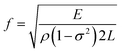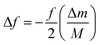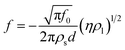Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biosensor-based detection of tuberculosis
Saurabh K.
Srivastava
*ab,
Cees J. M.
van Rijn
b and
Maarten A.
Jongsma
a
aPlant Research International, Wageningen UR, Droevendaalsesteeg 1, 6708 PB Wageningen, The Netherlands. E-mail: srivastava.saurabh@outlook.com
bLaboratory of Organic Chemistry, Wageningen UR, Dreijenplein 8, 6703 HB Wageningen, The Netherlands
First published on 5th February 2016
Abstract
Tuberculosis (TB), caused by Mycobacterium tuberculosis (M.tb.), is one of the most prevalent and serious infectious diseases worldwide with an estimated annual global mortality of 1.4 million in 2010. Diagnosis of TB in the developing world is very challenging due to the limited suitability of currently available techniques under tropical field conditions. M. tb. is a slowly growing Mycobacterium that takes around six to eight weeks to be detected via sensitive culture methods. There is also hardly any clinical symptom at an early stage of infection, thereby causing a delay in diagnosis and treatment, and the complexity of the disease is further increased by the emergence of multiple drug resistant (MDR) strains. A lot of work has been done over the last few decades to develop effective point of care diagnostic techniques that are cheap, robust and can be performed at high throughput in rural areas. However, despite considerable technical improvements reported from the lab, such economical fool-proof diagnostic assays are still lacking on the market. The objective of this review is to evaluate currently available biosensing techniques that are either already in use or under development for detection of TB. The focus of the review is on the emerging field of diagnostic biosensors that combine ligand capture and detection in a one-step assay. A comparison will also be made with conventional multistep techniques.
1. Introduction
1.1 Background
Tuberculosis (TB) is an age-old infectious disease of the human race,1,2 which has re-emerged globally as an important health-related issue, especially due to the emergence of multi-drug resistant (MDR) strains.3,4 The specific characteristics of the Gram-negative bacillus, Mycobacterium tuberculosis (M.tb.) causing TB5 make it currently one of the most prevalent diseases.6 TB bacteria can remain inactive or in a dormant state for years without causing symptoms or spreading to other subjects, but as soon as the immune system of the host becomes weakened, the bacteria become active and infects mainly the lungs along with other parts of body. TB cases are further aggravated by other illnesses that affect the immune system, such as HIV, which is very prevalent in resource-poor countries. Unfortunately, in spite of sustained efforts at national and global levels, only limited success has been achieved in detection and management of TB worldwide.7,8 The main obstacles facing successful field detection and treatment of TB are:(1) The omnipresence of the pathogen and disease across different continents, which complicates management and eradication programs.9 Most of the reported cases (∼98%) are concentrated in developing countries, due to which TB is also known as a disease of poverty, with two-thirds of cases estimated to occur among people aged 15–59, adults in their most productive years.6 However, also developed Western countries continue to report TB cases.10
(2) The high costs of the first line anti-TB drugs to treat TB (like isoniazid, rifampicin, pyrazinamide, ethambutol) bar their uses especially when multiple first line drugs need to be applied to suppress resistance.11–14 This follow-up is often lacking in several affected countries like India.15–17
(3) Emergence of resistance in M. tb. against major first line chemotherapeutic agents has hindered the treatment of TB cases.
(4) To date, most diagnostic procedures rely largely on immuno-assays that are not sufficiently specific. M. tb. shares antigens to many other Mycobacterium species resulting in false positive cases in ∼35% of patients with active TB.18–20 Other techniques that have been employed like flow-cytometry, radiometric detection, latex agglutination etc. have their own set of disadvantages which will be discussed in detail in the coming sections.
The objective of this study is to present a succinct review of the available biosensing techniques that are either already in use or under development in detection of TB. The focus of the review will be on emerging biosensor-approaches that are compared to conventional techniques. Special attention will be given to their potential for deployment under field conditions in resource-poor countries.
1.2 Epidemiology
The WHO Global TB Control Report 2012 reported that the WHO's Millennium Development Goal to halt the growth of the TB epidemic by 2015 is showing some success with a decrease of 2.2% observed between 2010 and 2011 for new TB cases, along with a 41% mortality rate decrease relative to 1990. Between 1995 and 2011, 51 million people were successfully treated for TB, saving 20 million lives. Despite this progress, the global burden of TB remains enormous with 8.7 million new cases of TB were registered in 2011, out of which 13% patients were co-infected with HIV. The mortality in 2011 due to TB was 1.4 million, including almost one million deaths among HIV-negative individuals and 0.4 million among people who were HIV-positive. The distribution of TB cases also shows a gender-dependence with men reporting more cases compared to females in all age groups and from every geographical region of the world. India and China together account for 40% whereas the African region contributes 24% of the world's TB cases. The statistics of the MDR cases in TB are alarming according to the WHO 2012 report. It reported resistance to more than one first line anti-TB drugs among 3.7% of new cases and 20% previously treated cases. In recognition of the problem, 26 nations have already listed MDR-TB as a top priority health programme and founded a global fund to support Directly Observed Therapy (DOT) to ensure the effectiveness of given medications.21–231.3 Current TB diagnostics
(1) Smear microscopy: microscopy based smear tests are rapid, inexpensive, simple and relatively easy to perform methods for the detection of acid-fast bacterium such as M. tb. bacterium. Conventionally the Ziehl-Neelsen staining is utilized requiring a minimum of 1 × 104 bacterium per ml of sputum.24 Fluorescence microscopy utilizing auramine-rhodamine staining was found to be more sensitive though expensive, as it requires a fluorescence microscope.25 The fluorescence-based method is more sensitive as slides can be examined at lower magnification.24 Results from this method can be obtained within h.
(2) Immunological assays like latex agglutination, ELISA, and Mantoux tests: in these tests, typically the binding of antibodies in serum to M. tb. antigens is tested.26,27 For example, in latex agglutination tests, the polystyrene (latex) beads are functionalized with antigens extracted from a pathogenic Mycobacterium, which are then reacted with serum samples.28 In case of a reaction, the latex beads coagulate showing a positive test. The sensitivity of these tests are not high enough.29 Hence, when the bacterial load is low, these tests will fail. Also, the serum could contain antibodies (due to vaccinations like Bacille-Calmette–Guérin/BCG) that might interfere with the tests adding further to the shortfalls of these assays.
(3) Radiometric detection test: in this type of test, the metabolic activity of M. tb. is detected radiologically.30 For example, M. tb. (and also some other Mycobacteria) are known to produce CO2 from carbon sources like glycerol or acetate. The important selective criterion is that M. tb. cannot form CO2 from glucose. This helps to differentiate M. tb. from other Mycobacterium sp. and bacteria. Utilizing this selective property, the capability of M. tb. in producing 14CO2 from 14C-U-glycerol or 14C-U-acetate, but not from 14C-U-glucose is measured. Due to the technical complexity this technique is not suitable for developing countries.
(4) Flow cytometry test: in the last few years, use of flow cytometry has increased rapidly in the detection of TB cases.31–33 This technique is based on the ability of viable M. tb. bacterium to absorb fluorescein diacetate (FDA) and to hydrolyse it into fluorescein, which upon accumulation in metabolically active bacterium could successfully be detected by flow cytometry. The reproducibility is high and it does not require active cell division of mycobacteria.34,35 The technique, however, needs logistic support from a specialized laboratory that makes it difficult to implement in developing countries.36
(5) Cultivation detection tests (like MB/Bact, Bactec MGIT 960 systems): in these techniques, biological samples are selectively cultured on solid media to detect and quantify the presence of M. tb.37–40 Although quite accurate, this technique is time consuming, as it requires a growth period of 9–42 days. It also requires a laboratory back up that complicates applications under field conditions in developing countries.40
(1) Polymerase chain reaction (PCR)-based techniques: PCR is one of the most sensitive methods to detect the presence of M. tb. strains, also sequencing of PCR amplified DNA fragments is the most direct technique of detecting codons responsible for resistance in M. tb.41–43 PCR amplified DNA fragments are generally analysed by the following two methods:
(a) Electrophoresis: this method mainly relies on the difference in electrophoretic mobility of mutated DNA fragments of M. tb. especially of resistant strains. By electrophoretic techniques, PCR amplified DNA can be compared with the electrophoretic mobility of a wild type/reference M. tb. DNA, and thus caution for any resistance.44 A hetero-duplex analysis method has also been developed in recent years where strands with a single-base mismatch can easily be identified and separated utilizing conformation-sensitive gel electrophoresis from the strands containing no mismatches.45,46
(b) Hybridization-based techniques: this type of technique relies on the hybridization of clinical DNA with complementary DNA and the binding is then compared with the results of wild/reference M. tb. DNA.47,48 In case the complementary strand is sufficiently homologous, the binding of clinical DNA can be detected using ELISA readers. The hybridization can be done user-friendly on strips, microtiter plate as well as microarrays.
(c) Real-time PCR: with the help of fluorescently labelled DNA strands, it is possible to visualize the increase of the product in “real time”.49–51 Different types of fluorescent labels have been applied in a diverse range of real-time PCR techniques, like TaqMan probes,50,52,53 Beacons54,55 and FRET probes.56,57
The main disadvantages of PCR-based tests are the relatively high costs of equipment and reagents. It also requires highly skilled personnel along with dedicated pre- and post-PCR rooms to avoid contamination. Even if it were less expensive, the technological and sample preparation complexities make it largely inappropriate for use in resource-poor settings.
2. Biosensors in TB detection
Considering the fact that 98% of all TB cases occur in developing countries without access to specialized laboratories, there is a strong need to develop alternative, simpler and lower cost techniques for TB diagnosis. Biosensors are analytical devices that transduce biochemical reactions/interactions of isolated enzymes, receptor proteins, antibodies, nucleic acids, organelles, whole cells or tissues with specific chemical compounds into an optical, thermal or electrical signal, which can be more easily measured and quantified. The main advantages of biosensors over conventional diagnostic techniques can be stated as follows:(1) Technical advantage: in biosensors often a high level of device and capture/detection integration is achieved allowing single step detection.
(2) Ease of use: many of the designed biosensors are tailored with user-friendly interfaces connecting them to advanced instrumentation.
(3) Quick response: response time is typically a few minutes for most biosensors enabling rapid and better control over the measurement.
A general framework of different classes of biosensors that could be used for the detection of TB is given in Fig. 1 to guide the reader through the upcoming sections of the review and will be discussed in more detail within relevant sections.
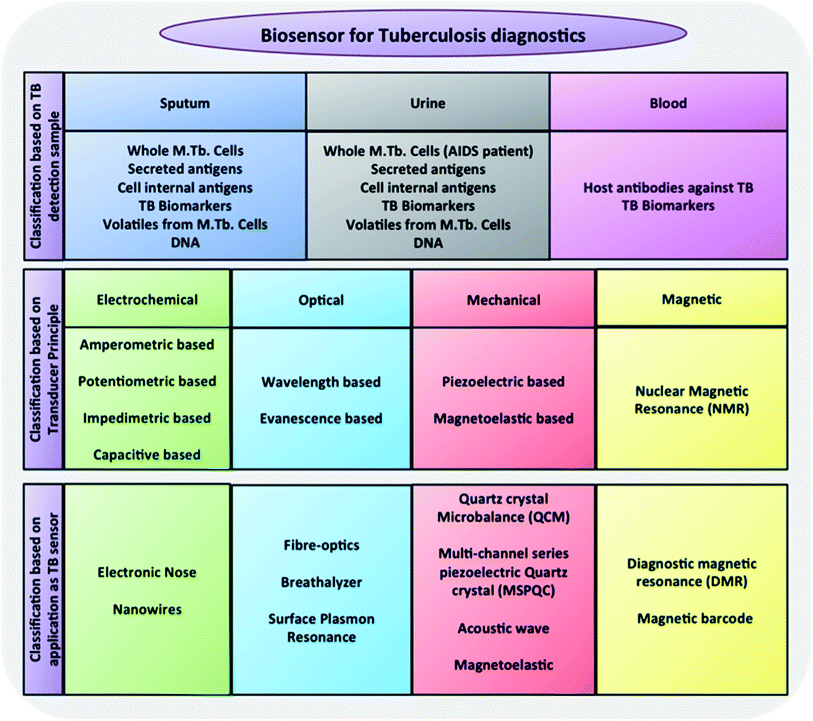 | ||
| Fig. 1 Generic classification of biosensors tested as TB diagnostics.58,59 | ||
2.1 Electrochemical and electrical biosensors
Electrochemical and electrical biosensors are among the most popular biosensors that are used today in detection of not only TB but also a large number of other diseases.60–62,129,130 The mechanism of detection relies on specific changes in electrical signals (conductance, impedance, potential, and capacitance) at a surface-functionalized electrode by either chemical reactions or physical interactions. For example, monoclonal antibodies raised against M. tb. cell wall components can be immobilized onto an electrode surface exposed to a suspension of M. tb. and detect the interaction between the M. tb. bacteria and the antibodies by a change of conductance.63 Similarly, an electrode-based DNA biosensor can also be made where certain probes that specifically bind to specific regions of M. tb. DNA are immobilized on the electrode.62,129,130 As electrodes, carbon has often been used.60,62,64–66,129 These sensors are sensitive, flexible and allow a high degree of multiplexing, but they also present some disadvantages. Firstly, a constant pH and ion concentration of the reaction compartment is very critical, as they directly affect the baseline conductance of the electrodes. Secondly, the obtained electrical signal strictly depends on many molecular factors, like the positive or negative net charge or neutrality of the analyte etc.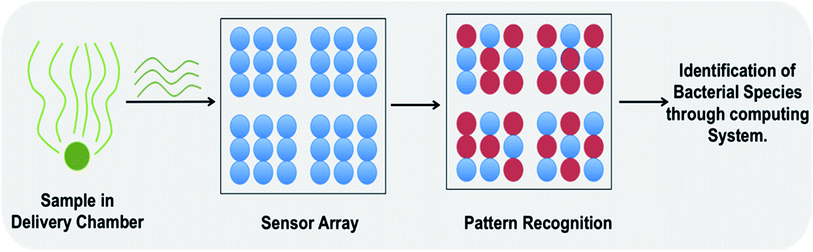 | ||
| Fig. 2 An electronic nose based biosensor consisting of three main parts i.e. a sample delivery chamber, a detection array/pattern recognition system and a computing system for data analysis and result interpretation. This is a sensor assay where a specific pattern is produced upon interaction with volatile/gaseous by-products from specific bacterial species.71 | ||
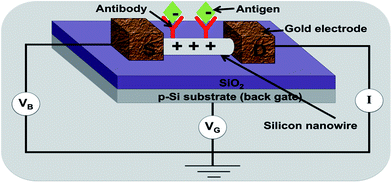 | ||
| Fig. 3 A silicon nanowire based biosensor: a change in conductance of an antibody-functionalized nanowire is measured upon binding of the antigen.81 | ||
The major advantage offered by this category of biosensors is its higher than normal sensitivity, because of two main factors i.e. (1) the size of the biological species/analyte to be detected is comparable to the size of the nanowire, hence the charge of the incoming species can sensitively control the conductivity of the charge carriers in the semiconductor material within the Debye length,82 (2) the wire represents a serial circuit where interaction at each receptor molecule can control the current flowing through the entire nanowire.83 The chemical nature of silicon and carbon wires and tubes allow a wide scope of surface functionalizations, which is an advantage compared to metal electrodes. However, the delicate surface functionalization and nature of silicon nanowires, makes it technologically much more challenging to actually design and implement these types of sensors84 thus limiting the technique merely to the proof of concept stage.
2.2 Optical biosensors
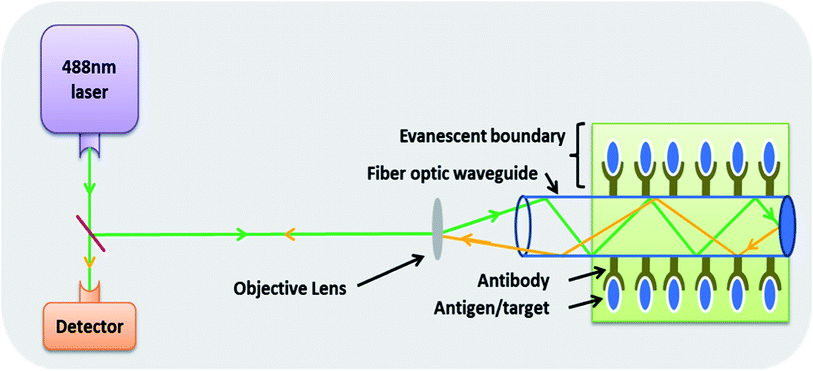 | ||
| Fig. 4 A fibre optics based biosensor where specific binding at the exterior of fibre optic waveguide via specific antibodies is measured by determining the change in the angle of reflection of the incident light. The system works using the principle of evanescence.89 | ||
Another interesting class of biosensors with good potential for TB detection is the so-called optical ring resonance (ORR) detectors. The principles of optical ring resonance are complex and fall beyond the scope of this review. In summary, these ORR systems are based on total internal reflection and constructive interference.90 Recently, such ORR detectors could be designed on lab-on-a-chip platforms91,92 with multiple parallel channels with one channel coated with specific antibody (test channel) and others coated with non-specific antibodies (reference channel). With binding of the antigens with their specific antibodies, there are changes observed in total internal reflection and simultaneous interference patterns which are recorded by a CCD camera and compared to the data obtained from reference channels. Pending the availability of highly specific antibodies to M. tb. antigens, these ORR detectors may be of potential in early detection of TB although the surface functionalization has the same challenges as in case of nanowire functionalization, because the core material of the fibre is mostly silicon based.
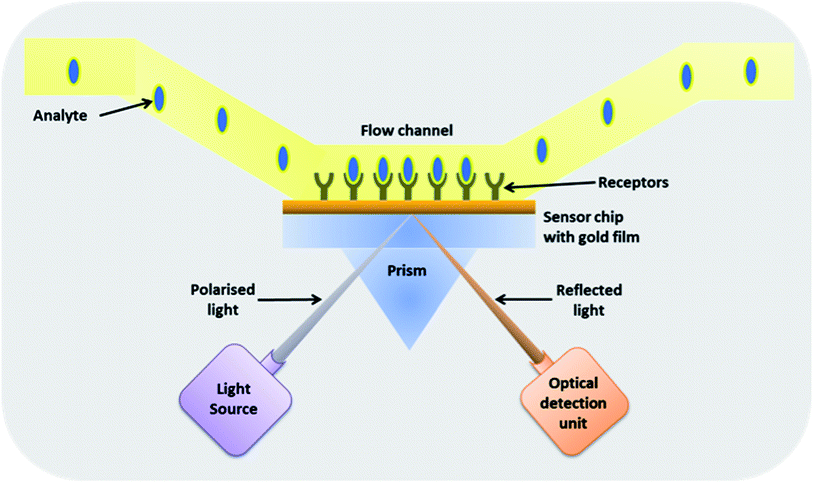 | ||
| Fig. 5 A surface plasmon resonance based biosensor. In this type of biosensor, a thin gold plate is functionalized with antibodies that upon binding to the antigen cause a change in the angle of reflection of the polarised light incident at the other side of the gold film.93 | ||
For a certain angle of incidence, the coupling between the light wave and the surface plasmon occurs, thereby generating a change in refractive index at the sensor surface giving rise to a change in angular position of the SPR dip, which is measured by position-sensitive photo-detector. SPR analysis is sensitive to environmental conditions like temperature and buffer concentration. Thus, a parallel background measurement is employed to correct for the change in refractive index caused by these background effects. Gold surfaces immobilized with antibodies that bind M. tb. antigens or complex was successfully used to detect TB.94–101 The sensitivity of the system to detect M. tb. antigen (CFP-10 antigen) in tissue fluid was 100 ng ml−1,102 and for M. tb. complex was 30 ng μl−1.97 The sensitivity of the SPR system could be further enhanced as demonstrated by Chen et al., where they detected the TB antigen CFP-10 in sandwich assay by coupling the secondary sandwich antibodies with nanoparticle thus obtaining sensitivity down to 0.1 ng ml−1.128 SPR based biosensors are already used for detection of various pathogens, including bacteria like Salmonella sp., however, the costs are high and trained personnel are required to operate them.103
2.3 Mechanical biosensors
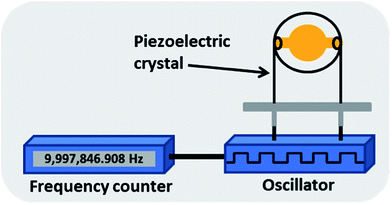 | ||
| Fig. 6 A piezoelectric quartz crystal based biosensor where the change in resonating frequency of a piezoelectric crystal is measured. The resonance frequency of the crystal shows a change after binding of antigens with specific antibodies immobilised on the crystal.110 | ||
2.3.1.1. Quartz crystal microbalance (QCM). In QCM systems applied to TB, the crystal electrode was first coated with protein A followed by binding of an anti-TB-cells antibody (rabbit IgG against M. tb.) and then incubated with weakened TB cells. Binding of M. tb. cells on the crystal electrodes was monitored in real time and the frequency shift was calculated. The sensitivity using this system was 105 CFU ml−1.110 The main advantage of QCM lies in the fact that it is fast, reusable (after washing), label-free, requires minimal sample preparation and is easily operated. The major disadvantage lies in the fact that the density, temperature, viscosity and electrical conductivity of the sample may affect the results and require calibration correction and that it is relatively insensitive.
2.3.1.2. Multi-channel series piezoelectric quartz crystal sensor (MSPQC). In the MSPQC system, there are eight sample detectors along with a microprocessor and a data output system. The system relies on detection of volatiles produced by the growth of M. tb. such as NH3 and CO2. These volatiles cause an impedance change when they are absorbed by a KOH absorbing solution, which in turn will change the oscillating frequency. The limit of detection with this system is 107 CFU ml−1 at the time of detection.111 Apart from being time consuming involving a laborious M. tb. culture process (eight days in case of a low M. tb. burden), another major drawback of this system is the sample pre-treatment required to remove contamination with other bacteria.
2.3.1.3. Acoustic wave biosensor. Acoustic wave biosensors rely upon acoustic waves propagating near the surface of a piezoelectric crystal. The sensor electrically produces a mechanical wave that is sensitive to any change or binding at the piezoelectric crystal and then transduces the mechanical output waves back into an electrical signal, which is compared with the input electrical signal for changes in amplitude, phase, and frequency.112 In one application apart from the acoustic signal also the conductivity change (impedance) of the medium over the crystal was measured. Here the conductivity change due to growth of M. tb. cells and by-product production was studied.113,114 The limit of detection with this bulk acoustic wave impedance biosensor was 2 × 103 CFU ml−1. The disadvantage of the system is the time consuming and laborious M. tb. culture process that reduced the applicability for field uses.
where E denotes modulus of elasticity, σ is the Poisson's ratio, ρ is the mass density of the sensor material, and L is the longitudinal dimension of the sensor. The change in resonant frequency also depends on mass change when the initial mass, temperature and humidity are constant and is given by the equation.
where f is the initial resonance frequency, M is the initial mass, Δm is the mass change, and Δf is the shift in the resonant frequency of the sensor.118 It is due to this dependence on mass change, that upon binding of antigens or M. tb. bacteria to the antibody, there is a shift in fundamental resonant frequency, which is often monitored wirelessly on a frequency counter.
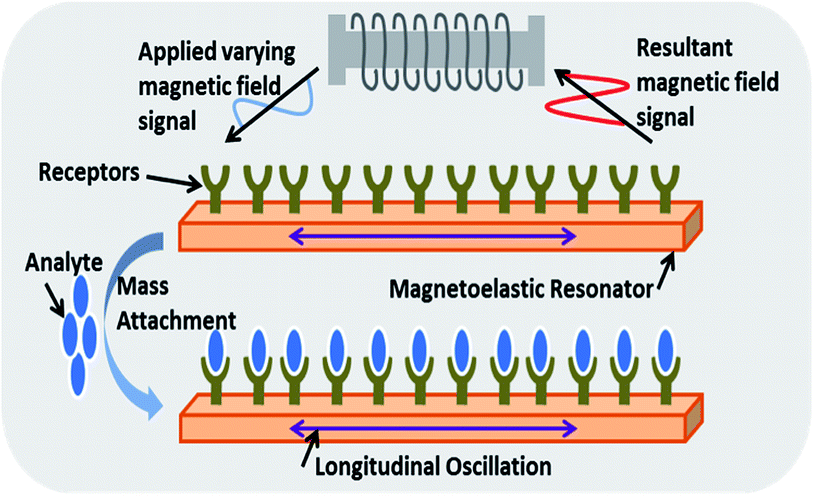 | ||
| Fig. 7 A magnetoelastic biosensor where the change in basal resonating frequency of a magnetoelastic strip as a result of binding of antigens with specific antibodies under varying magnetic fields is measured in order to detect analyte.119 | ||
The advantages of these types of sensors over SPR-based sensors are their ease of use as the sensors are (i) freely hanging inside a sample solution, (ii) do not require complicated integration with microfluidics pumps and (iii) do not produce a complex signal. The fundamental resonating frequency of the strip immersed in liquid (culture media) can also be altered due to the consumption of nutrients. M.tb. in the liquid media, upon proliferation decomposes macromolecule into by products (such as ammonia, carbon dioxide, organic acid) thereby causing a change in the physical properties of the culture that can then be detected with the help of such biosensors. A suitable calibration curve must be made to detect a specific M. tb. signature.117 The sensor resonant frequency shift upon immersion in liquid is given by the equation
2.4 Magnetic biosensors
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.125
000.125
3. Conclusions
TB remains one of the major unresolved global health problems, especially in the developing parts of the world, predominantly due to the complexity of a proper and economic affordable diagnosis and treatment in time along with recently arising issues like multiple drug resistance and other allied infections that decrease body immunity, like HIV. Current technologies for diagnosis are either too insensitive, too laboratory intensive or utilize expensive detection modules, which are all challenges in resource poor settings. Most of the biosensors (except Electronic nose and Breathalyzer) discussed in the present review are still at the developmental stage and lack clinical validation with real TB samples from patients. The test material in the reviewed published literature, mainly consists of genomic material, M.tb. protein, M. tb. by-products or whole M. tb. cells that are pure and tested under conditions far removed from patient samples. All sensor methods have their own merits and potential problems with respect to sample preparation, requirement of skilled personnel to handle the sample, sensitivity or cost. Moreover, since the sensitivity of each system is expressed in different units and not linked to the bacterial count, benchmarking these diagnostic platforms is not currently possible. An overview of the analyzed sample and detection limits of different biosensors is shown in Table 1. With WHO's aim to completely eliminate TB by 2050, development of techniques for early and accurate detection of TB is crucial. To build an effective biosensor for TB detection, criteria that need to be fulfilled are: (1) cost-effectiveness (2) high sensitivity (3) reliability (no false positives) (4) portability and (5) disposability. Unfortunately, designing a biosensor with all these advantages has not yet been completely successful, as each attempt has specific drawbacks. It is a challenge to satisfy all these needs in a single biosensing device. Nevertheless, the advances in nanosensors and other upcoming technologies reviewed here, suggest that biosensors to detect TB can be expected to play a larger role in the near future. Such platforms will also need to solve issues around sample collection and preparation. Currently most diagnosis techniques available utilize sputum samples as test sample, which due to its high viscosity and sticky nature is very difficult to work with. Hence other sources like blood or urine should also be considered as test samples. Lack of reliable and tested biomarkers in those samples is, however, an issue that needs attention.| # | Transducer | Sensing technique | Sample analysed | Detection limit | References |
|---|---|---|---|---|---|
| 2.1 | Electrochemical | Electronic nose | M.tb. produced volatile compounds | Not available | Pavlou et al.67 |
| Nanowire/nanotubes | M.tb. specific ssDNA | Not available | Das et al.66 | ||
| 2.2 | Optical | Fibre-optics | M.tb. produced organic compounds | Not available | Pariwono et al.132 |
| SPR | CFP 10 M.tb. antigen or protein complexes | 0.1 ng ml−1 | Chen et al.128 | ||
| 30 ng ml−1 | Duman et al.97 | ||||
| Breathalyzer | M.tb. cells | 50–75 CFU per ml | McNerney et al.104 | ||
| 2.3 | Mechanical | QCM | M.tb. cells | 105 cells per ml | He et al.108 |
| MSQC | Volatiles produced by M.tb. cells growth (NH3, CO2) | 107 CFU per ml | Ren et al.111 | ||
| Acoustic Wave | M.tb. cells | 2 × 103 CFU/ml | He et al.114 | ||
| Magnetoelastic | M.tb. cells by-products | 103 cells per ml | Pang et al.117 | ||
| 2.4 | Magnetic | DMR | M.tb. cells | 20 CFU ml−1 | Lee et al.123,126 |
| Magnetic barcode | RNA from M.tb. cells | 100 CFU ml−1 | Liong et al.125 |
4. Future outlook
For effective TB detection in resource-poor settings the field of biosensors has very strong potential. However, key to the development of such biosensors is a focus on both the sample preparation steps from biofluids (e.g. blood, sputum) and the functionalisation of non-fouling surfaces with antibodies, aptamers etc. that are sensitive enough to allow early diagnosis in the future. The existing portfolio of potential sensing techniques all have their own pros and cons and it is not yet possible unfortunately to select one as most promising, because they have not yet been developed and successfully validated into a marketed products that can be benchmarked.Acknowledgements
The authors acknowledge the financial support obtained from the Dutch Technology Foundation STW under the Project number 10058.References
- M. Gandy and A. Zumla, Soc. Sci. Med., 2002, 55, 385 CrossRef PubMed.
- J. M. Grange, M. Gandy, P. Farmer and A. Zumla, International Journal of Tuberculosis and Lung Disease, 2001, 5, 208 CAS.
- N. S. Shah, A. Wright, G. H. Bai, L. Barrera, F. Boulahbal, N. Martin-Casabona, F. Drobniewski, C. Gilpin, M. Havelkova, R. Lepe, R. Lumb, B. Metchock, F. Portaels, M. F. Rodrigues, S. Rusch-Gerdes, A. Van Deun, V. Vincent, K. Laserson, C. Wells and J. P. Cegielski, Emerging Infect. Dis., 2007, 13(3), 380–387 CrossRef CAS PubMed.
- J. C. Palomino, A. Martin, M. Camacho, H. Guerra, J. Swings and F. Portaels, Antimicrob. Agents Chemother., 2002, 46(8), 2720–2722 CrossRef CAS PubMed.
- N. A. Kruh, J. Troudt, A. Izzo, J. Prenni and K. M. Dobos, PLoS One, 2010, 5, e13938 Search PubMed.
- J. F. Murray, Am. J. Respir. Crit. Care Med., 2004, 169, 1181 CrossRef PubMed.
- P. Nahid, M. Pai and P. C. Hopewell, Proc. Am. Thorac. Soc., 2006, 3, 103 CrossRef CAS PubMed.
- Expert Group Meeting Report, WHO, 2014, p. 1 Search PubMed.
- S. Murray, Can. Med. Assoc. J., 2006, 174, 33 CrossRef PubMed.
- L. P. Ormerod, Br. Med. Bull., 2005, 73–74, 17 CrossRef CAS PubMed.
- C. Mitnick, J. Bayona, E. Palacios, S. Shin, J. Furin, F. Alcántara, E. Sánchez, M. Sarria, M. Becerra, M. C. Fawzi, S. Kapiga, D. Neuberg, J. H. Maguire, J. Y. Kim and P. Farmer, N. Engl. J. Med., 2003, 348, 119 CrossRef PubMed.
- J. A. Caminero, International Journal of Tuberculosis and Lung Disease, 2006, 10, 829 CAS.
- P. Isaakidis, H. S. Cox, B. Varghese, C. Montaldo, E. Da Silva, H. Mansoor, J. Ladomirska, G. Sotgiu, G. B. Migliori, E. Pontali, P. Saranchuk, C. Rodrigues and T. Reid, PLoS One, 2011, 6, e28066 CAS.
- C. Colijn, T. Cohen, A. Ganesh and M. Murray, PLoS One, 2011, 6, e18327 CAS.
- S. S. Shin, J. J. Furin, F. Alcantara, J. Bayona, E. Sanchez and C. D. Mitnick, Emerging Infect. Dis., 2006, 12, 687 CrossRef PubMed.
- C.-H. Lee, M.-C. Lee, H.-H. Lin, C.-C. Shu, J.-Y. Wang, L. N. Lee and K. M. Chao, PLoS One, 2012, 7, e37978 CAS.
- C. Mulenga, D. Mwakazanga, K. Vereecken, S. Khondowe, N. Kapata, I. C. Shamputa, H. Meulemans and L. Rigouts, BMC Public Health, 2010, 10, 756 CrossRef PubMed.
- A. Bekmurzayeva, M. Sypabekova and D. Kanayeva, Tuberculosis, 2013, 93, 381 CrossRef CAS PubMed.
- A. Somoskovi, J. E. Hotaling, M. Fitzgerald, V. Jonas, D. Stasik, L. M. Parsons and M. Salfinger, J. Clin. Microbiol., 2000, 38, 2743 CAS.
- S. Singh, J. Singh, S. Kumar, K. Gopinath, V. Balooni, N. Singh and K. Mani, PLoS One, 2012, 7(7), r40213 Search PubMed.
- S. C. Cavalcante, E. C. C. Soares, A. G. F. Pacheco, R. E. Chaisson and B. Durovni, International Journal of Tuberculosis and Lung Disease, 2007, 11, 544 CAS.
- A. Anuwatnonthakate, P. Limsomboon, S. Nateniyom, W. Wattanaamornkiat, S. Komsakorn, S. Moolphate, N. Chiengsorn, S. Kaewsa-Ard, P. Sombat, U. Siangphoe, P. A. Mock and J. K. Varma, PLoS One, 2008, 3, e3089 Search PubMed.
- A. D. Harries, A. Jahn, R. Zachariah and D. Enarson, PLoS Med., 2008, 5, e124 Search PubMed.
- J. C. Palomino, Eur. Respir. J., 2005, 26, 339 CrossRef CAS PubMed.
- K. R. Steingart, M. Henry, V. Ng, P. C. Hopewell, A. Ramsay, J. Cunningham, R. Urbanczik, M. Perkins, M. A. Aziz and M. Pai, Lancet Infect. Dis., 2006, 6, 570 CrossRef PubMed.
- K. Minton, Nat. Rev. Immunol., 2003, 3, 355 CAS.
- J. G. Fernández, I. Fernández-de-Mera, L. E. Reyes, M. C. Ferreras, V. Pérez, C. Gortazar, M. Fernández and J. F. García-Marín, J. Vet. Diagn. Invest., 2009, 21, 102 CrossRef.
- S. Bhaskar, J. N. Banavaliker and M. Hanif, FEMS Immunol. Med. Microbiol., 2003, 39, 235 CrossRef CAS PubMed.
- S. Bhaskar, J. N. Banavaliker, K. Bhardwaj and P. Upadhyay, J. Immunol. Methods, 2002, 262, 181 CrossRef CAS PubMed.
- D. Satana, A. Y. Coban and M. Uzun, J. Clin. Microbiol., 2010, 48, 4291 CrossRef PubMed.
- R. Dinser, M. Fousse, U. Sester, K. Albrecht, M. Singh, H. Köhler, U. Müller-Ladner and M. Sester, Rheumatology, 2008, 47, 212 CrossRef CAS PubMed.
- L. Cosmi, L. Maggi, V. Santarlasci, F. Liotta, F. Frosali, R. Angeli, M. Mazzetti, A. Vultaggio, A. Matucci, E. Maggi, S. Romagnani and F. Annunziato, Int. Arch. Allergy Immunol., 2007, 143(1), 1–9 CrossRef CAS PubMed.
- L. Li, D. Qiao, X. Fu, S. Lao, X. Zhang and C. Wu, PLoS One, 2011, 6, e20165 CAS.
- M. D. Bonecini-Almeida, Mem. Inst. Oswaldo Cruz, 2000, 95, 491 CrossRef CAS PubMed.
- M. Piuri, W. R. Jacobs Jr and G. F. Hatfull, PLoS One, 2009, 4, e4870 Search PubMed.
- C. Pina-Vaz, S. Costa-de-Oliveira and A. G. Rodrigues, J. Med. Microbiol., 2005, 54, 77 CrossRef CAS PubMed.
- J. J. Yan, A. H. Huang, S. H. Tsai, W. C. Ko, Y. T. Jin and J. J. Wu, Diagn. Microbiol. Infect. Dis., 2000, 37, 25 CrossRef CAS PubMed.
- H. Saitoh and N. Yamane, Rinsho Biseibutshu Jinsoku Shindan Kenkyukai Shi, 2000, 11, 19 CAS.
- F. Alcaide, M. A. Benítez, J. M. Escribà and R. Martín, J. Clin. Microbiol., 2000, 38, 398 CAS.
- M. Garrigo, L. M. Aragon, F. Alcaide, S. Borrell, E. Cardenosa, J. J. Galán, J. Gonzalez-Martín, N. Martin-Casabona, C. Moreno, M. Salvado and P. Coll, J. Clin. Microbiol., 2007, 45, 1766 CrossRef CAS PubMed.
- L. Y. Itani, M. A. Cherry and G. F. Araj, J. Med. Liban., 2005, 53, 208 Search PubMed.
- N. Kobayashi, T. G. Fraser, T. W. Bauer, M. J. Joyce, G. S. Hall, M. J. Tuohy and G. W. Procop, Arch. Pathol. Lab. Med., 2006, 130, 1053 Search PubMed.
- D. V. Cousins, S. D. Wilton, B. R. Francis and B. L. Gow, J. Clin. Microbiol., 1992, 30, 255 CAS.
- R. M. Shawar, F. A. el-Zaatari, A. Nataraj and J. E. Clarridge, J. Clin. Microbiol., 1993, 31, 61 CAS.
- G. A. Thomas, D. L. Williams and S. A. Soper, Clin. Chem., 2001, 47, 1195 CAS.
- A. M. Mohamed, D. R. Bastola, G. P. Morlock, R. C. Cooksey and S. H. Hinrichs, J. Clin. Microbiol., 2004, 42, 1016 CrossRef CAS PubMed.
- V. J. Tevere, P. L. Hewitt, A. Dare, A. Keen, J. P. Spadoro and K. K. Young, J. Clin. Microbiol., 1996, 34, 918 CAS.
- C. T. Michelon, F. Rosso, K. B. Schmid, R. D. Sperhacke, M. M. Oliveira, A. L. Kritski, L. Jr Rezende, E. R. Costa, A. W. Ribeiro, M. Verza, P. I. Cafrune, M. S. Silva, D. Kuhleis, A. Zaha and M. L. Rossetti, Mem. Inst. Oswaldo Cruz, 2011, 106, 194 CrossRef PubMed.
- C. Drosten, M. Panning and S. Kramme, Clin. Chem., 2003, 49, 1659 CAS.
- F. Broccolo, P. Scarpellini, G. Locatelli, A. Zingale, A. M. Brambilla, P. Cichero, L. A. Sechi, A. Lazzarin, P. Lusso and M. S. Malnati, J. Clin. Microbiol., 2003, 41, 4565 CrossRef CAS PubMed.
- K. Baba, S. Pathak, L. Sviland, N. Langeland, A. A. Hoosen, B. Asjo, A. M. Dyrhol-Riise and T. Mustafa, Diagn. Mol. Pathol., 2008, 17, 112 CrossRef CAS PubMed.
- J. H. Kim, Y. J. Kim, C.-S. Ki, J.-Y. Kim and N. Y. Lee, J. Clin. Microbiol., 2011, 49, 173 CrossRef PubMed.
- S. A. Bustin, J. Mol. Endocrinol., 2002, 29, 23 CrossRef CAS PubMed.
- J. Papaparaskevas, D. P. Houhoula, A. Siatelis and A. Tsakris, J. Clin. Microbiol., 2008, 46, 3177 CrossRef PubMed.
- P. Kumar, K. Nath, B. Rath, M. K. Sen, P. Vishalakshi, D. S. Chauhan, V. M. Katoch, S. Singh, S. Tyagi, V. Sreenivas and H. K. Prasad, J. Mol. Diagn., 2009, 11, 430 CrossRef CAS PubMed.
- M. J. Taylor, M. S. Hughes, R. A. Skuce and S. D. Neill, J. Clin. Microbiol., 2001, 39, 1272 CrossRef CAS PubMed.
- T. Luo, L. L. Jiang, W. M. Sun, G. Fu, J. Mei and Q. Gao, J. Clin. Microbiol., 2011, 49, 3132 CrossRef CAS PubMed.
- S. Sang, W. Zhang and Y. Zhao, Review on the Design Art of Biosensors, 2013 Search PubMed.
- R. Thusu, Sensors Online (Strong Growth Predicted for Biosensors Market), 2010 Search PubMed.
- Z. Dai, F. Yan, J. Chen and H. Ju, Anal. Chem., 2003, 75, 5429 CrossRef CAS PubMed.
- T. G. Drummond, M. G. Hill and J. K. Barton, Nat. Biotechnol., 2003, 21, 1192 CrossRef CAS PubMed.
- M. Das, G. Sumana, R. Nagarajan and B. D. Malhotra, Appl. Phys. Lett., 2010, 96, 133703 CrossRef.
- M. Diaz-Gonzalez, M. B. Gonzalez-Garcia and A. Costa-Garcia, Biosens. Bioelectron., 2005, 20, 2035 CrossRef CAS PubMed.
- C. Fernandez-Sanchez, M. B. Gonzalez-Garcia and A. Costa-Garcia, Biosens. Bioelectron., 2000, 14, 917 CrossRef CAS PubMed.
- O. Ouerghi, A. Senillou, N. Jaffrezic-Renault, C. Martelet, H. Ben Ouada and S. Cosnier, J. Electroanal. Chem., 2001, 501, 62 CrossRef CAS.
- M. Das, C. Dhand, G. Sumana, A. K. Srivastava, N. Vijayan, R. Nagarajan and B. D. Malhotra, Appl. Phys. Lett., 2011, 99, 143702 CrossRef.
- A. K. Pavlou, N. Magan, J. M. Jones, J. Brown, P. Klatser and A. P. Turner, Biosens. Bioelectron., 2004, 20, 538 CrossRef CAS PubMed.
- A. D. Wilson and M. Baietto, Sensors, 2011, 11, 1105 CrossRef CAS PubMed.
- R. Freeman, R. Goodacre, P. R. Sisson, J. G. Magee, A. C. Ward and N. F. Lightfoot, J. Med. Microbiol., 1994, 40, 170 CrossRef CAS PubMed.
- G. Keshri and N. Magan, J. Appl. Microbiol., 2000, 89, 825 CrossRef CAS PubMed.
- A. P. Turner and N. Magan, Nat. Rev. Microbiol., 2004, 2, 161 CrossRef CAS PubMed.
- T. M. H. Lee, Sensors, 2008, 8, 5535 CrossRef CAS.
- F. Shen, M. Tan, Z. Wang, M. Yao, Z. Xu, Y. Wu, J. Wang, X. Guo and T. Zhu, Environ. Sci. Technol., 2011, 45, 7473 CrossRef CAS PubMed.
- D. R. S. Jeykumari and S. S. Narayanan, J. Nanosci. Nanotechnol., 2009, 9, 5411 CrossRef CAS PubMed.
- L. X. Zhou, X. X. He, D. G. He, K. M. Wang and D. L. Qin, Clin. Dev. Immunol., 2011, 193963 Search PubMed.
- M. Pumera, Methods Mol. Biol., 2010, 625, 205 CAS.
- G. Zheng, X. P. A. Gao and C. M. Lieber, Nano Lett., 2010, 10, 3179 CrossRef CAS PubMed.
- J. Lund, R. Mehta and B. A. Parviz, Nanomedicine, 2006, 2, 230 CAS.
- S. N. Kim, J. F. Rusling and F. Papadimitrakopoulos, Adv. Mater., 2007, 19, 3214 CrossRef CAS PubMed.
- S. Chatterjee, M. Castro and J. F. Feller, J. Mater. Chem. B, 2013, 36, 4563 RSC.
- K.-I. Chen, B.-R. Li and Y.-T. Chen, Nano Today, 2011, 6, 131 CrossRef CAS.
- M. M. Gregersen, M. B. Andersen, G. Soni, C. Meinhart and H. Bruus, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2009, 79, 066316 CrossRef PubMed.
- G. F. Zheng, F. Patolsky, Y. Cui, W. U. Wang and C. M. Lieber, Nat. Biotechnol., 2005, 23, 1294 CrossRef CAS PubMed.
- N. Kazemi-Zanjani, E. Kergrene, L. J. Liu, T. K. Sham and F. Lagugne-Labarthet, Sensors, 2013, 13, 12744 CrossRef CAS PubMed.
- K. A. Denton, M. F. Kramer and D. V. Lim, J. Rapid Methods Autom. Microbiol., 2009, 17, 17 CrossRef.
- B. M. Hewitt, N. Singhal, R. G. Elliot, A. Y. H. Chen, J. Y. C. Kuo, F. Vanholsbeeck and S. Swift, Environ. Sci. Technol., 2012, 46, 5414 CrossRef CAS PubMed.
- R. J. Yan, N. S. Lynn, L. C. Kingry, Z. J. Yi, R. A. Slayden, D. S. Dandy and K. L. Lear, Appl. Phys. Lett., 2011, 98, 13702 CrossRef.
- A. Leung, P. M. Shankar and R. Mutharasan, Sens. Actuators, B, 2007, 125, 688 CrossRef CAS.
- C. A. J. Gouveia, J. M. Baptista and P. A. S. Jorge, Refractometric Optical Fiber Platforms for Label Free Sensing, 2013 Search PubMed.
- H. Zhu, I. M. White, J. D. Suter, P. S. Dale and X. Fan, Opt. Express, 2007, 15, 9139 CrossRef CAS PubMed.
- I. M. White, J. Gohring and X. Fan, Opt. Express, 2007, 15, 17433 CrossRef CAS PubMed.
- M. Baaske and F. Vollmer, ChemPhysChem, 2012, 13, 427 CrossRef CAS PubMed.
- M. A. Cooper, Nat. Rev. Drug Discovery, 2002, 1, 515 CrossRef CAS PubMed.
- A. Rachkov, S. Patskovsky, A. Soldatkin and M. Meunier, Talanta, 2011, 85, 2094 CrossRef CAS PubMed.
- S. C. Hsieh, C. C. Chang, C. C. Lu, C. F. Wei, C. S. Lin, H. C. Lai and C. W. Lin, Nanoscale Res. Lett., 2012, 7, 180 CrossRef PubMed.
- N. Prabhakar, K. Arora, S. K. Arya, P. R. Solanki, M. Iwamoto, H. Singh and B. D. Malhotra, Analyst, 2008, 133, 1587 RSC.
- M. Duman and E. Piskin, Biosens. Bioelectron., 2010, 26, 908 CrossRef CAS PubMed.
- J. G. Huang, C. C. K. Hung, H. C. Lai, C. K. Lee, S. M. Lin, P. Feng and C. W. Lin, IEEE Engineering in Medicine and Biology Magazine, 2004, 26, 2553 Search PubMed.
- M. Duman, M. O. Caglayan, G. Demirel and E. Piskin, Sens. Lett., 2009, 7, 535 CrossRef CAS.
- A. K. Trilling, H. de Ronde, L. Noteboom, A. van Houwelingen, M. Roelse, S. K. Srivastava, W. Haasnoot, M. A. Jongsma, A. Kolk, H. Zuilhof and J. Beekwilder, PLoS One, 2011, 6, e26754 CAS.
- S. K. Srivastava, V. J. B. Ruigrok, N. J. Thompson, A. K. Trilling, A. J. R. Heck, C. van Rijn, J. Beekwilder and M. A. Jongsma, PloS One, 2013, 8, e64040 CAS.
- S. C. Hong, H. Chen, J. Lee, H.-K. Park, Y. S. Kim, H. C. Shin, C. M. Kim and T. J. Park, Sens. Actuators, B, 2011, 156, 271 CrossRef CAS.
- J. Homola, S. S. Yee and G. Gauglitz, Sens. Actuators, B, 1999, 54, 3 CrossRef CAS.
- R. McNerney, B. A. Wondafrash, K. Amena, A. Tesfaye, E. M. McCash and N. J. Murray, BMC Infect. Dis., 2010, 10, 161 CrossRef PubMed.
- T. B. Begg, I. D. Hill and L. C. Nickolls, Br. Med. J., 1964, 1, 9 CrossRef CAS PubMed.
- A. S. Mustafa, F. A. Shaban, A. T. Abal, R. Al-Attiyah, H. G. Wiker, K. E. Lundin, F. Oftung and K. Huygen, Infect. Immun., 2000, 68, 3933 CrossRef CAS PubMed.
- X. Mi, F. He, M. Xiang, Y. Lian and S. Yi, Anal. Chem., 2011, 84, 939 CrossRef PubMed.
- F. J. He and L. D. Zhang, Anal. Sci., 2002, 18, 397 CrossRef CAS PubMed.
- F. J. He, L. D. Zhang, J. W. Zhao, B. L. Hu and J. T. Lei, Sens. Actuators, B, 2002, 85, 284 CrossRef CAS.
- A. Kumar and N. M. Ravindra, J. Miner. Met. Mater. Soc., 2000, 52(10), 14 CrossRef.
- J. L. Ren, F. J. He, S. L. Yi and X. Y. Cui, Biosens. Bioelectron., 2008, 24, 403 CrossRef CAS PubMed.
- T. M. Gronewold, Anal. Chim. Acta, 2007, 603, 119 CrossRef CAS PubMed.
- G. N. Ferreira, A. C. da-Silva and B. Tome, Trends Biotechnol., 2009, 27, 689 CrossRef CAS PubMed.
- F. J. He, J. W. Zhao, L. D. Zhang and X. N. Su, Talanta, 2003, 59, 935 CrossRef CAS PubMed.
- C. Ruan, K. Zeng, O. K. Varghese and C. A. Grimes, Anal. Chem., 2003, 75, 6494 CrossRef CAS PubMed.
- R. Zhang, M. I. Tejedor-Tejedor, C. A. Grimes and M. A. Anderson, Anal. Chem., 2007, 79, 7078 CrossRef CAS PubMed.
- P. Pang, Q. Cai, S. Yao and C. A. Grimes, Talanta, 2008, 76, 360 CrossRef CAS PubMed.
- Q. Y. Cai, A. Cammers-Goodwin and C. A. Grimes, J. Environ. Monit., 2000, 2, 556 RSC.
- W. Shen, S. Li, M.-K. Park, Z. Zhang, Z. Cheng, V. Petrenko and B. A. Chin, J. Electrochem. Soc., 2012, 159(10), B818 CrossRef CAS.
- D. Issadore, C. Min, M. Liong, J. Chung, R. Weissleder and H. Lee, Lab Chip, 2011, 11, 2282 RSC.
- J. B. Haun, C. M. Castro, R. Wang, V. M. Peterson, B. S. Marinelli, H. Lee and R. Weissleder, Sci. Transl. Med., 2011, 3, 71 Search PubMed.
- A. L. Chun, Nat. Nanotechnol., 2009, 4, 698 CrossRef CAS PubMed.
- H. Lee, E. Sun, D. Ham and R. Weissleder, Nat. Med., 2008, 14, 869 CrossRef CAS PubMed.
- H. J. Chung, C. M. Castro, H. Im, H. Lee and R. Weissleder, Nat. Nanotechnol., 2013, 8, 369 CrossRef CAS PubMed.
- M. Liong, A. N. Hoang, J. Chung, N. Gural, C. B. Ford, C. Min, R. R. Shah, R. Ahmad, M. Fernandez-Suarez, S. M. Fortune, M. Toner, H. Lee and R. Weissleder, Nat. Commun., 2013, 4, 1752 CrossRef PubMed.
- H. Lee, T.-J. Yoon and R. Weissleder, Angew. Chem., Int. Ed., 2009, 48, 5657 CrossRef CAS PubMed.
- H. Mukundan, S. Kumar, D. N. Price, S. M. Ray, Y. J. Lee, S. Min, S. Eum, J. Kubicek-Sutherland, W. K. Grace, A. S. Anderson, S. H. Hwang, S. N. Cho, L. E. Via, C. III Barry, R. Sakamuri and B. I. Swanson, Tuberculosis, 2012, 92(5), 407 CrossRef CAS PubMed.
- H. Chen, F. Lie, K. Koh, J. Lee, Z. Ye, T. Yin and L. Sun, Microchim. Acta, 2013, 180(5), 431 CrossRef CAS.
- A. Miodek, N. Mejri, M. Gomgnimbou, C. Sola and H. Korri-Youssoufi, Anal. Chem., 2015, 87(18), 9257 CrossRef CAS PubMed.
- C. Liu, D. Jiang, G. Xiang, L. Liu, F. Liu and X. Pu, Analyst, 2014, 139(21), 5460 RSC.
- P. A. Marin, L. E. Botero, J. A. Robledo, A. M. Murillo, R. A. Torres, Y. J. Montagut, E. Pabon and M. Jaramillo, Acta Biol. Colomb., 2015, 20(1), 40731 Search PubMed.
- A. M. Pariwono, T. Lo, C. S. Lim, S. X. Wang and Y. W. Chan, Journal of Biomedical & Pharmaceutical Engineering, 2007, 1, 27 Search PubMed.
- R. Fend, A. H. J. Kolk, C. Bessan, P. Buijtels, P. R. Klatser and A. C. Woodman, J. Clin. Microbiol., 2006, 44(6), 2039 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2016 |