Following Ramachandran: exit vector plots (EVP) as a tool to navigate chemical space covered by 3D bifunctional scaffolds. The case of cycloalkanes†
Abstract
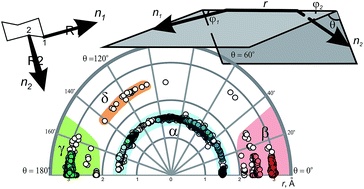
An approach to the analysis and visualization of chemical space covered by disubstituted scaffolds, which is based on exit vector plots (EVPs), is used for analysis of the simplest disubstituted cyclic cores – cycloalkanes – deposited in the Cambridge Structural Database (CSD). It is shown that four clearly defined regions are found in EVPs of the cycloalkanes, similar to those observed in Ramachandran plots for peptides. These results can be used for directed design of more complex scaffolds, classification of conformational space for the disubstituted scaffolds, rational scaffold replacement, or SAR studies.

 Please wait while we load your content...
Please wait while we load your content...