings provide a better understanding of the biodegradation mechanism of CNTs and pre-estimating the biodegradation potential of CNTs and are useful when developing more promising CNT-degrading enzymes.
Introduction
Carbon nanotubes (CNTs) consisting of cylindrical graphite sheets exhibit diverse properties, including physical strength, light weight and electroconductivity.1–4 Researchers have been stimulated to use CNTs in a wide range of fields such as environmental remediation,5–9 drug delivery agents, sensors,10–12 and hydrogen storage.13 Despite wide applications, little is known about the structural dynamics of enzyme–CNT interactions when CNTs are subjected to different enzyme-catalyzed fates (degradation and nondegradation). Can molecular dynamics provide clues to enzyme-catalyzed fates of CNTs? Why do the same CNTs have different fates when facing enzymes with similar catalytic cycles at the molecular level? These questions are yet to be answered.Protein–CNT interaction mechanism remains largely unclear. In 2012, Calvaresi et al. pointed out that only a small number of studies investigated the protein–CNT interactions at a molecular level.14 Probing enzyme–CNT interactions can broaden our understanding of protein–CNT interactions, as enzymes are proteins in essence.15 It has been observed that some proteins such as lysozyme interacted with CNTs.14,16 Shams et al.17 investigated the interaction of actin with SWCNT through MD simulations, finding that actin formed hydrophobic interactions with SWCNT. To improve the understanding of protein–CNT interactions, He et al. probed the interactions of 20 standard amino acids with CNT, finding that four types of amino acids (Phe, Tyr, Trp and Arg) had the highest binding affinity for CNT.18
Increasing use of CNTs and other pollutants in society are bringing risks to the environment and human health.19–23 Thus, it is necessary to remove and degrade CNTs released into the environment. Unfortunately, the high aspect ratio, the aromatic structure and the size of SWCNTs make degradation of CNTs rather challenging.24 It has been demonstrated by several previous studies that biodegradation was a good choice for the removal of CNTs and other pollutants.25,26 Zhao et al. investigated the degradation of carboxylated and nitrogen-doped multiwalled carbon nanotubes (MWCNTs) by horseradish peroxidase with H2O2.27 After 80 days, carboxylated MWCNTs were partly degraded, whereas nitrogen-doped MWCNTs were decomposed completely. Sparassis latifolia mushroom could decompose the thermally-treated and raw grade carboxylated SWCNTs by lignin peroxidase (LiP).28 Lactoperoxidase from the airways together with H2O2 and NaSCN was capable of degrading oxidized SWCNTs with or without a pulmonary surfactant.29 SWCNTs were also found to be degraded by eosinophil peroxidise.30 Interestingly, Zhang et al. studied the degrading potential of ligninolytic enzymes for SWCNTs.31 They found that manganese peroxidase (MnP) from Phanerochaete chrysosporium could degrade pristine SWCNTs, but lignin peroxidase (LiP) from P. chrysosporium could not. MnP and LiP belong to the heme-containing peroxidases and have similar catalytic cycles.32,33 For efficient degradation of CNTs to reduce the adverse impact of CNTs incautiously released into the workplace on human health and the environment, it is necessary to explore the structural dynamics of CNT-degrading enzymes and CNT-nondegrading enzymes when interacting with the same CNT. Due to the similar properties and completely different catalytic effects on SWCNT of LiP and MnP from P. chrysosporium, they were a pair of ideal model systems for the present study.
In this study, we aim to analyze the interactions of a CNT-degrading enzyme and a CNT-nondegrading enzyme with the SWCNT by multiple molecular dynamics (MD) simulations using two ligninolytic enzymes, LiP and MnP, as representatives. The distinction of structural dynamics between complexes of CNT with a CNT-degrading enzyme and a CNT-nondegrading enzyme could be helpful in estimating the potential for enzymatic decomposition of CNTs and for developing more promising CNT-degrading enzymes.
Materials and methods
Molecular dynamics (MD) simulation gives a detailed overview of the interaction process between an enzyme and SWCNT at a molecular level.14 Comparison between LiP–SWCNT (LiP tends not to degrade CNT31) and MnP–SWCNT (MnP tends to degrade CNT31) could provide the initial cues to the enzyme-catalyzed fate of carbon nanotubes, because binding is an initial step for enzyme catalysis according to the induced fit theory. The starting configurations of LiP–SWCNT and MnP–SWCNT were constructed using PatchDock, a molecular docking tool taking shape complementarity into account.34 The best structures were further produced by FireDock.35 PatchDock and FireDock have been confirmed useful in the docking of CNT to a protein.14 The crystal structures of LiP (PDB code: 1LLLP36) and MnP (PDB code: 3M5Q37) from P. chrysosporium were downloaded from the Protein Data Bank.38 Ligands and water molecules were removed from these enzyme structures. SWCNT (5,5) was constructed by Nanotube Builder in Visual Molecular Dynamics (VMD).39We carried out separate simulations for LiP–water, LiP–SWCNT–water, MnP–water and MnP–SWCNT–water systems. The initial configurations are shown in Figure S1.† Single enzyme or enzyme–SWCNT complexes were positioned at the center of a cubic box solvated with SPC water model. The side length of the box is 8.98788 nm for LiP–water and LiP–SWCNT–water systems, whereas it is 7.82558 nm for MnP–water and MnP–SWCNT–water systems. Gromacs 4.6 package40,41 was used to carry out MD simulations with the OPLS-AA force field42 under periodic boundary conditions. The systems were subjected to steep descent minimization with an energy step size of 0.01, followed by 400 ps of NVT and 400 ps of NPT simulations. The time step was 2 fs. Na+ was added into the solvated box to neutralize the system. After equilibrium, a 30 ns simulation was applied to explore the structural dynamics of LiP, MnP, LiP–SWCNT and MnP–SWCNT in water solution. The algorithms for long-range electrostatics, holonomic constraints, temperature coupling and pressure coupling were Particle Mesh Ewald,43 LINCS,44 V-rescale45 and Parrinello–Rahman,46 respectively. Temperature (300 K) and pressure (1 atm) were held constant during simulation. Trajectories and energies were saved every 10 ps.
Results
Our goals were to investigate the atomic level interactions of the same CNT with CNT-degrading enzyme and CNT-nondegrading enzyme. On this basis, we analyzed the molecular basis leading to different enzyme-catalyzed fates of CNT (degradation and nondegradation). We selected CNT (5,5) for the present study, which is consistent with the previous study done by Shams et al.17 MD simulations were performed for enzyme–SWCNT complexes and the control groups that only contain enzymes to shed light on the conformational changes of simulated enzymes in the presence of the SWCNT.Binding regions
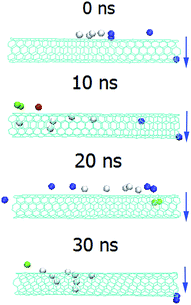
The initial conformations of LiP–SWCNT and MnP–SWCNT are shown in Figure S1.† SWCNT located only adjacent to loop regions of LiP, whereas SWCNT was positioned proximal to both α-helical and loop regions of MnP. We further extracted the binding conformations of LiP–SWCNT and MnP–SWCNT at 0 ns, 10 ns, 20 ns and 30 ns, showing that the characteristics of the binding regions of LiP–SWCNT and MnP–SWCNT were consistent with those of their respective initial ones (Fig. 1). LiP bound to SWCNT by loop regions, whereas MnP interacted with SWCNT using helices in addition to loop regions. | ||
| Fig. 1 Images of SWCNT interacting with LiP and MnP at 0, 10, 20 and 30 ns. 0 and 10 ns: front view; 20 and 30 ns: side view. | ||
To observe the variations in binding regions of LiP and MnP to SWCNT during the simulation, we retained the residues within 3 Å of SWCNT. We termed the region consisting of these residues the “3 Å-region”. We found that hydrophobic and hydrophilic residues were always within the 3 Å-region of LiP at 0, 10, 20 and 30 ns, and that charged residues only disappeared at 10 ns (Fig. 2). It was noteworthy that atoms in hydrophobic residues in the 3 Å-region were relatively more abundant than atoms in other types of residue. Our study showed that LiP residues near the SWCNT were not fixed (Table S1†). For example, at 0 ns, residues in the 3 Å-region were HIS30, PRO296, GLY297, ASN298, GLY299, PRO300, LEU328, PRO329, ILE338, PRO339, HIS341 and LYS342. After 10 ns, residues in the 3 Å-region became GLN33, GLY35, THR196, ILE199, PRO296, GLY297, GLY299, PHE303, LEU328, PRO329, ALA336 and ILE338. Interestingly, four residues (PRO296, GLY297, LEU328 and PRO329) were common at 0, 10, 20 and 30 ns, implying their important contribution to stabilizing the LiP–SWCNT interaction. Similarly, residues in the 3 Å-region of MnP also changed during the simulation (Fig. 3) and their types varied from 3 ARG, 2 ALA and 1 PHE at 0 ns to 1 ARG, 1 ALA, 2 PHE, 1 CYS, 1 ILE and 1 SER at 30 ns (Table S2†). ARG8, PHE264 and ALA267 were always found in the 3 Å-region of MnP at 0, 10, 20 and 30 ns. Thus, it was inferred that these three residues were critical to the interaction of MnP with SWCNT. In general, atoms in hydrophobic and charged residues in the 3 Å-region of MnP were more numerous than those in hydrophilic residues.
 | ||
| Fig. 2 Residue variation within 3 Å of SWCNT for LiP. Atoms in hydrophobic residues are colored in white, hydrophilic in green, and charged in red and blue. | ||
 | ||
| Fig. 3 Residue variation within 3 Å of SWCNT for MnP. Atoms in hydrophobic residues are colored in white, hydrophilic in green, and charged in red and blue. | ||
Interaction energy
Interaction energies (sum of short range coulomb and short range Lennard-Jones energies) were estimated and are shown in Fig. 4. Mean interaction energies were −394.4 and −339.6 kJ mol−1 for LiP–SWCNT and MnP–SWCNT, respectively. In general, the interaction energy of LiP–SWCNT was lower than that of MnP–SWCNT, and fluctuated within a narrow range. Fig. 4 shows that unlike LiP–SWCNT, the interaction energy of MnP–SWCNT did not quickly stabilize. The lowest and highest interaction energies of MnP–SWCNT were −443.704 and −184.73 kJ mol−1, respectively, implying a large fluctuation in interaction energy of MnP–SWCNT.Conformational transitions
One of this study's aims was to examine whether the dynamic behaviors of a CNT-degrading enzyme and a CNT-nondegrading enzyme were different when they were subjected to the same SCWNT. Comparison between trends of LiP complexed with SWCNT and free LiP based on radius of gyration (Rg), root-mean-square deviation (RMSD) and root-mean square fluctuation (RMSF) showed that LiP did not maintain its native conformation upon complexation with the SWCNT and became more stable (Fig. 5). However, MnP had the opposite tendency. | ||
| Fig. 5 Conformational transitions of LiP and MnP in the presence and absence of SWCNT. Left: LiP; Right: MnP. | ||
RMSD analysis indicated that SWCNT tended to stabilize the LiP conformation (average RMSD: 0.164 nm for LiP backbone with SWCNT and 0.201 nm for free LiP backbone), whereas MnP backbone RMSD exhibited a larger fluctuation (average RMSD = 0.301 nm) and became more unstable than free MnP backbone (0.242 nm) in the presence of SWCNT. Rg, an indicator of structural compactness,26 showed a similar trend between free LiP and unfree LiP up to about 20 ns. After 20 ns, Rg of free LiP started to vary, but Rg of unfree LiP still followed its original trend. In other words, native conformational change in free LiP did not occur in unfree LiP due to the presence of SWCNT. In contrast, SWCNT almost did not affect the Rg pattern of MnP, because the Rg lines for MnP protein with and without SWCNT basically overlapped. RMSFs are often used to describe the residue flexibility in a protein.26,47 Cα-RMSFs fluctuated remarkably around the regions consisting of residues 54–64, 175–191, 212–228 and 318–343 in free LiP during the simulation. These flexibilities were significantly reduced in LiP with SWCNT. Although the residue flexibility differed in MnP in the presence and absence of SWCNT, SWCNT did not inhibit the residue flexibility. Even in some regions, such as residues 209–227, 341–349 and 356–357, SWCNT enhanced the residue flexibility in MnP.
Secondary structure and residue–residue distance
Secondary structures of LiP in the presence and absence of SWCNT were investigated to reveal how different secondary structures varied between them (Fig. 6). For residues 1–10, the secondary structure pattern in these two complexes almost did not vary in the first period; in the period between about 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 20
000 and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ps, the secondary structural composition was transformed into bend and coil and the structural composition afterwards became coil, bend, turn and 3-helix in free LiP. For residues 11–20, in the period between 17
000 ps, the secondary structural composition was transformed into bend and coil and the structural composition afterwards became coil, bend, turn and 3-helix in free LiP. For residues 11–20, in the period between 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 30
000 and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ps, some residues tended to maintain an α-helix structure in free LiP, rather than a turn structure in unfree LiP. For other regions of LiP, secondary structural transitions were also often observed. Another common feature for LiP with and without SWCNT was that α-helix, turn, bend and coil were relatively abundant structural forms.
000 ps, some residues tended to maintain an α-helix structure in free LiP, rather than a turn structure in unfree LiP. For other regions of LiP, secondary structural transitions were also often observed. Another common feature for LiP with and without SWCNT was that α-helix, turn, bend and coil were relatively abundant structural forms.
 | ||
| Fig. 6 Changes in secondary structures of LiP ((A) with SWCNT; (B) without SWCNT) and MnP ((C) with SWCNT; (D) without SWCNT) during the simulation. | ||
Furthermore, we analyzed secondary structure plots of MnP with and without SWCNT (Fig. 6). In residues 1–50, secondary structure changes exhibited a similar pattern overall between unfree and free MnP with many minor differences. For example, secondary structures of residues 40–50 in free MnP changed frequently between turn and α-helix in the later stage of the simulation, but these residues' secondary structures in MnP with SWCNT slightly varied and were α-helix during most of the simulation time. Some regions in MnP with and without SCWNT were conserved in secondary structures such as the regions composed of residues 120–130. Turn, α-helix, bend and coil were relatively common secondary structural forms in MnP, as observed in LiP. In summary, SWCNT affected the secondary structures of both LiP and MnP, leading to many local differences observed between free and unfree forms of LiP and MnP, respectively.
Fig. 7 shows the residue–residue contact maps based on mean smallest distance. We found that the contact map of LiP with SWCNT was similar to that of LiP without SWCNT. Only very small differences were found between them. The same was true for MnP with and without SWCNT.
 | ||
| Fig. 7 Residue–residue contact maps of LiP ((A) with SWCNT; (B) without SWCNT) and MnP ((C) with SWCNT; (D) without SWCNT) based on mean smallest distance. | ||
Discussion
CNTs are acting as highly promising materials for wide applications in various fields, such as biosensors48,49 and environmental remediation.6,50–52 It was estimated that the demand for SWCNTs increased from 90 million US$ in 2009 to 600 million US$ in 2014.19 However, the increasing use of CNTs accelerated the probabilities of CNTs released into the environment. More and more studies have shown that CNTs are toxic and posed significant threats to the environment and human health.53 For example, it has been reported that CNTs could bring various harmful impacts on human health, including cancer (e.g. skin and lung cancer), inflammation, mutagenicity, epithelioid granulomas and genotoxicity.19,53 It is thus desired that efficient technologies are developed for achieving the removal of CNTs from the environment. In this regard, the application of biodegradation technology for CNTs has been confirmed to be successful, as multiple types of enzymes have been found to have the ability to degrade CNTs from the previous studies, including MnP,31 horseradish peroxidise,27 lactoperoxidase,29 and eosinophil peroxidise.30 However, to date, no studies have investigated the molecular basis of CNT degradation and nondegradation by enzymes. In this study, we analyzed the effects of the same SWCNT (5,5) on structural dynamics in the CNT-degrading enzyme (MnP) and the CNT-nondegrading enzyme (LiP), looking for the initial clues to enzyme-catalyzed fates of CNTs through MD simulations. Previously, MD simulations have been confirmed to be an efficient method for the exploration of the interactions of CNTs with enzyme,14 DNA,4 antibodies,54 and other types of proteins.17Binding regions of SWCNT to LiP and MnP were different in secondary structure. SWCNT tended to be wrapped by the loop regions of LiP and remained close to the loop and helical regions of MnP during the simulation (Fig. 1). Hydrophobic residues were generally more abundant than hydrophilic residues in the 3 Å-regions of SWCNT (Fig. 2 and 3). Shams et al. mentioned that the dominance of hydrophobic residues in contact with SWCNT might be attributed to the nonpolarity and electric neutrality of SWCNT.17 In addition to hydrophobic residues, hydrophilic and charged residues also might contribute to the interactions of LiP and MnP with SWCNT. Our results showed that the interacting residues of LiP and MnP with SWCNT were not fixed during the simulation. Despite the high variations in these residues, some residues were always within 3 Å-regions of SWCNT at 0, 10, 20 and 30 ns, including PRO296, GLY297, LEU328 and PRO329 of LiP and ARG8, PHE264 and ALA267 of MnP. We suggested these residues were potentially important to the interactions of SWCNT with LiP and MnP.
Native conformational variation in free LiP was impeded by the SWCNT based on RMSD, RMSF and Rg results (Fig. 5). According to the induced fit theory,55,56 conformational transition is necessary for enzymatic degradation. Thus, LiP was incapable of degrading SWCNT, as previously observed in experimental research.31 In MnP–SWCNT, SWCNT did not prevent MnP from maintaining its native conformational changes. In addition, it appeared that SCWNT enhanced the conformational change in MnP on the basis of RMSD and RMSF results. This might be one of the reasons why pristine SWCNT could be degraded by MnP in the experimental study.31 This finding, related to different dynamic behavior of CNT-degrading and CNT-nondegrading enzymes in the presence of CNTs, was useful for pre-estimation of the potential for enzymatic degradation of CNTs, selection of suitable enzymes or microbes for bioremediation of CNT-contaminated environments and the design of more efficient CNT-degrading enzymes.
Interaction energy between LiP and SWCNT was generally lower than that between MnP and SWCNT (Fig. 4), implying a stronger LiP–SWCNT interaction. The strong interaction between SWCNT and CNT-nondegrading enzymes might have potential applications such as enzyme immobilization to enhance the stability and catalytic activity.57,58
An interesting phenomenon was the transitions in secondary structures and residue–residue mean smallest distance (Fig. 6 and 7). Dominant secondary structure forms were α-helix, turn, bend and coil in LiP and MnP regardless of whether SWCNT was present or not. SWCNT influenced the secondary structural patterns and residue–residue mean smallest distances in LiP and MnP, leading to slight differences between free and unfree proteins. These findings implied that SWCNT allowed minor secondary structural changes and residue fluctuations in the CNT-degrading enzyme but it impeded the native conformational transitions in CNT-nondegrading enzyme.
Conclusions
Our study results revealed the molecular basis of SWCNT degradation and nondegradation by enzymes through molecular dynamics. The SWCNT binding region is located adjacent to helical and loop regions of MnP (a CNT-degrading enzyme) and loop regions of LiP (a CNT-nondegrading enzyme). PRO296, GLY297, LEU328 and PRO329 of LiP and ARG8, PHE264 and ALA267 of MnP were potentially important to the binding of SWCNT to LiP and MnP. Conformational transition in free CNT-nondegrading enzyme, but not in CNT-degrading enzyme, was impeded by the presence of SWCNT. Our study is beneficial for understanding the CNT-biodegrading mechanism and finding and developing more efficient enzymes for remediation of CNT-contaminated environments.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
The study was financially supported by the National Natural Science Foundation of China (51508177, 51521006), the Program for Changjiang Scholars and Innovative Research Team in University (IRT-13R17) and the Fundamental Research Funds for the Central Universities.References
- R. R. Johnson, A. C. Johnson and M. L. Klein, Nano Lett., 2008, 8, 69–75 CrossRef CAS PubMed.
- H. Liu, D. Nishide, T. Tanaka and H. Kataura, Nat. Commun., 2011, 2, 309 CrossRef PubMed.
- B. Pan, D. Zhang, H. Li, M. Wu, Z. Y. Wang and B. S. Xing, Environ. Sci. Technol., 2013, 47, 7722–7728 CrossRef CAS PubMed.
- D. Roxbury, J. Mittal and A. Jagota, Nano Lett., 2012, 12, 1464–1469 CrossRef CAS PubMed.
- X. Gui, Z. Zeng, Z. Lin, Q. Gan, R. Xiang, Y. Zhu, A. Cao and Z. Tang, ACS Appl. Mater. Interfaces, 2013, 5, 5845–5850 CAS.
- S. Vadahanambi, S.-H. Lee, W.-J. Kim and I.-K. Oh, Environ. Sci. Technol., 2013, 47, 10510–10517 CAS.
- V. K. Gupta, R. Kumar, A. Nayak, T. A. Saleh and M. Barakat, Adv. Colloid Interface Sci., 2013, 193, 24–34 CrossRef PubMed.
- J.-L. Gong, B. Wang, G.-M. Zeng, C.-P. Yang, C.-G. Niu, Q.-Y. Niu, W.-J. Zhou and Y. Liang, J. Hazard. Mater., 2009, 164, 1517–1522 CrossRef CAS PubMed.
- P. Xu, G. M. Zeng, D. L. Huang, C. L. Feng, S. Hu, M. H. Zhao, C. Lai, Z. Wei, C. Huang and G. X. Xie, Sci. Total Environ., 2012, 424, 1–10 CrossRef CAS PubMed.
- S. Cui, H. Pu, G. Lu, Z. Wen, E. C. Mattson, C. Hirschmugl, M. Gajdardziska-Josifovska, M. Weinert and J. Chen, ACS Appl. Mater. Interfaces, 2012, 4, 4898–4904 CAS.
- P. W. Barone, S. Baik, D. A. Heller and M. S. Strano, Nat. Mater., 2005, 4, 86–92 CrossRef CAS PubMed.
- Q. Cao and J. A. Rogers, Adv. Mater., 2009, 21, 29–53 CrossRef CAS.
- E. Tylianakis, G. K. Dimitrakakis, F. J. Martin-Martinez, S. Melchor, J. A. Dobado, E. Klontzas and G. E. Froudakis, Int. J. Hydrogen Energy, 2014, 39, 9825–9829 CrossRef CAS.
- M. Calvaresi, S. Hoefinger and F. Zerbetto, Chem.–Eur. J., 2012, 18, 4308–4313 CrossRef CAS PubMed.
- Q. Hu, P. S. Katti and Z. Gu, Nanoscale, 2014, 6, 12273–12286 RSC.
- E. Wu, M. O. Coppens and S. Garde, Langmuir, 2015, 31, 1683–1692 CrossRef CAS PubMed.
- H. Shams, B. D. Holt, S. H. Mahboobi, Z. Jahed, M. F. Islam, K. N. Dahl and M. R. K. Mofrad, ACS Nano, 2014, 8, 188–197 CrossRef CAS PubMed.
- Z. J. He and J. Zhou, Carbon, 2014, 78, 500–509 CrossRef CAS.
- A. A. Shvedova, A. Pietroiusti, B. Fadeel and V. E. Kagan, Toxicol. Appl. Pharmacol., 2012, 261, 121–133 CrossRef CAS PubMed.
- G. Zeng, M. Chen and Z. Zeng, Science, 2013, 340, 1403 CrossRef CAS PubMed.
- G. Zeng, M. Chen and Z. Zeng, Nature, 2013, 499, 154 CrossRef CAS PubMed.
- X. Qin, Stochastic Environmental Research and Risk Assessment, 2012, vol. 26, pp. 43–58 Search PubMed.
- X. Qin, Y. Xu and J. Su, Environ. Eng. Sci., 2011, 28, 573–584 CrossRef CAS.
- T. D. Berry, T. R. Filley and R. A. Blanchette, Environ. Pollut., 2014, 193, 197–204 CrossRef CAS PubMed.
- M. Chen, P. Xu, G. Zeng, C. Yang, D. Huang and J. Zhang, Biotechnol. Adv., 2015, 33, 745–755 CrossRef CAS PubMed.
- M. Chen, G. M. Zeng, C. Lai, J. Li, P. Xu and H. P. Wu, RSC Adv., 2015, 5, 52307–52313 RSC.
- Y. Zhao, B. L. Allen and A. Star, J. Phys. Chem. A, 2011, 115, 9536–9544 CrossRef CAS PubMed.
- G. Chandrasekaran, S. K. Choi, Y. C. Lee, G. J. Kim and H. J. Shin, J. Ind. Eng. Chem., 2014, 20, 3367–3374 CrossRef CAS.
- K. Bhattacharya, R. El-Sayed, F. T. Andon, S. P. Mukherjee, J. Gregory, H. Li, Y. C. Zhao, W. Seo, A. Fornara, B. Brandner, M. S. Toprak, K. Leifer, A. Star and B. Fadeel, Carbon, 2015, 91, 506–517 CrossRef CAS.
- F. T. Andón, A. A. Kapralov, N. Yanamala, W. Feng, A. Baygan, B. J. Chambers, K. Hultenby, F. Ye, M. S. Toprak and B. D. Brandner, Small, 2013, 9, 2721–2729 CrossRef PubMed.
- C. Zhang, W. Chen and P. J. Alvarez, Environ. Sci. Technol., 2014, 48, 7918–7923 CrossRef CAS PubMed.
- T. D. Bugg, M. Ahmad, E. M. Hardiman and R. Rahmanpour, Nat. Prod. Rep., 2011, 28, 1883–1896 RSC.
- Z. Huang, C. Liers, R. Ullrich, M. Hofrichter and M. A. Urynowicz, Fuel, 2013, 112, 295–301 CrossRef CAS.
- D. Schneidman-Duhovny, Y. Inbar, R. Nussinov and H. J. Wolfson, Nucleic Acids Res., 2005, 33, W363–W367 CrossRef CAS PubMed.
- E. Mashiach, D. Schneidman-Duhovny, N. Andrusier, R. Nussinov and H. J. Wolfson, Nucleic Acids Res., 2008, 36, W229–W232 CrossRef CAS PubMed.
- T. Choinowski, W. Blodig, K. H. Winterhalter and K. Piontek, J. Mol. Biol., 1999, 286, 809–827 CrossRef CAS PubMed.
- M. Sundaramoorthy, M. H. Gold and T. L. Poulos, J. Inorg. Biochem., 2010, 104, 683–690 CrossRef CAS PubMed.
- P. W. Rose, C. Bi, W. F. Bluhm, C. H. Christie, D. Dimitropoulos, S. Dutta, R. K. Green, D. S. Goodsell, A. Prlić and M. Quesada, Nucleic Acids Res., 2013, 41, D475–D482 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- B. Hess, C. Kutzner, D. van Der Spoel and E. Lindahl, J. Chem. Theory Comput., 2008, 4, 435–447 CrossRef CAS PubMed.
- S. Pronk, S. Pall, R. Schulz, P. Larsson, P. Bjelkmar, R. Apostolov, M. R. Shirts, J. C. Smith, P. M. Kasson, D. van der Spoel, B. Hess and E. Lindahl, Bioinformatics, 2013, 29, 845–854 CrossRef CAS PubMed.
- G. A. Kaminski, R. A. Friesner, J. Tirado-Rives and W. L. Jorgensen, J. Phys. Chem. B, 2001, 105, 6474–6487 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- B. Hess, H. Bekker, H. J. C. Berendsen and J. G. E. M. Fraaije, J. Comput. Chem., 1997, 18, 1463–1472 CrossRef CAS.
- G. Bussi, D. Donadio and M. Parrinello, J. Chem. Phys., 2007, 126, 014101 CrossRef PubMed.
- M. Parrinello and A. Rahman, J. Appl. Phys., 1981, 52, 7182–7190 CrossRef CAS.
- K. M. Kumar, A. Anbarasu and S. Ramaiah, Mol. BioSyst., 2014, 10, 891–900 RSC.
- R. J. Chen, S. Bangsaruntip, K. A. Drouvalakis, N. W. S. Kam, M. Shim, Y. Li, W. Kim, P. J. Utz and H. Dai, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 4984–4989 CrossRef CAS PubMed.
- W. Yang, K. R. Ratinac, S. P. Ringer, P. Thordarson, J. J. Gooding and F. Braet, Angew. Chem., Int. Ed., 2010, 49, 2114–2138 CrossRef CAS PubMed.
- W.-W. Tang, G.-M. Zeng, J.-L. Gong, Y. Liu, X.-Y. Wang, Y.-Y. Liu, Z.-F. Liu, L. Chen, X.-R. Zhang and D.-Z. Tu, Chem. Eng. J., 2012, 211, 470–478 CrossRef.
- W.-W. Tang, G.-M. Zeng, J.-L. Gong, J. Liang, P. Xu, C. Zhang and B.-B. Huang, Sci. Total Environ., 2014, 468, 1014–1027 CrossRef PubMed.
- R. Kumar, M. A. Khan and N. Haq, Crit. Rev. Environ. Sci. Technol., 2014, 44, 1000–1035 CrossRef CAS.
- C.-w. Lam, J. T. James, R. McCluskey, S. Arepalli and R. L. Hunter, Crit. Rev. Toxicol., 2006, 36, 189–217 CrossRef CAS PubMed.
- F. De Leo, J. Sgrignani, D. Bonifazi and A. Magistrato, Chem.–Eur. J., 2013, 19, 12281–12293 CrossRef CAS PubMed.
- D. E. Koshland Jr, Proc. Natl. Acad. Sci. U. S. A., 1958, 44, 98–104 CrossRef.
- P. Csermely, R. Palotai and R. Nussinov, Trends Biochem. Sci., 2010, 35, 539–546 CrossRef CAS PubMed.
- A. P. Tavares, C. G. Silva, G. Dražić, A. M. Silva, J. M. Loureiro and J. L. Faria, J. Colloid Interface Sci., 2015, 454, 52–60 CrossRef CAS PubMed.
- N. M. Mubarak, J. R. Wong, K. W. Tan, J. N. Sahu, E. C. Abdullah, N. S. Jayakumar and P. Ganesan, J. Mol. Catal. B: Enzym., 2014, 107, 124–131 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra21814d |
| This journal is © The Royal Society of Chemistry 2016 |

