Nano hydroxyapatite particles promote osteogenesis in a three-dimensional bio-printing construct consisting of alginate/gelatin/hASCs
Xiao-Fei Wang,
Pei-Jun Lu*,
Yang Song,
Yu-Chun Sun,
Yu-Guang Wang and
Yong Wang*
Center of Digital Dentistry, Faculty of Prosthodontics, Peking University School and Hospital of Stomatology, National Engineering Laboratory for Digital and Material Technology of Stomatology, Research Center of Engineering and Technology for Digital Dentistry of Ministry of Health, Beijing 100081, China. E-mail: kqlpj@bjmu.edu.cn; kqcadc@bjmu.edu.cn
First published on 12th January 2016
Abstract
To design a hydrogel material containing nano hydroxyapatite particles for three-dimensional (3D) bio-printing of human adipose-derived stem cells (hASCs) and to explore whether nano hydroxyapatite particles can promote osteogenic differentiation of a 3D bio-printing construct consisting of hASCs in vivo and in vitro. A 3D reticular printing structure was designed. Sodium alginate/gelatin/hASCs (AG group) was considered as the control group, and sodium alginate/gelatin/nano hydroxyapatite/hASCs (AGH group) was considered as the experimental group. Immunofluorescence microscopy was used to observe the cell viability and cell adhesion, and cell proliferation was analyzed by comparison of viable cell numbers in printed constructs at 1 day and 7 days after printing. After 14 days of osteogenic induction for the AG group and AGH group, real-time quantitative PCR and immunofluorescence were used to analyse the expression of the osteogenesis-related genes Runt-related transcription factor 2 (RUNX2), osterix (OSX), and osteocalcin (OCN). New bone formation in printed constructs was observed using micro-CT, HE staining, Masson trichrome staining, and OCN immunohistochemical staining 8 weeks after being implanted. The cells in the AG group and AGH group were evenly distributed in the 3D printed constructs. The number of viable cells and cell viability both in the AG group and AGH group at 7 days after printing were higher than those at 1 day after printing (p < 0.05); however, the difference between the AG group and AGH group was not significant. At 14 days after osteogenic induction in vitro, real-time PCR results showed that the expression of osteogenesis-related genes in the AGH OM group was significantly higher than that in the AGH PM group, AG PM group, and AG OM group (p < 0.05). At 8 weeks after bio-printed construct implantation, the results of micro-CT, HE staining, Masson trichrome staining, and OCN immunohistochemical staining showed that the new bone formation in the AGH group was higher than that in the AG group (p < 0.05). The in vivo and in vitro results demonstrated that nano hydroxyapatite particles dispersed in a sodium alginate/gelatin matrix could promote osteogenic differentiation of hASCs in a 3D bio-printed construct, and this scaffold material could be considered to repair large bone tissue defects.
Introduction
Bone defects caused by caries, periodontal disease, trauma, tumours, and other conditions are commonly encountered by dentists.1 A small scale of bone defect could be resolved by the self-healing function of bone tissue, but large bone defects can only be resolved through bone tissue transplantation. The currently studied method for repairing a bone tissue defect involves bone tissue engineering, due to the difficulties faced in obtaining materials for an autogenous bone graft.2 As one of the bone tissue engineering technologies, 3D bio-printing technology has been researched in recent years.3–7 This bio-fabrication approach that is able to generate a 3D blueprint of the patient's specific disorder is needed in order to restore the functionality of the tissue and repair the defect using autologous cells. Compared with traditional bone tissue engineering, the 3D bio-printing has great potential to fabricate tissues with multiple bio-composite materials and cell types, all of which are extremely important for the advancement of bone tissue engineering. Moreover, 3D porous scaffold designs are conducive to cell–matrix interactions and more efficient blood vessels in growth.8,9In current research, the stem cells used for bone tissue defect repair are mainly bone marrow mesenchymal stem cells (BMMCs) and human adipose derived stem cells (hASCs). Compared with BMMCs, hASCs have the advantages of minimal invasive capacity, ease of access, and faster proliferation in vitro, which make these stem cells an ideal source for tissue engineering therapies.10–13 Researches have shown that hASCs in hydrogel materials still have proliferation and osteogenic differentiation potentials after 3D bio-printing, indicating their promise as a type of pluripotent stem cells.14
With the development of 3D bio-printing technology, there have been more studies on the application of hydrogel scaffolds in 3D printing.15–18 Currently, the widely studied matrix materials in bone tissue engineering are sodium alginate and gelatin hydrogel. Sodium alginate can provide 3D growth space and meets the diversity requirement of tissue engineering morphology.19 Gelatin is a commonly used cytokine carrier, and it can form a porous structure after cross-linking to facilitate the migration of cytokines.20–23 Because of the porosity of hydrogel material that is susceptible to degradation in vivo,24 the osteogenic factor coated in the hydrogel material is extremely vulnerable to enzymatic degradation in vivo, which can reduce its efficacy. For instance, the commonly used biological factor bone morphogenetic protein 2 (BMP-2) not only is easily degradable in vivo, but also requires complex in vitro synthesis, which involves high costs. Although BMP-2 already has been used in the clinical treatment of bone tissue defects and can promote stem cell osteogenic differentiation,13,25 its osteogenic promotion effect in vivo still needs improvement. Currently, many studies are being performed on reducing BMP-2 degradation in vivo.26
If we can add a bioactive molecule into the sodium alginate/gelatin that does not affect material properties or cell viability and can also promote stem cell osteogenic differentiation, it will greatly facilitate the application of 3D bio-printing in bone tissue engineering and replace the use of osteogenic factors. Hydroxyapatite is a basic component of bone tissue; nano hydroxyapatite can be easily prepared in vitro, has lower raw material cost, and has been well studied in bone tissue engineering.27–29 Researches have shown that hydroxyapatite promotes osteoblast proliferation and bone tissue formation in vivo, and inhibits the absorption of bone tissue.30,31 Another study confirmed that hydroxyapatite particles can promote bone marrow mesenchymal stem cell proliferation and osteogenic differentiation and that hydroxyapatite particle-coated titanium implants have better osteogenic effects than titanium implants without coating.32 In addition, the lesser fragility of chitosan with additive hydroxyapatite is more conducive to the formation of 3D bio-printing constructs.33 Although many studies have performed on hydroxyapatite, no studies have been performed on nano hydroxyapatite particles added to hydrogel used in 3D bio-printing systems, and no in vivo studies have been performed on whether nano hydroxyapatite can promote hASCs osteogenesis in 3D bio-printing construct.
In this study, we prepared a 3D bio-printing matrix material by mixing nano hydroxyapatite particles and sodium alginate/gelatine together. We used hASCs as seed cells and constructed a 3D structure with uniform pores through the 3D bio-printing technology. The effects of nano hydroxyapatite on hASCs osteogenic differentiation in 3D structures were observed through in vivo and in vitro experiments, which provide a basis for the application of 3D bio-printing technology in bone tissue engineering.
Materials and methods
This study was approved by the Ethics Committee of the Peking University Health Science Center, Beijing, China (LA2014227).hASCs culture and osteogenic induction
hASCs were purchased from ScienCell Company (San Diego, CA, USA). All materials were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise stated. Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), and 100× penicillin and streptomycin mixture for cell culture were purchased from Gibco (Grand Island, NY, USA).P3 hASCs were cultured in proliferation medium (PM) containing DMEM with 100 U mL−1 penicillin G and 100 mg mL−1 streptomycin and 10% FBS at 37 °C in an incubator with an atmosphere comprising 95% air and 5% CO2 and with 100% relative humidity.1 All cell-based experiments were repeated three times using hASCs from the three patients.
For inducing osteogenesis, osteogenic-induction medium (OM) comprising 10 mM β-glycerophosphate, 100 nM dexamethasone, and 200 mM ascorbic acid was used. The level of alkaline phosphatase (ALP) activity was examined on day 7 using an ALP kit according to the manufacturer's protocol. Mineralization in OM cultures was determined by staining with Alizarin red S on day 14.
Preparation of the hydrogel
The applicable concentration of hydrogel material is typically 10–20% of the mass volume ratio.34 The sodium alginate/gelatin ratio used in this study was 2%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8% (A2G8), eventually forming a 10% hydrogel material. The reason for using this concentration was mainly determined by the previous experiment results: in the five groups A1G9, A2G8, A3G7, A4G6, and A5G5, cell proliferation activity gradually increased with increase in gelatin concentration, whereas with the increase in sodium alginate concentration, cell proliferation activity gradually decreased. When sodium alginate concentration was too low, hydrogel material formation during the 3D printing process was difficult; therefore, the A2G8 group was selected based on pre-experiment results. Those concentrations of alginate and gelatin gave a suitable viscosity for 3D printing.
8% (A2G8), eventually forming a 10% hydrogel material. The reason for using this concentration was mainly determined by the previous experiment results: in the five groups A1G9, A2G8, A3G7, A4G6, and A5G5, cell proliferation activity gradually increased with increase in gelatin concentration, whereas with the increase in sodium alginate concentration, cell proliferation activity gradually decreased. When sodium alginate concentration was too low, hydrogel material formation during the 3D printing process was difficult; therefore, the A2G8 group was selected based on pre-experiment results. Those concentrations of alginate and gelatin gave a suitable viscosity for 3D printing.
Hydrogels consisting of AG and AGH were fabricated. We used 2 wt% alginate and 8 wt% gelatin dissolved in NaCl solution for cell suspension under constant stirring at 40 °C. This yielded 10 wt% AG hydrogel. Then, 1 wt% of nano hydroxyapatite was added in the AG hydrogel, and AGH hydrogel was formed after mixing. The two prepared solutions were sterilized and mixed with hASCs at 3 × 106 cells per mL. The used nanocrystalline hydroxyapatite (Sigma-Aldrich) was synthesized through a wet chemical process and was rod-like in shape, which size was less than 200 nm.35
3D bio-printing
The 3D Bioplotter (Envision Tec, Germany) was used for building 3D construct and a cuboid model was designed. The distance between each printed line was 1.5 mm. The hydrogel scaffolds were fabricated by layer-by-layer deposition. When each print was completed, the final constructs were then ionically crosslinked in CaCl2 (200 Mm L−1) for 5 min. The printed tissue constructs were seeded in a 6 cm dish containing 2 mL OM or 2 mL PM. Medium was changed three times a week.hASCs in the 3D bio-printed construct
Cell conditions in the AG group and AGH group and nano hydroxyapatite particle distribution in the AGH group were observed through an inverted light microscope immediately after the 3D bio-printing. Before SEM observation, samples were fixed overnight in 4% paraformaldehyde at 4 °C. The 3D constructs were dehydrated with a graded series of ethanol, dried in a critical point dryer (Micro Modul YO-230, Thermo Scientific, Waltham, MA, USA), mounted onto aluminum stubs, sputter coated with gold, and viewed under a field emission SEM (FESEM, Hitachi, S4800, Japan).36 Phalloidin was applied to perform immunofluorescent staining of cytoskeletal proteins within the 3D printed tissue construct according to the manufacturer's protocol. Next, cells were counterstained with DAPI for nuclear staining and visualized under a Confocal Zeiss Axiovert 650 microscope (Carl Zeiss Microimaging, Oberkochen, Germany) using the laser with wavelengths of 488 nm (green, FITC-labeled phalloidin) and 405 nm (blue, DAPI).Cell viability and adhesion
At 1 day and 7 days after printing, the hASCs cell viability and viable cell count in printed constructs were examined in the AG group and AGH group separately. 3D printed constructs were washed with phosphate-buffered saline (PBS) 3 times, incubated with saline containing 5 μmol L−1 calcein-AM (CAM) and 3 μmol L−1 propidium iodide (PI) (Dojindo, Japan) at 37 °C, incubated in 5% carbon dioxide in the incubator for 45 minutes, and washed with PBS for 3 times again. Cell viability within the structure was observed with a laser scanning confocal microscope at wavelengths of 488 nm (green, living cells) and 543 nm (red, dead cells). The numbers of green and red dots were counted using Image pro plus 6.0 software, the changes in cell number at Day 1 and Day 7 were observed, and the hASCs viability within the 3D bio-printed constructs in each group was calculated.| Cell viability = 100% × total number of living cells/(total number of living cells + total number of dead cells). |
At 1 day after printing, the AG and AGH printed 3D constructs were rinsed three times with PBS, and immunofluorescent staining for vinculin was performed according to the manufacturer's protocol (Cell Signaling Technology, Danvers, MA, USA). After staining for vinculin, the cells were counterstained with DAPI for nuclear staining and visualized under a Confocal Zeiss Axiovert 650 microscope using a laser with wavelengths of 543 nm (red, vinculin) and 405 nm (blue, DAPI).
Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR)
The printed AG and AGH 3D constructs cultured for 14 days were analyzed for gene expression (n = 3). Total RNA was extracted and reverse transcribed according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA). Real-time quantitative PCR assays were performed according to the manufacturer's protocol (KAPA Biosystems, USA). The primers for RUNX2, OSX, and OCN were synthesized by Invitrogen, which are listed in Table 1. β-actin was used as an internal standard.| Genes | Forward primer | Reverse primer |
|---|---|---|
| RUNX2 | CGCATTCCTCATCCCAGTAT | AGGGGTAAGACTGGTCATAGGA |
| OCN | CTGTATCAATGGCTGGGAGC | GCCTGGAGAGGAGCAGAACT |
| OSX | GTGCAAGGCACTATGCTAGATC | CGTTACAGGAAAGGCACGAA |
| β-actin | AGCACAATGAAGATCAAGATCAT | ACTCGTCATACTCCTGCTTGC |
Immunofluorescent staining for OCN and RUNX2
After 14 days of culture, the AG and AGH printed 3D constructs cultured in different groups were rinsed three times with phosphate-buffered saline (PBS), and immunofluorescent staining for OCN and RUNX2 was performed according to the manufacturer's protocol (Cell Signaling Technology, Danvers, MA, USA). After staining for OCN and RUNX2, the cells were counterstained with DAPI for nuclear staining and visualized under a Confocal Zeiss Axiovert 650 microscope using a laser with wavelengths of 543 nm (red, RUNX2), 488 (green, OCN), and 405 nm (blue, DAPI).38Animal experiments in vivo
The 3D bio-printed constructs cultured in OM after 7 days were implanted into the back sub-cutaneous area of nude mice for in vivo study, as reported by Hall et al.39 Implants were generally divided into four groups, including AG 3D bio-printed constructs without cells, AGH 3D bio-printed constructs without cells, AG 3D bio-printed constructs with hASCs, and AGH 3D bio-printed constructs with hASCs. The 8 week-old male BalB/c nude mice were anaesthetized with pentobarbital, and the above bio-printed constructs were placed aseptically into the dorsal subcutaneous area. At 8 weeks after surgery, the bio-printed constructs were harvested (10 implants for each group).Micro-computed tomography (micro-CT) and image analysis
To analyse ectopic bone formation, micro-CT scans were performed using a high resolution Inveon Micro-CT (Siemens, Munich, Germany). The following experimental settings were used: an X-ray voltage of 60 kVp, anode current of 200 mA and an exposure time of 400 ms for each of the 360 rotational steps. The images were used to reconstruct tomograms with a Feldkamp algorithm, using a commercial software package (Cobra EXXIM, EXXIM Computing Corp., Livermore, CA). Quantification of micro-CT images was then performed. New bone volume was evaluated using Inveon Research Workplace (Siemens, Germany).HE staining, Masson trichrome staining and immunohistochemistry
All constructs were decalcified for 7 days in 10% EDTA (pH 7.4), dehydrated and subsequently embedded in paraffin. Sections (5 mm thickness) were observed under light microscope after H.E. and Masson trichrome staining. Osteogenesis was evaluated by immunohistochemical (IHC) analysis for OCN.Statistical analysis
Data are expressed as the mean ± standard deviation and were analyzed using SPSS 17.0 software. One-way analysis of variance followed by Fisher's least significant difference test was performed. For all tests, statistical significance was accepted at p values lower than 0.05.Results
hASC cultivation and evaluation of osteogenic differentiation ability
P3 hASCs showed fibroblast-like adherent growth (Fig. 1A) with regular morphology. Cell counts of hASCs reached 4 × 106 cells in all cell culture dishes (diameter: 10 cm). After 7 days in OM, hASCs showed positive ALP staining. After 14 days in OM, Alizarin red S staining revealed the formation of mineralization nodules. The PM group did not show positive results for all tests (Fig. 1B).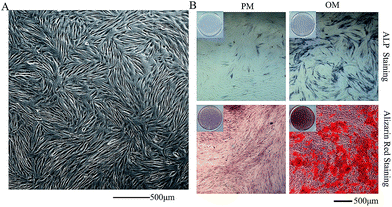 | ||
| Fig. 1 Evaluation of the osteogenic differentiation capacity of hASCs. (A) P3 hASCs; (40×) (B) ALP staining and Alizarin red staining (100×). | ||
Distribution of hASCs within 3D constructs
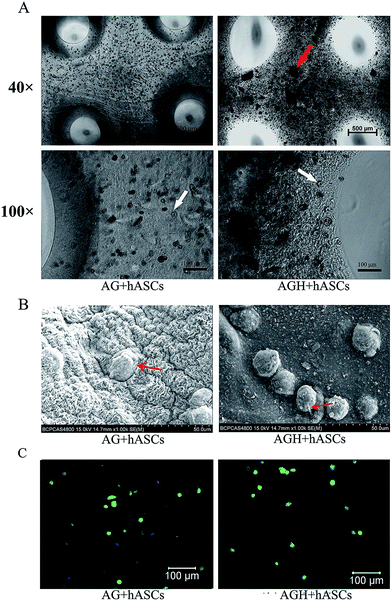
The uniform cell distribution of 3D bio-printed constructs in the AG group and AGH group could be observed using an inverted light microscope immediately after printing. Nano hydroxyapatite particles were widely dispersed around the cells in AGH hydrogel, as shown in Fig. 2A. At 24 h, the images of SEM showed that cells were successfully seeded on both AG and AGH scaffolds and extended into pseudopods (Fig. 2B). FITC-phalloidin staining helped to view the morphology of the adhered cells. Most of the cells seeded on both AG and AGH scaffolds exhibited an oval morphology, and some cells showed fibroblast-like morphology (Fig. 2C). hASCs morphology showed no significant difference between the AG group and AGH group.hASC viability and adhesion
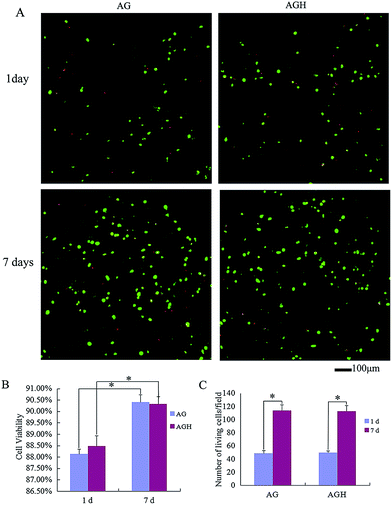
According to CAM/PI staining results, at 1 day after printing, the viable cell count of the AG group and AGH group were both significantly lesser than that at 7 days after in vitro culture, indicating that hASCs could proliferate in both printed constructs (Fig. 3A). After statistical analysis of the cell viability on day 1 and day 7 in each group, we found that the cell viability in the AG group and AGH group were 88.13% ± 0.21% and 88.48% ± 0.45%, respectively on day 1, and were 90.41% ± 0.32% and 90.33% ± 0.29%, respectively on day 7; these differences between AG group and AGH group were not statistically significant. The cell viability and viable cell count between day 1 and day 7 in the AG group showed a statistically significant difference (p < 0.05), and the cell viability and viable cell count between day 1 and day 7 in the AGH group also showed a statistically significant difference (p < 0.05) (Fig. 3B and C).Vinculin expression detected by immunofluorescence showed that cells seeded on both AG and AGH 3D scaffolds had the same vinculin-positive staining results, indicating good cell adhesion and good cell compatibility (Fig. 4).
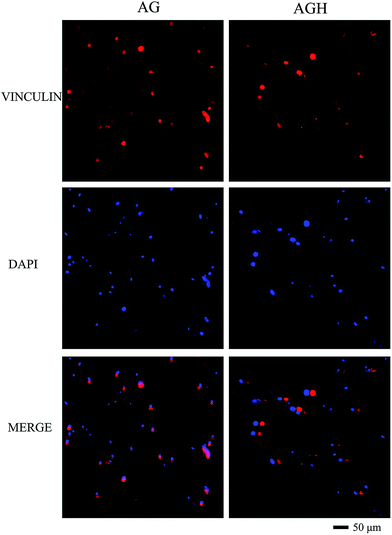 | ||
| Fig. 4 Immunofluorescent staining for vinculin in hASCs cultured in both AG and AGH 3D scaffolds (200×). Vinculin is coloured red and nuclei are coloured blue. | ||
Osteogenic differentiation of hASCs in AG and AGH 3D scaffolds
Gene expression of osteogenesis-related genes (i.e. RUNX2, OCN, and OSX) was detected after 14 days of culture. The relative RUNX2 expression in the AGH group with OM was higher than that in the AGH group with PM and AG group with both PM and OM (p < 0.05). The relative RUNX2 expression in the AGH group with PM was higher than that in the AG group with PM (p < 0.05) but lower than that in the AG group with OM (Fig. 5A). Analysis of the relative expression of OCN (Fig. 5B) and OSX (Fig. 5C) showed the same results.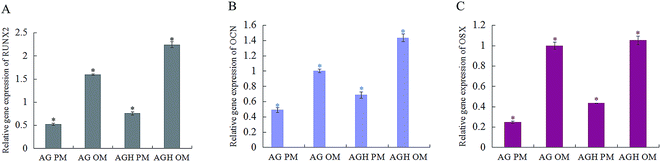 | ||
| Fig. 5 The expression of osteogenic genes in hASCs cultured for 14 days in different groups. *p < 0.05. | ||
OCN expression detected by immunofluorescence showed that, after 14 days of culture, both the AG group and the AGH group with OM had stronger OCN-positive staining than the other groups. Sporadic green fluorescence could be seen in the AGH group with PM and lesser green staining could be seen in the AG group with PM (Fig. 6). RUNX2 immunofluorescence staining showed a similar tendency (Fig. 7).
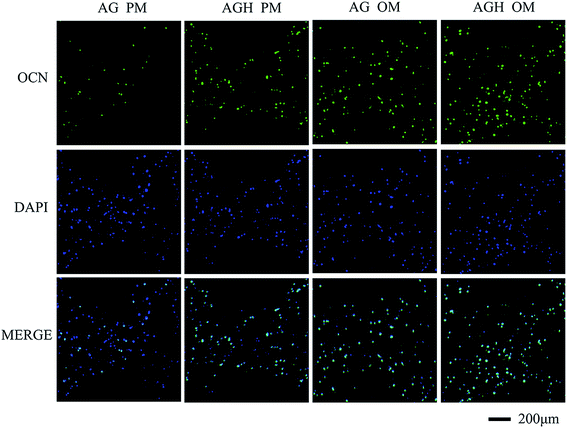 | ||
| Fig. 6 Immunofluorescent staining for OCN in hASCs cultured for 14 days in different groups (100×). OCN is coloured green and nuclei are coloured blue. | ||
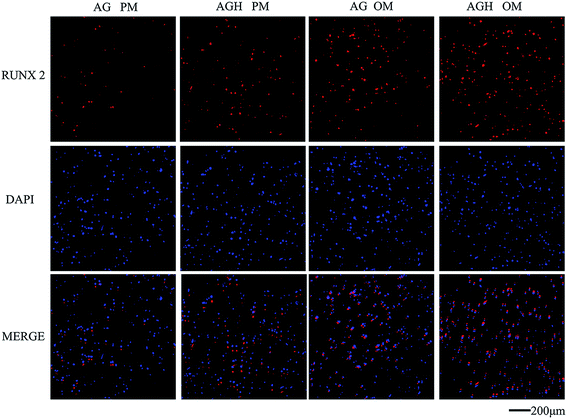 | ||
| Fig. 7 Immunofluorescent staining for RUNX2 in hASCs cultured for 14 days in different groups (100×). RUNX2 is coloured red and nuclei are coloured blue. | ||
Ectopic bone formation in vivo
The printed constructs were harvested 8 weeks after implantation, and we found that the printed construct's structures of the no-cells-added AG group and no-cells-added AGH group were relatively loose. The hydrogel material was partially degraded, which resulted in a lesser volume than that before implantation, and the texture of them was soft and brittle, which made the constructs difficult to hold. The printed constructs of the AG group with hASCs and the AGH group with hASCs basically maintained the original volume, with slight changes in shape; the texture was relatively tough, and vascular tissue growth was visible through the pores (Fig. 8).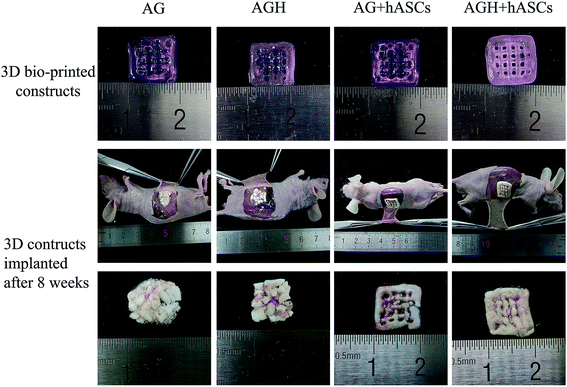 | ||
| Fig. 8 Results of morphological analysis for 3D construct in each group before and after implantation. | ||
Radiological assessment of bone formation
To assess new bone formation, micro-CT was performed. Quantification of micro-CT images provided evidence that significantly more bone was formed in the AGH group than in the AG group (p < 0.05) (Fig. 9A and B).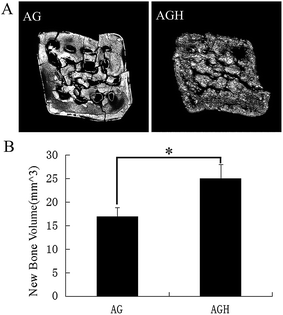 | ||
| Fig. 9 Radiological assessment of bone formation. (A) Micro-CT images of bone formation in each group after 8 weeks. (B) Quantitative comparison of new bone volume. *p < 0.05. | ||
Histological assessment of bone formation
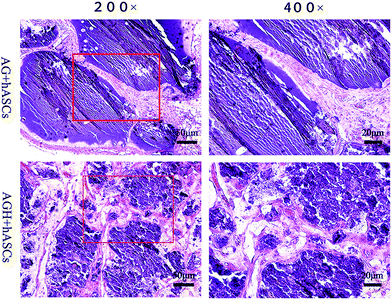
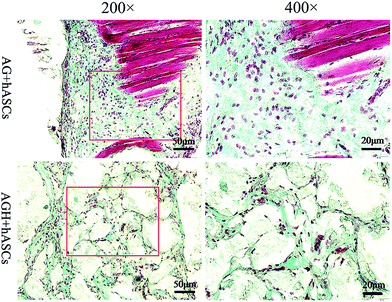
HE staining of representative sections from each group is shown (Fig. 10). In the AG group, new bone was mainly produced around the pores of the printed construct, while in the AGH group, the new bone formed a reticular structure in the entire printed construct, indicating that the new bone formation was not limited to the region around the pores of the printed construct and that high levels of bone formation could also occur inside the construct. The results of Masson-trichrome staining were consistent with those of HE staining (Fig. 11). The results of IHC staining showed that the osteogenic marker OCN was highly expressed in the new bone area (Fig. 12).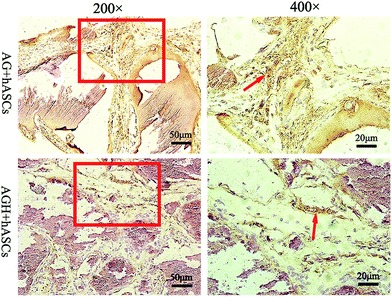 | ||
| Fig. 12 Immunohistochemical staining for the osteogenic marker OCN (200×, 400×); arrow indicates OCN protein staining. | ||
Discussion
In the present study, we found that nano hydroxyapatite particles could be mixed with sodium alginate/gelatin to form a 3D bio-printing matrix material, which had no adverse effects on the adhesion or proliferation of hASCs. Some studies reported that nano hydroxyapatite particles could promote cell proliferation and adhesion,40,41 but the cell proliferation and adhesion in the AG group and AGH group were not significantly different in this study. Possible reasons for this might be that this study used a 3D matrix material, while immunofluorescence observation used two-dimensional display; therefore, some fusiform cells only exhibited a morphology with an elliptical cross-section, and not the long fusiform shape exhibited through full cell extension reported in other studies. However, vinculin staining results proved that vinculin was expressed around almost all cells, which also indicated that hASCs could show adhesive growth in both AG matrix and AGH matrix. The viable cell count on day 7 was significantly greater than that on day 1, indicating that the cells still had proliferative capacity in the matrix material.The cell viability after the 3D bio-printing is a key indicator in evaluating the cell printing effect, and CAM/PI staining is a typical fluorescent dye used for cell activity evaluation.34,42 CAM diffused into viable cells and was hydrolysed by intracellular non-specific esterase. The product was excited by excitation light with a wavelength of 488 nm to emit green fluorescence; PI diffused into the cells with incomplete membranes (dead cells), combined with the intracellular DNA to form a compound that was excited by excitation light with a wavelength of 543 nm, and emitted red fluorescence. In this study, CAM/PI double staining results showed that the cell viability of the AG group and AGH group on day 1 of cell printing were 88.13% ± 0.21% and 88.48% ± 0.45%, respectively, and the viability of the AG group and AGH group on day 7 of cell printing were 90.41% ± 0.32% and 90.33% ± 0.29%, respectively; the differences were not statistically significant. Compared with other literature reporting cell bio-printing technology, the cell viability was higher.34,43 The reasons why the cell activity on day 7 was higher than that on day 1 might be the temperature change during the printing process (from 37 °C down to the gelatin cross-linking temperature, 4 °C) and the printer head extrusion on the cells, together with other possible reasons that had an impact on cell viability. The results in our study showed the nano hydroxyapatite particles added did not affect cell viability after 3D bio-printing. The hydroxyapatite particles used in this study had a diameter of less than 200 nm, and they were uniformly distributed in the material. Therefore, future research can be based on such hydroxyapatite particles.
In our study of the osteogenic effects of nano hydroxyapatite on the 3D hASCs construct, we found that nano hydroxyapatite could promote hASCs 3D constructs to osteogenic differentiation, which was consistent with the previous studies.30,44 The genes related to osteogenic differentiation include RUNX2, OCN, OSX, and COL-1, and RUNX2 has always been considered as an important factor in early osteogenic differentiation,45 while OSX and OCN are marker proteins in late osteogenic differentiation.46,47 The relative expression of the osteogenic differentiation-related genes RUNX2, OCN, and OSX in the AGH OM group was significantly higher than that in all the other groups in this study, and the expression in the AGH PM group was higher than that in the AG PM group. Immunofluorescence staining of OCN and RUNX2 protein showed the same results. Although few clear examples have been demonstrated and little is known about the mechanism of these cases. Some studies48,49 showed that nano hydroxyapatite had the capacity to produce sustained changes in the expression of the osteoblast differentiation marker genes by DNA methylation, which was capable of being passed to daughter cells during division. This could explain why osteogenic genes in the AGH OM group was significantly up regulated than that in all the other groups in this study. Nano-hydroxyapatite could alter osteoblast behavior through specific molecular and then promoted differentiation of stem cells and osteogenic precursor cells into osteo-blasts. However, more researches still needed to be done in the future to explore the detailed mechanism of nano hydroxyapatite on stem cells.
The subcutaneous ectopic osteogenesis in nude mice in our in vivo experiments is the key measure of testing whether there is new bone formation. The results were consistent with the preliminary studies: the amount of new bone formed by the 3D bio-printed construct containing nano hydroxyapatite particles was larger,30,44 and hASCs were once again proven to be seed cells suitable for 3D bio-printing in bone tissue engineering. In the present study, we found that the volume of material in the control group with no cell addition reduced and an irregular shape was observed; a white block shape and brittle texture were observed before fixation with 4% paraformaldehyde. After fixation, it became translucent, with a texture similar to that before implantation. This change might occur because the dehydrated hydrogel material reabsorbed moisture. The morphology of the printed construct in the two experiment groups did not change considerably, and the original morphology and texture could be maintained after fixation. Fibroid tissues within the pores on the printed constructs could be macroscopically observed, and vascular ingrowth could be seen.
Previous studies have confirmed that micro-CT is a repeatable technique for assessing the formation of new bone through analysis of mineralization and microarchitecture.50 The quantitative analysis results of micro-CT imaging in this study showed that the amount of new bone formation in AGH group was significantly higher than that in the AG group. HE staining and Masson staining results were consistent: the AG group had less new bone formation, most of which was around the pores; and the new bone formed in the AGH group was reticular in shape, and was widely distributed in the matrix material. Analysis showed two possible reasons for this: the first reason might be that there was no osteogenic growth inducing factor added to the AG group matrix, making it impossible for the internal cells to contact corresponding stimulation and thus only cells around and close to the pores had the ability to differentiate into osteogenic tissues. Although the AGH group matrix had no osteogenic induced factor, it contained widely distributed nano hydroxyapatite particles. These particles were in contact with the surrounding cells, played an osteogenic induction role, and subsequently promoted osteogenic differentiation. The nano-hydroxyapatite solubility is usually poor in biologic environment, but osteoblasts can absorb hydroxyapatite particles, then secrete bone matrix, and further promote the differentiation of other cells. In the opposite, the cellular uptake of the hydrogel metabolites is very slow, and therefore the second reason might be that the degradation of nano hydroxyapatite particles was much faster than that of alginate/gelatin in vivo, and could lead to degradation and formation of pores after promoting cell differentiation, which could facilitate nutrient transfer, thus promoting bone formation. The specific mechanism still needs further study. OCN is an important marker protein in the late osteogenic stage. Large amounts of OCN immunohistochemical staining could be seen in the new bone formation regions in both the AG group and AGH group, which further proved the occurrence of formation of new bone.
Although our studies have demonstrated the 3D construct consisting of hASCs owned regular pores for blood vessel ingrowth, and the alginate/hydroxyapatite had a well-interconnected porous structure for cells' growth and proliferation as other studies showed,51 the limitations for alginate/gelatin/hydroxyapatite used in 3D bio-printing technology still existed, which mainly was the mechanical properties of the hydrogel materials. 3D construct with large height could also be builded using the AGH composition. However, the distance between layers was not accurate in the vertical direction, due to the poor strength of the materials, which was same as the results in other studies.24,42 As long as this problem was solved, our proposed construct and the composition chosen in this study will satisfy the essential requirements for bone tissue engineering well.
In this study, the construction of 3D bio-printing material with cell viabilities up to 90% was achieved, and it was proved that the nano hydroxyapatite particles could promote osteogenic differentiation of the hASCs/alginate/gelatin 3D structure; however, there are still some limitations. First, the reason for selecting nano hydroxyapatite particle concentration as 1% was that the limitation of the printer head diameter determined we could not use the design with a higher concentration. In subsequent experiments, we have tried self-synthesizing nano hydroxyapatite particles, and modified the printer head diameter, to attempt using a nano hydroxyapatite mixture with multiple concentrations for printing in order to obtain the best and applicable ratio. Second, further research is still needed on the specific mechanisms of how nano hydroxyapatite could promote hASCs 3D printed construct osteogenesis. Studies have shown that this might due to osteoblast stimulation of DNA methylation and promotion of the expression of osteoblast-related genes.48 Based on the results of this study, future studies will be done to explore the mechanisms of the promotion of osteogenesis and compare nano hydroxyapatite with osteogenic inducing factors in the effects on hASCs osteogenic differentiation.
Conclusions
This study has demonstrated that the nano hydroxyapatite particles could promote osteogenic differentiation of the alginate/gelatin/hASCs 3D bio-printed construct in vitro and in vivo. Hydrogel materials containing nano hydroxyapatite particles could be used as the matrix material of 3D bio-printing technology for bone tissue engineering.Acknowledgements
The authors are grateful for the support of the Project of the Chinese Ministry of Education, China (No. 113002A).References
- Y. Liu, Y. Zhou, H. Feng, G. Ma and Y. Ni, Biomaterials, 2008, 29, 3338–3345 CrossRef CAS PubMed.
- G. H. Wu and S. H. Hsu, J. Med. Biol. Eng., 2015, 35, 285–292 CrossRef PubMed.
- J. A. Inzana, D. Olvera, S. M. Fuller, J. P. Kelly, O. A. Graeve, E. M. Schwarz, S. L. Kates and H. A. Awad, Biomaterials, 2014, 35, 4026–4034 CrossRef CAS PubMed.
- C. H. Lee, J. Hajibandeh, T. Suzuki, A. Fan, P. Shang and J. J. Mao, Tissue Eng., Part A, 2014, 20, 1342–1351 CrossRef CAS PubMed.
- P. Yilgor, R. A. Sousa, R. L. Reis, N. Hasirci and V. Hasirci, Macromol. Symp., 2008, 269, 92–99 CrossRef CAS.
- J. Li, P. Habibovic, M. Vandendoel, C. Wilson, J. Dewijn, C. Vanblitterswijk and K. Degroot, Biomaterials, 2007, 28, 2810–2820 CrossRef CAS PubMed.
- A. Nandakumar, A. Barradas, J. de Boer, L. Moroni, C. van Blitterswijk and P. Habibovic, Biomatter, 2014, 3, e23705 CrossRef PubMed.
- F. Obregon, C. Vaquette, S. Ivanovski, D. W. Hutmacher and L. E. Bertassoni, J. Dent. Res., 2015, 94, 143S–152S CrossRef CAS PubMed.
- N. E. Fedorovich, J. R. De Wijn, A. J. Verbout, J. Alblas and W. J. A. Dhert, Tissue Eng., Part A, 2008, 14, 127–133 CAS.
- L. Lv, Y. Liu, P. Zhang, X. Zhang, J. Liu, T. Chen, P. Su, H. Li and Y. Zhou, Biomaterials, 2015, 39, 193–205 CrossRef CAS PubMed.
- Y. Liu, M. Ou, H. Liu, M. Gu, L. Lv, C. Fan, T. Chen, X. Zhao, C. Jin, X. Zhang, Y. Ding and Y. Zhou, Biomaterials, 2014, 35, 4489–4498 CrossRef CAS PubMed.
- G. I. Im, Histol. Histopathol., 2013, 28, 557–564 Search PubMed.
- W. Bi, Z. Gu, Y. Zheng, X. Zhang, J. Guo and G. Wu, PLoS One, 2013, 8, e78198 CAS.
- T. Debnath, S. Ghosh, U. S. Potlapuvu, L. Kona, S. R. Kamaraju, S. Sarkar, S. Gaddam and L. K. Chelluri, PLoS One, 2015, 10, e120803 Search PubMed.
- W. Lee, C. Y. Bae, S. Kwon, J. Son, J. Kim, Y. Jeong, S. S. Yoo and J. K. Park, Adv. Healthcare Mater., 2012, 1, 635–639 CrossRef CAS PubMed.
- L. Moroni, R. Schotel, D. Hamann, J. R. de Wijn and C. A. van Blitterswijk, Adv. Funct. Mater., 2008, 18, 53–60 CrossRef CAS.
- P. Schumann, D. Lindhorst, C. von See, N. Menzel, A. Kampmann, F. Tavassol, H. Kokemuller, M. Rana, N. C. Gellrich and M. Rucker, J. Biomed. Mater. Res., Part A, 2014, 102, 1652–1662 CrossRef PubMed.
- M. Rücker, M. W. Laschke, D. Junker, C. Carvalho, A. Schramm, R. Mülhaupt, N. Gellrich and M. D. Menger, Biomaterials, 2006, 27, 5027–5038 CrossRef PubMed.
- B. Sibaja, E. Culbertson, P. Marshall, R. Boy, R. M. Broughton, A. A. Solano, M. Esquivel, J. Parker, L. L. Fuente and M. L. Auad, Carbohydr. Polym., 2015, 134, 598–608 CrossRef CAS PubMed.
- K. Yue, S. G. Trujillo-de, M. M. Alvarez, A. Tamayol, N. Annabi and A. Khademhosseini, Biomaterials, 2015, 73, 254–271 CrossRef CAS PubMed.
- B. Huber, K. Borchers, G. E. Tovar and P. J. Kluger, J. Biomater. Appl., 2016, 30, 699–710 CrossRef PubMed.
- X. Wang, E. Tolba, H. C. Schroder, M. Neufurth, Q. Feng, B. Diehl-Seifert and W. E. Muller, PLoS One, 2014, 9, e112497 Search PubMed.
- S. Das, F. Pati, Y. J. Choi, G. Rijal, J. H. Shim, S. W. Kim, A. R. Ray, D. W. Cho and S. Ghosh, Acta Biomater., 2015, 11, 233–246 CrossRef CAS PubMed.
- J. Jia, D. J. Richards, S. Pollard, Y. Tan, J. Rodriguez, R. P. Visconti, T. C. Trusk, M. J. Yost, H. Yao, R. R. Markwald and Y. Mei, Acta Biomater., 2014, 10, 4323–4331 CrossRef CAS PubMed.
- X. Zhang, J. Guo, Y. Zhou and G. Wu, Tissue Eng., Part B, 2014, 20, 84–92 CrossRef CAS PubMed.
- M. T. Poldervaart, H. Wang, J. van der Stok, H. Weinans, S. C. G. Leeuwenburgh, F. C. Oner, W. J. A. Dhert and J. Alblas, PLoS One, 2013, 8, e72610 CAS.
- K. Pandi and N. Viswanathan, Carbohydr. Polym., 2015, 134, 732–739 CrossRef CAS PubMed.
- H. S. Roh, S. W. Myung, S. C. Jung and B. H. Kim, J. Nanosci. Nanotechnol., 2015, 15, 5585–5588 CrossRef CAS PubMed.
- Y. Liu, R. Zhou and H. Wu, Huaxi Kouqiang Yixue Zazhi, 2015, 33, 301–305 CAS.
- Y. He, Y. Dong, F. Cui, X. Chen and R. Lin, PLoS One, 2015, 10, e135366 Search PubMed.
- J. Huang, J. Xiong, J. Liu, W. Zhu, J. Chen, L. Duan, J. Zhang and D. Wang, Bio-Med. Mater. Eng., 2015, 26(suppl. 1), S197–S205 CrossRef PubMed.
- H. W. Yang, M. H. Lin, Y. Z. Xu, G. W. Shang, R. R. Wang and K. Chen, Int. J. Clin. Exp. Med., 2015, 8, 257–264 CAS.
- J. Chen, P. Pan, Y. Zhang, S. Zhong and Q. Zhang, Colloids Surf., B, 2015, 134, 401–407 CrossRef CAS PubMed.
- N. E. Fedorovich, W. Schuurman, H. M. Wijnberg, H. Prins, P. R. van Weeren, J. Malda, J. Alblas and W. J. A. Dhert, Tissue Eng., Part C, 2012, 18, 33–44 CrossRef CAS PubMed.
- M. Sato, M. A. Sambito, A. Aslani, N. M. Kalkhoran, E. B. Slamovich and T. J. Webster, Biomaterials, 2006, 27, 2358–2369 CrossRef CAS PubMed.
- Y. Liu, X. Zhang, Y. Liu, X. Jin, C. Fan, H. Ye, M. Ou, L. Lv, G. Wu and Y. Zhou, PLoS One, 2014, 9, e97741 Search PubMed.
- W. Ge, L. Shi, Y. Zhou, Y. Liu, G. Ma, Y. Jiang, Y. Xu, X. Zhang and H. Feng, Stem Cells, 2011, 29, 1112–1125 CrossRef CAS PubMed.
- L. Lv, Y. Liu, P. Zhang, X. Zhang, J. Liu, T. Chen, P. Su, H. Li and Y. Zhou, Biomaterials, 2015, 39, 193–205 CrossRef CAS PubMed.
- J. Hall, R. G. Sorensen, J. M. Wozney and U. M. Wikesjo, J. Clin. Periodontol., 2007, 34, 444–451 CrossRef CAS PubMed.
- Y. Xia, S. S. Peng, L. Z. Xie, X. J. Lian, X. J. Zhang, H. Cui, T. X. Song, F. M. Zhang, N. Gu and F. Z. Cui, J. Biomater. Appl., 2014, 29, 59–71 CrossRef CAS PubMed.
- G. Gao, A. F. Schilling, T. Yonezawa, J. Wang, G. Dai and X. Cui, Biotechnol. J., 2014, 9, 1304–1311 CrossRef CAS PubMed.
- T. Billiet, E. Gevaert, T. De Schryver, M. Cornelissen and P. Dubruel, Biomaterials, 2014, 35, 49–62 CrossRef CAS PubMed.
- M. Gruene, M. Pflaum, A. Deiwick, L. Koch, S. Schlie, C. Unger, M. Wilhelmi, A. Haverich and B. N. Chichkov, Biofabrication, 2011, 3, 15005 CrossRef CAS PubMed.
- J. Y. Park, C. Yang, I. H. Jung, H. C. Lim, J. S. Lee, U. W. Jung, Y. K. Seo, J. K. Park and S. H. Choi, Biomater. Res., 2015, 19, 7 CrossRef PubMed.
- S. Gronthos, S. Q. Chen, C. Y. Wang, P. G. Robey and S. T. Shi, J. Bone Miner. Res., 2003, 18, 716–722 CrossRef CAS PubMed.
- M. Ozeki, S. Kuroda, K. Kon and S. Kasugai, J. Biomater. Appl., 2011, 25, 663–684 CrossRef CAS PubMed.
- J. B. Lian, G. S. Stein, A. Javed, A. J. van Wijnen, J. L. Stein, M. Montecino, M. Q. Hassan, T. Gaur, C. J. Lengner and D. W. Young, Rev. Endocr. Metab. Disord., 2006, 7, 1–16 CrossRef CAS PubMed.
- S. W. Ha, H. L. Jang, K. T. Nam and G. J. Beck, Biomaterials, 2015, 65, 32–42 CrossRef CAS PubMed.
- C. L. Salgado, L. Grenho, M. H. Fernandes, B. J. Colaco and F. J. Monteiro, J. Biomed. Mater. Res., Part A, 2016, 104, 57–70 CrossRef PubMed.
- L. M. De Rycke, M. N. Boone, A. I. Van Caelenberg, M. Dierick, L. Van Hoorebeke, H. van Bree and I. M. Gielen, Am. J. Vet. Res., 2012, 73, 227–232 CrossRef PubMed.
- H. Lin and Y. Yeh, J. Biomed. Mater. Res., Part B, 2004, 71, 52–65 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2016 |