Open Access Article
Open Access ArticleCharacterisation of 6-DMATSMo from Micromonospora olivasterospora leading to identification of the divergence in enantioselectivity, regioselectivity and multiple prenylation of tryptophan prenyltransferases†
Julia
Winkelblech
ab,
Xiulan
Xie
c and
Shu-Ming
Li
*ab
aPhilipps-Universität Marburg, Institut für Pharmazeutische Biologie und Biotechnologie, Robert-Koch-Straße 4, 35037 Marburg, Germany. E-mail: shuming.li@staff.uni-marburg.de
bZentrum für Synthetische Mikrobiologie, Philipps-Universität Marburg, Hans-Meerwein-Straße, D-35032 Marburg, Germany
cPhilipps-Universität Marburg, Fachbereich Chemie, Hans-Meerwein-Straße, 35032 Marburg, Germany
First published on 20th September 2016
Abstract
Prenylated secondary metabolites including indole derivatives usually demonstrate improved biological and pharmacological activities, which make them promising candidates for drug discovery and development. The transfer reactions of a prenyl moiety from a prenyl donor, e.g. dimethylallyl diphosphate (DMAPP), to an acceptor is catalysed by prenyltransferases. One special group of such enzymes uses DMAPP and tryptophan as substrates with dimethylallyltryptophans as reaction products and functions therefore as dimethylallyltryptophan synthases (DMATSs). Sequence homology search with known tryptophan prenyltransferases from Streptomyces led to identification of a putative prenyltransferase gene MolI14.36 in Micromonospora olivasterospora. Expression and biochemical investigations revealed that MolI14.36 acts as a tryptophan C6-prenyltransferase (6-DMATSMo). Study on substrate specificity of 6-DMATSMo displayed a significantly high activity towards D-tryptophan, which prompted us to carry out comparative studies on enantioselectivity, regioselectivity and multiple prenylation ability of additional DMATSs including FgaPT2, 5-DMATS, 5-DMATSSc, 6-DMATSSv, 6-DMATSSa and 7-DMATS towards L- and D-isomers of tryptophan and their analogues. The relative activities of the tested enzymes towards D-tryptophan differ clearly from each other. Incubation of L-, D-isomers or the racemates of 5-, 6- and 7-methyltryptophan revealed distinctly different preferences of the DMATS enzymes. Interestingly, 6-DMATSMo and 5-DMATSSc accepted 5-methyl-D-tryptophan much better than the L-enantiomer. Furthermore, the conversion yields of the D-isomers were strongly inhibited in the reactions with racemates. More interestingly, the regioselectivities of FgaPT2, 5-DMATSSc and 7-DMATS towards D-tryptophan and its C5-methylated derivative differed clearly from those of the L-forms. In addition, both mono- and diprenylated products were clearly detected for 5-DMATSSc with L- and D-enantiomers of tryptophan and their methylated derivatives.
Introduction
Chiral molecules such as D- and L-amino acids play an important role in biological and chemical processes of life.1 Moreover, the chirality of small molecules is of high importance for their application as drugs. The biological and pharmacological activities of D- and L-isomers as well as toxicity and metabolism could strongly differ from each other.2 The natural occurrence of α-amino acids is clearly predominated by the L-form, but also the D-form is widely distributed in nature, fulfilling essential roles in biological systems.3 For example, D-amino acids are found in the cell walls of bacteria to provide protease resistance or as neurotransmitters in the nervous system of animals.4–6 Moreover, D-amino acids are of great interest for pharmaceutical application. Peptides containing D-amino acids are used as antibiotics or considered as potential agents for treatment of Alzheimer's disease, HIV and cancers.3,7–9 Amino acids like tryptophan are also key precursors for a number of secondary metabolites including prenylated indole derivatives.10 These isoprenoid-derived natural products are widely distributed in nature, e.g. in the fungi of ascomycetes and bacteria of actinomycetes.10 The connection of the indole and isoprenoid moieties is usually catalysed by prenyltransferases.11 Prenylation of aromatic compounds could improve their biological activity.10,12–15 For instance, studies on the antifungal activity of substituted indole analogues revealed the importance of the allyl side chain of 6-prenylindole for its bioactivity.16One special group of prenyltransferases uses dimethylallyl diphosphate (DMAPP) as the donor and L-tryptophan as the acceptor and functions thus as dimethylallyltryptophan synthase (DMATS). For example, FgaPT2 from the ascomycetous fungus Aspergillus fumigatus (A. fumigatus) catalyses the transfer of a dimethylallyl moiety from DMAPP to C-4 of L-tryptophan and is involved in the biosynthesis of ergot alkaloids.17 Later on, at least six additional fungal DMATSs were identified, which act as C4-, C5-, and C7-prenylating enzymes.11 In contrast with DMATSs from fungi, a few members of the bacterial tryptophan prenyltransferases were only recently characterised biochemically. Therefore, biochemical characterisation of new DMATSs from bacteria will contribute to our understanding on the catalytic features of these enzymes from different origins. The known bacterial DMATSs are from actinomycetes, catalyse the transfer of the dimethylallyl moiety to C-5, C-6 or C-7 of the indole ring, and are involved in the biosynthesis of prenylated indole derivatives.11 For example, IptA from Streptomyces sp. SN-593 functions as a 6-DMATS and is involved in the biosynthesis of 6-dimethylallylindole-3-carbaldehyde.18 Three IptA orthologues, IptAAm from Actinoplanes missouriensis,19 6-DMATSSa from Streptomyces ambofaciens (S. ambofaciens), and 6-DMATSSv from S. violaceusniger have been recently identified and characterised.20SCO7467 from S. coelicolor A3(2) belongs to a gene cluster being responsible for the biosynthesis of 5-dimethylallylindole-3-acetonitrile and the encoded protein acts as a 5-DMATS (5-DMATSSc).21,22 Recently, Wu et al. have identified a biosynthetic gene cluster for a new antibiotic 7-prenylisatin in Streptomyces MBT28-91. Thereby, the prenyltransferase IsaA catalyses the prenylation of L-tryptophan at C-7.23
In the present study, we continue to expand our knowledge on DMATSs from bacteria by cloning, expression and biochemical investigations on a putative prenyltransferase MolI14.36 from Micromonospora olivasterospora (M. olivasterospora). Biochemical investigations revealed that MolI14.36 acts as a tryptophan C6-prenyltransferase (6-DMATSMo). Previous studies on DMATSs revealed that L-tryptophan was much better accepted than D-tryptophan.24 In comparison, D-tryptophan was very well accepted by 6-DMATSMo. This finding promoted us to carry out systematic investigation on the acceptance and enzyme products of L- and D-tryptophan and their methylated derivatives including 5-, 6-, and 7-methyltryptophan by DMATSs from bacteria and fungi. These include three fungal (FgaPT2, 5-DMATS, and 7-DMATS) and four bacterial DMATSs (5-DMATSSc, 6-DMATSSa, 6-DMATSSv, and 6-DMATSMo). Evaluation of the enzyme products demonstrated a clear difference in substrate specificity, regioselectivity of the prenyl transfer reactions as well as the ability for multiple prenylation.
Results and discussion
Identification and characterisation of a new 6-DMATS from M. olivasterospora
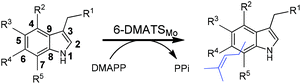
BLAST search by using known bacterial tryptophan prenyltransferases led to the identification of a putative prenyltransferase from M. olivasterospora. MolI14.36 comprises 376 amino acids and has a predicted molecular mass of 40.6 kDa. MolI14.36 shares clearly different sequence identities with known tryptophan C6-prenyltransferases on the amino acid level, e.g. 68% with IptAAm from A. missouriensis,19 44% with IptA from Streptomyces sp. SN-593,18 42% with 6-DMATSSv from S. violaceusniger,20 and 38% with 6-DMATSSa from S. ambofaciens.20 The differences of the sequence identities raised the question about the function of MolI14.36.MolI14.36 was then cloned from genomic DNA into the expression vector pHIS8 and overexpressed in E. coli. The recombinant His8-tagged protein with a molecular mass of 43.5 kDa was purified to near homogeneity with a yield of 16 mg per litre culture (Fig. S1, ESI†). Size exclusion chromatography revealed that the enzyme acts as a monomer.
To prove its function and substrate specificity, the purified recombinant MolI14.36 (1 μM) was incubated with 0.5 mM of L-tryptophan (1a), D-tryptophan (1b), and eight analogues thereof (2a, 3a, 4–8, and 9a) in the presence of 1 mM DMAPP. To show the relationships of the enantiomers, we use Arabic numbers for racemates, numbers with a for L-isomers and with b for D-isomers. After incubation at 37 °C for 1 h, the reaction mixtures were analysed on HPLC under the conditions listed in Table S1 in the ESI.† As shown in Table 1 and the ESI (Fig. S2 and S3†), eight of them, 1a, 1b, 2a, 3a, 4, 5, 8, and 9a, were well accepted by this enzyme, with 1a as the best substrate. In the presence of L-tryptophan (1a), MolI14.36 also used geranyl diphosphate as the prenyl donor, but with a significantly lower product yield than with DMAPP (about 10% of that of DMAPP). Farnesyl diphosphate was not accepted by MolI14.36 (Table S2, ESI†). HPLC analysis of the reaction mixtures of MolI14.36 with seven tryptophan-containing cyclic dipeptides showed product formation, but with much lower conversion yields than with tryptophan and its analogues (less than 8% of that of 1a) (Table S3, ESI†). These results provided evidence for the function of MolI14.36 as a dimethylallyl diphosphate:L-tryptophan transferase.
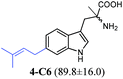
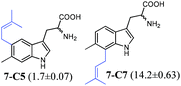
To confirm the prenylation in their structures and particularly the prenylation positions, the enzyme products of tryptophan and its analogues were isolated from large-scale incubation mixtures on preparative HPLC and subsequently analysed by MS and NMR including homonuclear correlation spectroscopy (1H–1H COSY) for 5-C5–5-C7, 6-C6 and 6-C7 (Tables S4–S7 and Fig. S4–S22, ESI†). For better understanding, the enzyme products were termed by addition of the prenylation position like C4, C5, C6 or C7 to the number of the substrate. Inspection of the NMR spectra of the isolated peaks confirmed the unique C6-prenylated products 1a-C6, 1b-C6, 2a-C6, 3a-C6, 4-C6, 8-C6, and 9a-C6 from the reaction mixtures of 1a, 1b, 2a, 3a, 4, 8, and 9a, whereas three products with the prenyl moiety attached to C-5 (5-C5), C-6 (5-C6) and C-7 (5-C7) in a ratio of 0.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.6 were identified in the reaction mixture of 5. Two products either 6-C6 and 6-C7 or 7-C5 and 7-C6 were detected in those of 6 and 7 (see the ESI† for detailed structure elucidation, Tables S5–S7, Fig. S4–S22†).
0.6 were identified in the reaction mixture of 5. Two products either 6-C6 and 6-C7 or 7-C5 and 7-C6 were detected in those of 6 and 7 (see the ESI† for detailed structure elucidation, Tables S5–S7, Fig. S4–S22†).
In conclusion, MS and NMR analyses of the isolated enzyme products prove that MolI14.36 acts as a L-tryptophan C6-prenyltransferase and is termed 6-DMATSMo, in analogy to the notation of 6-DMATSSa and 6-DMATSSv.20 6-DMATSMo catalyses a unique or predominant C6-prenylation at the indole ring of tryptophan and its analogues (Table 1). If the position 6 is blocked by a methyl group as in the case of 7, a switch of the prenylation site to C-7 was detected, as observed for IptA and 6-DMATSSa, previously.18,20
Additional biochemical characterisation revealed that metal ions are not essential for the enzyme activity as observed for other members of the DMATS superfamily (Fig. S23, ESI†).18,20,25 Furthermore, the function of 6-DMATSMo as a tryptophan prenyltransferase was justified by kinetic studies. The KM value of 0.014 ± 0.002 mM and a turnover number kcat of 0.07 ± 0.002 s−1 were determined for 1a. The kinetic parameters for the prenyl donor DMAPP were found to be 0.037 ± 0.007 mM and 0.08 ± 0.004 s−1, respectively (Fig. S24, ESI†). For comparison of the substrate preferences of 6-DMATSMo and its orthologues 6-DMATSSa and 6-DMATSSv, enzyme assays of 1a, 1b, 2a, 3a, 4–8 and 9a were carried out for the three 6-DMATSs under similar conditions and the relative enzyme activities to 1a were compared with each other (Fig. S3, ESI†). In summary, with the exception for 3a, 6-DMATSSa and 6-DMATSSv seem to share similar substrate preferences towards the tested tryptophan analogues, whereas 6-DMATSMo shows more distinct preferences from those of the two other 6-DMATSs. 1b, 4, 5, and 8 were better accepted by 6-DMATSMo, whereas 6, 7, and 9a were better substrates for the other two 6-DMATS enzymes. The most remarkable feature is the high acceptance of 1b by 6-DMATSMo, with a relative activity of approximately 50% of that of 1a (Table 1 and Fig. S3, ESI†). To the best of our knowledge, such high conversion of 1b has not been reported for other prenyltransferases prior to this study. In comparison, less than 25% relative activities of that of 1a were detected for 6-DMATSSa and 6-DMATSSv with 1b under these conditions. These results prompted us to have detailed insights into the enantioselectivity of the DMATS enzymes towards tryptophan.
DMATSs showed different preferences towards D-tryptophan
To gain deeper insights into the substrate preferences of DMATSs towards the stereoisomers of tryptophan, FgaPT2, 5-DMATS, 5-DMATSSc, 6-DMATSSa, 6-DMATSSv, 6-DMATSMo, and 7-DMATS were overproduced in E. coli and purified to near homogeneity as reported previously.20,25–28 As aforementioned, these enzymes use the same substrates 1a and DMAPP, but catalyse prenylations at different positions of the indole ring. By size exclusion chromatography, these enzymes were determined to have different quaternary structures. The native forms of FgaPT2 and 5-DMATS were found to be homodimer, whereas 5-DMATSSc and 7-DMATS were reported to be active as monomers.17,25,28 In this study, the molecular mass of 6-DMATSSa was determined to be 36.9 kDa and the other two enzymes 6-DMATSSv and 6-DMATSMo to be 46.0 kDa. This proved that they are active as monomers, being consistent with their orthologue IptA.18 Crystal structure analysis of several fungal prenyltransferases including FgaPT2 revealed that each single subunit contains one active centre and forms one catalytic unit.29–32 To ensure adequate comparability, the following analyses on substrate specificity and kinetic parameters of the recombinant proteins all refer to a unique protein subunit.For investigations on substrate preferences, the seven DMATSs were incubated with the prenyl donor DMAPP and 1a, 1b, or their racemate 1. The relative activities to 1a were determined by HPLC analysis on the CHIRALPAK® Zwix(+) column. The prenylation of the enzyme products was proven by LC-MS analysis (Table 2 and Fig. 1, S25–S48, ESI†). As given in Table 2, clearly different enantioselectivities were observed for the tested DMATSs. 5-DMATS and 6-DMATSMo accepted 1b much better than other enzymes, with relative activities of 34.7 and 45.7% of those of L-tryptophan (1a), respectively. In contrast, 1b was a very poor substrate for FgaPT2 and 7-DMATS with relative conversion yields of approximately 5 and 6%, respectively, corresponding well to the data reported previously.17,27,28,33 In the reaction mixtures of racemates, the conversion of 1b was strongly reduced, indicating an inhibition. For better understanding of the observed acceptance of 1a and 1b by the tested DMATSs, kinetic parameters were determined by nonlinear regression using GraphPad Prism 4.0 (Fig. S49–S55, ESI†). All investigated reactions apparently followed the Michaelis–Menten kinetics. The calculated KM values of the seven DMATSs for 1a varied from 0.012 mM to 0.055 mM, whereas those for 1b were found in the range of 0.10 to 1.76 mM (Table 3). The significantly higher affinity of the enzymes to the L-form is justified by their native functions as L-tryptophan prenyltransferases and also explained in parts the very low conversion of 1b in the reaction with the racemate. It is plausible that in the initial phase of the reactions with racemates, only 1a was used as substrate by the enzymes. However, the higher affinity and turnover numbers of the tested enzymes towards 1a than 1b could not explain the observed very low conversion of 1b. Under the conditions used for the conversion yields given in Table 2, 1a was almost completely converted. Therefore, we speculated that the products of 1a should also contribute to the inhibition of 1b reactions.
 | ||
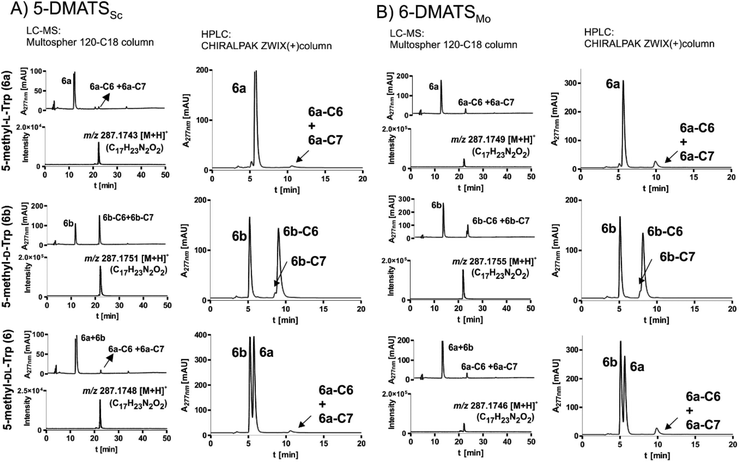
| Fig. 1 Evaluation of the enantioselectivity of (A) 5-DMATS and (B) 6-DMATSMo. The enzymes were incubated with 1 mM DMAPP and 0.5 mM of L-tryptophan, D-tryptophan or a combination of both (1 mM) and the reaction products were profiled by LC-MS and HPLC. 1 μM of the indicated enzyme was incubated with the indicated substrate(s). UV detection was carried out with a diode array detector and illustrated for absorption at 277 nm. Additional chromatograms and MS analyses for all tested enzymes and substrates are provided as Fig. S25–S48 in the ESI.† | ||
| Substrate | Relative conversion yields [%] | |||||||
|---|---|---|---|---|---|---|---|---|
| FgaPT2 | 5-DMATS | 5-DMATSSc | 6-DMATSSa | 6-DMATSSv | 6-DMATSMo | 7-DMATS | ||
The enzyme assays contained 0.5 mM of the L- or D-isomers or 1 mM of the racemates and 1 mM DMAPP were incubated at 37 °C for 1.5 h with 1 μM purified protein. The conversion yields of L-tryptophan (1a) with FgaPT2 at 97.3%, 5-DMATS at 98.4%, 5-DMATSSc at 96%, 6-DMATSMo at 82.7%, 6-DMATSSv at 96.1%, 6-DMATSSa at 97.0%, and with 7-DMATS at 88.9% were defined as 100% relative activity, respectively. The conversion yields of D- or L-enantiomers in the reaction mixtures with racemates were calculated separately by considering the respective enantiomer as the substrate.a Diprenylated products, with a ratio of 0.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to the monoprenylated product, were detected.b Diprenylated products, with a ratio of 0.1 1 to the monoprenylated product, were detected.b Diprenylated products, with a ratio of 0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to the monoprenylated product, were detected. 1 to the monoprenylated product, were detected. |
||||||||
| L-Tryptophan | 1a | 100.0 ± 2.5 | 100.0 ± 0.7 | 100.0 ± 0.04a | 100.0 ± 2.7 | 100.0 ± 0.9 | 100.0 ± 12.2 | 100.0 ± 4.3 |
| D-Tryptophan | 1b | 5.2 ± 0.9 | 34.7 ± 4.4 | 20.2 ± 2.6b | 10.4 ± 0.02 | 16.8 ± 1.1 | 45.7 ± 2.9 | 6.4 ± 0.2 |
| DL-Tryptophan | 1a | 96.3 ± 2.1 | 96.7 ± 4.8 | 103.5 ± 4.7 | 99.4 ± 8.2 | 87.1 ± 2.6 | 99.9 ± 1.0 | 98.9 ± 1.5 |
| 1b | ≤0.5 | 12.6 ± 2.2 | ≤0.5 | 1.6 ± 0.3 | 2.5 ± 0.01 | 1.3 ± 0.1 | ≤0.5 | |
| 5-Methyl-L-tryptophan | 6a | 43.0 ± 1.8 | 2.5 ± 0.1 | 2.3 ± 0.2 | 102.5 ± 0.2 | 100.5 ± 0.7 | 12.9 ± 0.2 | 84.9 ± 3.8 |
| 5-Methyl-D-tryptophan | 6b | ≤0.5 | ≤0.5 | 57.3 ± 1.8 | 11.4 ± 0.6 | 22.4 ± 0.4 | 66.6 ± 0.5 | 2.5 ± 0.6 |
| 5-Methyl-DL-tryptophan | 6a | 22.4 ± 4.0 | 3.1 ± 1.1 | 3.8 ± 0.6 | 76.2 ± 6.5 | 100.7 ± 4.1 | 13.0 ± 0.6 | 97.9 ± 4.3 |
| 6b | ≤0.5 | ≤0.5 | ≤0.5 | 2.4 ± 1.1 | 4.7 ± 0.1 | ≤0.5 | ≤0.5 | |
| 6-Methyl-L-tryptophan | 7a | 84.0 ± 1.2 | 85.5 ± 5.7 | 15.0 ± 1.7 | 9.1 ± 1.7 | 35.4 ± 2.3 | 5.5 ± 0.2 | 17.5 ± 1.1 |
| 6-Methyl-D-tryptophan | 7b | ≤0.5 | ≤0.5 | 1.1 ± 0.1 | ≤0.5 | ≤0.5 | 1.2 ± 0.2 | ≤0.5 |
| 6-Methyl-DL-tryptophan | 7a | 61.2 ± 0.8 | 82.5 ± 5.1 | 15.1 ± 0.2 | 8.2 ± 0.4 | 33.2 ± 0.6 | 5.0 ± 0.6 | 16.6 ± 0.9 |
| 7b | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | |
| 7-Methyl-L-tryptophan | 8a | 97.8 ± 1.7 | 75.7 ± 0.5 | 8.2 ± 2.3 | 4.3 ± 0.2 | 7.4 ± 1.6 | 72.4 ± 8.5 | ≤0.5 |
| 7-Methyl-D-tryptophan | 8b | ≤0.5 | 3.7 ± 1.4 | 1.9 ± 0.2 | ≤0.5 | ≤0.5 | 13.8 ± 3.8 | ≤0.5 |
| 7-Methyl-DL-tryptophan | 8a | 68.2 ± 5.0 | 70.6 ± 4.2 | 5.9 ± 0.2 | 3.8 ± 0.4 | 7.5 ± 0.7 | 72.5 ± 13.1 | ≤0.5 |
| 8b | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | |
| DMATS | L-Tryptophan | D-Tryptophan | 5-Methyl-L-tryptophan | 5-Methyl-D-tryptophan | ||||
|---|---|---|---|---|---|---|---|---|
| 1a | 1b | 6a | 6b | |||||
| K M [mM] | k cat [s−1] | K M [mM] | k cat [s−1] | K M [mM] | k cat [s−1] | K M [mM] | k cat [s−1] | |
| —, Not determined. | ||||||||
| FgaPT2 | 0.034 ± 0.003 | 0.67 ± 0.01 | 0.10 ± 0.007 | 0.012 ± 0.0002 | — | — | — | — |
| 5-DMATS | 0.055 ± 0.002 | 0.39 ± 0.001 | 0.62 ± 0.08 | 0.066 ± 0.004 | — | — | — | — |
| 5-DMATSSc | 0.020 ± 0.002 | 0.19 ± 0.004 | 1.47 ± 0.08 | 0.046 ± 0.001 | 0.009 ± 0.001 | 0.005 ± 0.0001 | 0.03 ± 0.01 | 0.035 ± 0.005 |
| 6-DMATSSa | 0.012 ± 0.001 | 0.10 ± 0.002 | 1.02 ± 0.07 | 0.021 ± 0.001 | — | — | — | — |
| 6-DMATSSv | 0.022 ± 0.002 | 0.19 ± 0.004 | 0.77 ± 0.08 | 0.021 ± 0.001 | — | — | — | — |
| 6-DMATSMo | 0.014 ± 0.002 | 0.07 ± 0.002 | 0.47 ± 0.04 | 0.066 ± 0.002 | 0.008 ± 0.001 | 0.012 ± 0.0002 | 0.30 ± 0.05 | 0.042 ± 0.003 |
| 7-DMATS | 0.043 ± 0.004 | 0.12 ± 0.002 | 1.76 ± 0.19 | 0.013 ± 0.001 | — | — | — | — |
The calculated turnover numbers (kcat) for 1a from 0.07 to 0.67 s−1 are in almost all cases much higher than those for 1b between 0.012 and 0.066 s−1. In comparison to 1b reactions with other enzymes, relative high affinity and turnover numbers were determined for those with 5-DMATS and 6-DMATSMo. The turnover numbers of 6-DMATSMo towards 1a and 1b are nearly identical at approximately 0.07 s−1. These data supported the high conversion yields of 1b by 5-DMATS and 6-DMATSMo given in Table 2.
Different regioselectivities of DMATSs towards 1a and 1b
Previous investigations have shown that the characterised DMATSs are highly regiospecific for 1a, i.e. C4-prenylation by FgaPT2,17C5-prenylation by 5-DMATS and 5-DMATSSc,22,25,26C6-prenylation by IptA, 6-DMATSSa, 6-DMATSSv and 6-DMATSMo18,20 and C7-prenylation by 7-DMATS and 7-DMATSNeo.28,34 In the course of the biochemical characterisation, the enzyme activities of DMATSs towards 1b were usually demonstrated by HPLC analysis using achiral columns.18,20,25,27,28,34–36 No product of 1b has been isolated and characterised. As mentioned above, isolation and structure elucidation of the enzyme products confirmed the same C6-prenylation of 6-DMATSMo for 1a and 1b (Tables 4, S5 and Fig. S4, S5, ESI†). In contrast, inspection of the NMR spectrum of the enzyme products of 5-DMATSSc with 1b revealed the presence of a mixture of three compounds (Table 4, Fig. 2 and Table S8 and Fig. S56, ESI†). Comparison of the chemical shifts and coupling patterns with the previously published data of the prenylated derivatives of 1a led to the identification of these compounds to be C5-prenylated 1b-C5, C6-prenylated 1b-C6, and C7-prenylated 1b-C7 in a ratio of 0.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.4. In comparison, the C5-monoprenylated derivative 1a-C5 is the unique product in the reaction mixture of 1a with 5-DMATSSc.
0.4. In comparison, the C5-monoprenylated derivative 1a-C5 is the unique product in the reaction mixture of 1a with 5-DMATSSc.
 | ||
| Fig. 2 HPLC identification of the enzyme products of DMATSs and 1a or 1b. The enzymes were incubated with 1 mM DMAPP and 1 mM of L-tryptophan (1a) or D-tryptophan (1b). Detailed conditions of the enzyme assays are given in Table S9 in the ESI.† For HPLC analysis, an Eclipse XDB-C18 column was used (condition 4 in Table S1†). Detection was carried out on a diode array detector and illustrated for absorption at 277 nm. | ||
| Substrate structures | Enzyme products and their ratios | |||||||
|---|---|---|---|---|---|---|---|---|
| FgaPT2 | 5-DMATS | 5-DMATSSc | 6-DMATSSa | 6-DMATSSv | 6-DMATSMo | 7-DMATS | ||
| The ratio of the enzyme products was evaluated from HPLC chromatograms.a The ratio of the enzyme products was evaluated from 1H NMR spectra.b Product formation was detected by LC-MS analysis. | ||||||||
| 1a |

|
C4 | C5 | C5 | C6 | C6 | C6 | C7 |
| 1b |
|
C4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C5 C5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C7 C7 |
C5 |
C5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C6 C6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C7 C7 |
C6 | C6 | C6 |
C6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C7 C7 |
0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.04 0.04 |
0.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.4a 0.4a |
0.05![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
||||||
| 6a |
|
C4 |
C6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C7 C7 |
C6 | C6 |
C6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C7 C7 |
C7 | |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.05 0.05 |
|||||||
| 6b |

|
C4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C6/C7 C6/C7 |
C6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C7 C7 |
C6 | C6 |
C6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C7 C7 |
C6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C7 C7 |
|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.4 0.4 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2a 0.2a |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1a 0.1a |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2 0.2 |
|||||
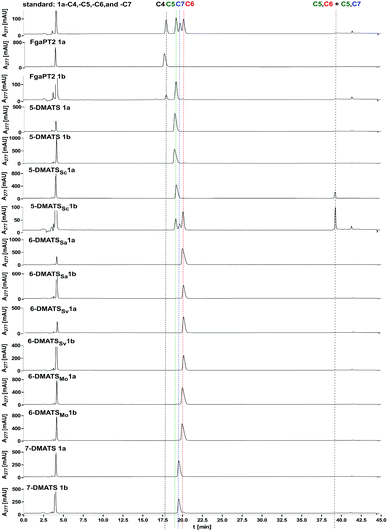
To prove the prenylation positions, the seven DMATSs were incubated with 1a and 1b and analysed on an achiral XDB-C18 column under the improved HPLC condition 4 (Fig. 2 and Tables S1, S9, ESI†). Under this condition, the enantiomeric pairs 1a and 1b have the same retention times, which is also true for their derivatives with the same prenylation positions. More importantly, C4-, C5-, C6-, and C7-prenylated tryptophan were well separated from each other. The results in Fig. 2 confirmed the previously published regiospecific prenylation of 1a by the tested DMATSs. The enzyme products of 1b with 5-DMATS and the three C6-prenyltransferases 6-DMATSSa, 6-DMATSSv and 6-DMATSMo had the same prenylation positions as those of 1a. Interestingly, the main product of 1b with FgaPT2 was prenylated at position C-5 instead of C-4. C4- and likely C7-prenylated derivatives were detected as minor products (Table 4 and Fig. 2). As aforementioned, three products 1b-C5, 1b-C6, and 1b-C7 with a ratio of 0.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0
1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.4 were detected in the reaction mixture of 1b with 5-DMATSSc (Fig. 2, Tables 4, S8 and Fig. S56, ESI†).20,25,37 HPLC analysis of the reaction mixture of 1b with 7-DMATS proved the main product to be 7-DMA-D-tryptophan (1b-C7) and the minor product the C6-prenylated derivative 1b-C6 in a ratio of 1
0.4 were detected in the reaction mixture of 1b with 5-DMATSSc (Fig. 2, Tables 4, S8 and Fig. S56, ESI†).20,25,37 HPLC analysis of the reaction mixture of 1b with 7-DMATS proved the main product to be 7-DMA-D-tryptophan (1b-C7) and the minor product the C6-prenylated derivative 1b-C6 in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.05 (Fig. 2 and Table 4). The observed changes in regioselectivity could indicate different orientations of the two enantiomers in the active sites. Crystal structures of such bacterial tryptophan prenyltransferases could provide detailed insights into their reaction chambers.
0.05 (Fig. 2 and Table 4). The observed changes in regioselectivity could indicate different orientations of the two enantiomers in the active sites. Crystal structures of such bacterial tryptophan prenyltransferases could provide detailed insights into their reaction chambers.
DMATSs also catalyse the diprenylation of 1a and 1b with different activities
HPLC analysis on a chiral column of the reaction mixture of 1a with 5-DMATSSc revealed the presence of two product peaks with retention times at 9.2 and 18.5 min in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.6 (Fig. S32, ESI†). The main peak was identified as C5-monoprenylated tryptophan by Ozaki et al.,21 and also confirmed in this study, whereas the second peak was not mentioned in the previous study. LC-MS analysis led to the identification of the second product peak as diprenylated derivative(s). Isolation and structure elucidation with the help of 1H NMR confirmed the diprenylation of 1a and provided evidence for the presence of a mixture of two products (Table S8 and Fig. S57, ESI†). Detailed interpretation of the spectra and comparison of the coupling patterns with those of the previously published data led to identification of 5,6- and 5,7-di-dimethylallyl-L-tryptophan (1a-C5,C6, 1a-C5,C7) in a ratio of approximately 1
0.6 (Fig. S32, ESI†). The main peak was identified as C5-monoprenylated tryptophan by Ozaki et al.,21 and also confirmed in this study, whereas the second peak was not mentioned in the previous study. LC-MS analysis led to the identification of the second product peak as diprenylated derivative(s). Isolation and structure elucidation with the help of 1H NMR confirmed the diprenylation of 1a and provided evidence for the presence of a mixture of two products (Table S8 and Fig. S57, ESI†). Detailed interpretation of the spectra and comparison of the coupling patterns with those of the previously published data led to identification of 5,6- and 5,7-di-dimethylallyl-L-tryptophan (1a-C5,C6, 1a-C5,C7) in a ratio of approximately 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.20,25,37 The characteristic singlets for H-4, H-7 and H-2 of 1a-C5,C6 were observed at 7.45, 7.14 and 7.08 ppm, respectively. For 1a-C5,C7, the signals of H-4 and H-6 were detected as broad singlets. The two products were well separated from each other under an improved HPLC condition (Fig. 3). In the HPLC chromatogram of the reaction mixture of 1b with 5-DMATSSc (Fig. 2), four product peaks were detected. As mentioned above, three monoprenylated derivatives were identified as C5-, C6-, and C7-prenylated D-tryptophan. LC-MS confirmed the additional product peak to be diprenylated D-tryptophan. An improved HPLC condition (condition 6) allowed the separation of this peak into one predominant peak and one minor product peak (Fig. 3), with similar retention times to those observed for the diprenylated products of 1a.
1.20,25,37 The characteristic singlets for H-4, H-7 and H-2 of 1a-C5,C6 were observed at 7.45, 7.14 and 7.08 ppm, respectively. For 1a-C5,C7, the signals of H-4 and H-6 were detected as broad singlets. The two products were well separated from each other under an improved HPLC condition (Fig. 3). In the HPLC chromatogram of the reaction mixture of 1b with 5-DMATSSc (Fig. 2), four product peaks were detected. As mentioned above, three monoprenylated derivatives were identified as C5-, C6-, and C7-prenylated D-tryptophan. LC-MS confirmed the additional product peak to be diprenylated D-tryptophan. An improved HPLC condition (condition 6) allowed the separation of this peak into one predominant peak and one minor product peak (Fig. 3), with similar retention times to those observed for the diprenylated products of 1a.
 | ||
| Fig. 3 HPLC analysis of the reaction mixtures of 5-DMATSSc with L- and D-enantiomers of tryptophan and methylated derivatives. 1 μM of the enzyme was incubated with 1 mM DMAPP and 0.5 mM of the indicated substrate(s) at 37 °C for 16 h. The reaction mixtures were analysed on an Eclipse Plus-C18 column (condition 6, Table S1, ESI†). The structures of the enzyme products of 1a, 1b, and 6b have been elucidated by NMR and MS analyses. The main products of 7a, 7b, 8a, and 8b are expected to be C5-prenylated derivatives. Detection was carried out with a diode array detector and illustrated for absorption at 277 nm. | ||
Structure elucidation of the isolated diprenylated peak by NMR analysis confirmed the C5,C6-diprenylation in the main product 1b-C5,C6 (Table S8 and Fig. S58, ESI†) with three singlets at 7.45, 7.15 and 7.10 ppm for H-4, H-7 and H-2, respectively. In this spectrum, signals of aromatic protons for C5,C7-diprenylated derivatives 1b-C5,C7 were also detected, with a low intensity of 5% of that of 1b-C5,C6. To the best of our knowledge, this is the first report on diprenylation of tryptophan by a tryptophan prenyltransferase. By UV detection mentioned above, diprenylated products of 1a, 1b and 1 were only observed for 5-DMATSSc after incubation for 1.5 h. By using the extracted ion chromatogram (EIC) mode, diprenylated products were also detected for almost all the DMATSs with 1a, with an exception for FgaPT2 (Fig. S25–S48, ESI†). However, the product yields were less than 0.5% of those of monoprenylated derivatives.
Separation and identification of L- and D-enantiomers of 5-, 6-, and 7-methyltryptophan
To expand our knowledge on the enantioselectivity and regioselectivity of DMATSs, we initiated to investigate their behaviours towards enantiomer pairs of methylated tryptophan derivatives. Unfortunately, these compounds are commercially not available, or available but very expensive. Therefore, we separated L- and D-enantiomers from the racemates 5-methyl-DL-tryptophan (6), 6-methyl-DL-tryptophan (7) and 7-methyl-DL-tryptophan (8) on HPLC by using the CHIRALPAK® Zwix(+) column (Fig. S59, ESI†). Inspection of the HPLC chromatograms of 1a, 1b and their racemate (1) revealed that the D-enantiomer 1b was eluted from the column prior to the L-enantiomer 1a. This elution order is also true for C5-, C6-, and C7-methylated tryptophan, which was proven by determination of their CD-spectra. Two opposite cotton effects were observed for the isolated enantiomers, which corresponded very well to those of 1a and 1b (Fig. S59, ESI†), respectively. The enantiomers in the racemate 4-methyl-DL-tryptophan (5) (Fig. S60, ESI†) could not be separated in this study.Substrate preferences of DMATSs for L- and D-enantiomers of methylated tryptophan
The isolated L- and D-enantiomers of C5-, C6- and C7-methylated tryptophan (6a, 6b, 7a, 7b, 8a, 8b) and their racemates (6, 7, 8) were incubated with the seven DMATSs under similar conditions (Fig. 4 and S25–S48, ESI†). The observed substrate preferences of the DMATSs differed clearly from each other (Table 2). As reported previously20,25,26,28 and demonstrated in this study, 6 was a poor substrate for the C5-prenyltransferases 5-DMATS and 5-DMATSSc and the C6-methylated derivative 7 was accepted by the C6-prenyltransferases 6-DMATSSa and 6-DMATSMo with low conversion yields. Higher conversion yields were found for their orthologue 6-DMATSSv (Table 2). 8 was not accepted by 7-DMATS. The low acceptance of tryptophan analogues, which were blocked at the prenylation position for 1a by a methyl group, was also demonstrated by using the pure L-enantiomers (Table 2). For methylated tryptophan analogues, L-enantiomers were in general better or in most cases much better accepted by DMATSs than D-enantiomers as in the case of tryptophan.It seems that FgaPT2 had a much higher enantioselectivity than other tested enzymes and accepted no D-enantiomers 6b, 7b, or 8b. 7b and 8b were no substrates for 6-DMATSSa, 6-DMATSSv, and 7-DMATS. No product formation was detected in the reaction mixture of 7b with 5-DMATS. From Table 2, it is obvious that the reactions of L-enantiomers were almost not inhibited by D-enantiomers and nearly the same activities were detected for the L-enantiomers in the racemates. Interestingly, 6b was a much better substrate for 5-DMATSSc and 6-DMATSMo than 6a. Relative conversion yields of 57.3 ± 1.8 and 66.6 ± 0.5% of that of 1a were calculated for the reactions of 6b with 5-DMATSSc and 6-DMATSMo, respectively, whereas the values for 6a are 2.3 ± 0.2 and 12.9 ± 0.2%, respectively (Table 2 and Fig. 4).
More interestingly, 6b in the racemate 6 was not converted by these two enzymes, while similar conversion yields for 6a were observed. This indicated an inhibition of the 6b reactions in the presence of 6a, which could be explained by determination of their kinetic parameters. Kinetic parameters of 6-DMATSMo and 5-DMATSSc were then determined for 6a and 6b (Table 3 and Fig. S61–S62, ESI†). Substrate inhibition was observed for the 5-DMATSSc reaction with 6b at concentrations higher than 0.05 mM. Therefore, the KM value was determined from graphical nonlinear evaluation for concentrations less than 0.05 mM. For calculation of kcat, the maximal velocity vmax at 0.05 mM 6b was used (Fig. S61, ESI†). For other reactions, kinetic parameters were calculated from nonlinear regression (Fig. S61 and S62, ESI†) and are given in Table 3. KM values of 0.009 ± 0.001 and 0.03 ± 0.01 mM and kcat values of 0.005 ± 0.0001 and 0.035 ± 0.005 s−1 were calculated for 5-DMATSSc reactions with L- and D-isomers, respectively. For the 6-DMATSMo reaction with 6a, a KM value of 0.008 ± 0.001 mM and a kcat of 0.012 ± 0.0002 s−1 were determined. In comparison, a significantly higher KM value of 0.30 ± 0.05 mM and larger kcat of 0.042 ± 0.003 s−1 were calculated for the 6-DMATSMo reaction with 6b. In summary, 5-DMATSSc and 6-DMATSMo also display higher affinities to the L- than the D-enantiomer. The turnover numbers for the D-enantiomer are, however, clearly higher than those of the L-enantiomer, confirming the observed conversion yields of the reactions with pure enantiomers and the inhibition of the 6b reactions in the presence of 6a in the racemates. Due to the high affinity of the DMATSs towards the L-form, the active site of the enzyme will be occupied by the L-enantiomer, so that no or very low prenylation of 6b is observed in the reaction with the racemate.
Regioselectivity of DMATSs towards L- and D-enantiomers of methylated tryptophan derivatives
For determination of the prenylation positions, DMATS enzymes, which accepted D-enantiomers (Table 2), were incubated with 6a and 6b, 7a and 7b as well as 8a and 8b. As shown in Fig. S63 in the ESI† and given in Table 4, the three C6-prenyltransferases 6-DMATSSa, 6-DMATSSv and 6-DMATSMo catalysed the same unique or predominant C6-prenylation of 6a and 6b. In the reaction mixtures of 6-DMATSMo with 6a and 6b, C7-prenylated derivatives were identified as minor products (Tables 4 and S10, Fig. S63 and S66, ESI†). Different regioselectivities were observed for the reactions of 6a and 6b with FgaPT2, 5-DMATSSc and 7-DMATS. C4-prenylated 6a was detected as the unique product of the FgaPT2 reaction, while an additional product (C6- or C7-prenylated) was detected for the 6b reaction with FgaPT2 by LC-MS (Fig. S26, ESI†). In comparison, 5-DMATSSc displayed a low regioselectivity towards 6a, with C6- and C7-prenylated derivatives 6a-C6 and 6a-C7 in a ratio of approximately 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 as enzyme products. For the 5-DMATSSc reaction with 6b, 6b-C6 was detected as the main and 6b-C7 as a minor product in a ratio of 1
1 as enzyme products. For the 5-DMATSSc reaction with 6b, 6b-C6 was detected as the main and 6b-C7 as a minor product in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2 (Table S10 and Fig. S63, ESI†). 1H NMR spectra of the product mixture confirmed these results (Fig. S66, ESI†). For the 7-DMATS reaction, a switch from a unique C7-prenylation with the L-isomer to a predominant C6-prenylation with the D-isomer of 5-methyltryptophan was observed. 6b-C7 was detected as a minor product with a relative product yield of 20% to that of 6b-C6 (Fig. S63, ESI†). These results demonstrated again that stereochemistry of the substrates has influence on the regioselectivity of the used enzymes, at least in several cases. For 6-methyl- (7a and 7b) and 7-methyltryptophan (8a and 8b), accurate determination of the prenylation positions of the enzyme products was not always possible, because the products could not be separated completely from each other under the HPLC conditions used in this study (Fig. S64 and S65, ESI†). C5- and C7-prenylations of 7a and 7b with 6-DMATSMo confirmed the previous results (Table S1†). Two product peaks each of the 5-DMATSSc reactions with 7a and 7b indicate C5- and C7-prenylations. In the HPLC chromatograms of the reaction mixtures of 8a and 8b with 5-DMATS, 5-DMATSSc, and 6-DMATSMo, one monoprenylated product peak with similar retention times could also be observed.
0.2 (Table S10 and Fig. S63, ESI†). 1H NMR spectra of the product mixture confirmed these results (Fig. S66, ESI†). For the 7-DMATS reaction, a switch from a unique C7-prenylation with the L-isomer to a predominant C6-prenylation with the D-isomer of 5-methyltryptophan was observed. 6b-C7 was detected as a minor product with a relative product yield of 20% to that of 6b-C6 (Fig. S63, ESI†). These results demonstrated again that stereochemistry of the substrates has influence on the regioselectivity of the used enzymes, at least in several cases. For 6-methyl- (7a and 7b) and 7-methyltryptophan (8a and 8b), accurate determination of the prenylation positions of the enzyme products was not always possible, because the products could not be separated completely from each other under the HPLC conditions used in this study (Fig. S64 and S65, ESI†). C5- and C7-prenylations of 7a and 7b with 6-DMATSMo confirmed the previous results (Table S1†). Two product peaks each of the 5-DMATSSc reactions with 7a and 7b indicate C5- and C7-prenylations. In the HPLC chromatograms of the reaction mixtures of 8a and 8b with 5-DMATS, 5-DMATSSc, and 6-DMATSMo, one monoprenylated product peak with similar retention times could also be observed.
Diprenylation of methylated tryptophan derivatives by DMATSs
As mentioned above, diprenylation of tryptophan was observed for most of the tested DMATSs by the EIC mode in LC-MS. With exceptions for 5-DMATSSc, the product yields of less than 0.5% were so low that UV detection was almost not possible for 1.5 h incubation mixtures. Even after extension of the incubation time to 16 h, UV detection of the diprenylated derivatives of 1a and 1b was only observed for 5-DMATSSc (data not shown). HPLC analysis of the 16 h reaction mixture of 7a with 5-DMATSSc revealed the presence of two mono- and one diprenylated product peaks (Fig. 3). Low product formation was observed in the reaction mixtures of 5-DMATSSc with 7b, 8a and 8b. Similar retention times of the products of the D-form with those of the L-form suggest identical diprenylated products for both enantiomers. The diprenylated features of these products were confirmed by LC-MS analysis. These results highlight the increased capability of 5-DMATSSc for diprenylation of tryptophan and its derivatives. Diprenylation of tryptophan and its analogues by tandem reactions of two DMATSs has been described for FgaPT2 and 7-DMATS.38 However, diprenylation by one single tryptophan prenyltransferase has not been reported previously.Conclusion
This study deals with comparative investigation on the divergence of microbial dimethylallyltryptophan synthases regarding their enantio- and regioselectivities. Different preferences were clearly observed for the seven tested DMATSs towards D- and L-enantiomers of tryptophan and methylated derivatives. High flexible enantioselectivity was found for 5-DMATS, 5-DMATSSc, and 6-DMATSMo. Remarkably high conversion yields were detected for D-tryptophan with 6-DMATSMo and 5-DMATS. 5-Methyl-D-tryptophan was even better accepted by 5-DMATSSc and 6-DMATSMo than the L-enantiomer. In other cases, the L-forms were the best accepted substrates. Interestingly, lower or in most cases, no product formation was observed for D-enantiomers in the presence of L-enantiomers in the racemates. Kinetic studies of 5-DMATSSc and 6-DMATSMo suggest that the very high affinities towards the L-enantiomers led, in most cases, to the repression of the D-enantiomer reaction under the used conditions.In contrast with the highly regiospecific prenylation on the L-tryptophan of the tested enzymes, the L-tryptophan C4-prenyltransferase FgaPT2 and C5-prenyltransferase 5-DMATSSc displayed a reduced regioselectivity towards D-tryptophan with C5- and C6-prenylated derivatives as the main products, respectively. Clearly different regioselectivities were also identified for the enantiomers of 5-methyltryptophan with FgaPT2, 5-DMATSSc and 7-DMATS. Furthermore, diprenylation of tryptophan by 5-DMATSSc was the first example that a tryptophan prenyltransferase catalyses two successive prenylation steps. These results expand our knowledge on the similarity and differences in the biochemical features of the tryptophan prenyltransferases. Moreover, the presented biochemical properties of these enzymes make them potential biocatalysts in chemical synthesis for prenylated indole derivatives, which represent promising candidates for drug discovery and development in the pharmaceutical industry. As mentioned in the Introduction, D-amino acids and their derivatives are interesting drug candidates and could be used for treatment of different diseases.3,39–44 Testing the bioactivity of these compounds is intended in the near future.
Experimental section
Chemicals
DMAPP, GPP, and FPP were synthesised according to the method described for GPP reported elsewhere.45 Aromatic substrates used for the enzyme assays were purchased from TCI, Sigma-Aldrich, Bachem, Roth, and Fluka and were of the highest available purity.Bacterial strains and plasmids
pGEM-T Easy was obtained from Promega (Mannheim, Germany). pHIS8 was kindly provided by Prof. Joseph P. Noel (Salk Institute for Biological Studies). Escherichia coli strains XL1 Blue MRF’ (Stratagene, Amsterdam, the Netherlands), M15 (pREP4) (Qiagen), and BL21 (DE3) pLysS (AMS Biotechnology, Abingdon, UK) were used for cloning and expression experiments. Micromonospora olivasterospora DSM 43868 was purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany).Cloning and expression of MolI14.36
For genomic DNA isolation by phenol-chloroform extraction,46 cultivation of M. olivasterospora was carried out in a 300 ml cylindrical flask containing 50 ml liquid YMG medium (0.4% (w/v) yeast extract, 1% (w/v) malt extract, and 0.4% (w/v) glucose) at 160 rpm on a rotary shaker at 28 °C for three days. Standard procedures for DNA manipulation in E. coli were performed as described previously.47With the isolated genomic DNA as the template, a PCR fragment of 1143 bp containing the coding sequence of MolI14.36 was amplified by using the Expand high fidelity kit (Roche Diagnostic, Mannheim, Germany) and the primers Mol_SacI_fw: (5′-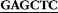 ATGGCCGGCTTGTCCG-3′) and Mol_HindIII_Rev: 5′-
ATGGCCGGCTTGTCCG-3′) and Mol_HindIII_Rev: 5′- TCAGGGTCGGGTACGGC-3′). Bold underlined letters represent the restriction sites for SacI and HindIII, located immediately before the predicted start and at the stop codon in the primer sequences. PCR amplification was performed on an iCycler from Bio-Rad. The PCR product was cloned via the pGEM-T Easy vector into pHIS8
TCAGGGTCGGGTACGGC-3′). Bold underlined letters represent the restriction sites for SacI and HindIII, located immediately before the predicted start and at the stop codon in the primer sequences. PCR amplification was performed on an iCycler from Bio-Rad. The PCR product was cloned via the pGEM-T Easy vector into pHIS8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 48 resulting in the expression construct pJW32. E. coli BL21 [DE3] cells harbouring pJW32 were cultivated in 1 L liquid lysogeny broth (LB) medium supplemented with kanamycin (50 μg ml−1) until an OD600 of 0.6. For induction of gene expression, IPTG was added to a final concentration of 1 mM. After further incubation at 30 °C and 220 rpm for 16 h, the recombinant His8-tagged protein was purified on Ni-NTA agarose according to the manufacturer's protocol.
48 resulting in the expression construct pJW32. E. coli BL21 [DE3] cells harbouring pJW32 were cultivated in 1 L liquid lysogeny broth (LB) medium supplemented with kanamycin (50 μg ml−1) until an OD600 of 0.6. For induction of gene expression, IPTG was added to a final concentration of 1 mM. After further incubation at 30 °C and 220 rpm for 16 h, the recombinant His8-tagged protein was purified on Ni-NTA agarose according to the manufacturer's protocol.
Overproduction and purification of the recombinant DMATS enzymes
Escherichia coli (E. coli) M15 [pREP4] (Qiagen) and BL21 (DE3) pLysS (AMS Biotechnology, Abingdon, UK) were used for expression. Culture conditions for gene expression and purification of FgaPT2, 5-DMATS, 5-DMATSSc, 6-DMATSSa, 6-DMATSSv and 7-DMATS were carried out as described previously.20,21,25,27,28Protein analyses
The purified proteins were analysed on 12% (w/v) SDS-PAGE49 and stained with Coomassie Brilliant Blue G-250 (Fig. S1, ESI†). The protein content was determined on a NanoDrop 2000c (Thermo Scientific) at an absorption of 280 nm. The molecular masses of the native recombinant His8-6-DMATSMo, His8-6-DMATSv and 6-DMATSa-His6 were determined by size exclusion chromatography on a HiLoad 16/60 Superdex 200 column (GE Healthcare). 50 mM Tris-HCl buffer (pH 7.5) containing 150 mM NaCl was used to equilibrate the column and elute proteins. The column was calibrated with blue dextran 2000 (2000 kDa), aldolase (158 kDa), conalbumin (75 kDa), chymotrypsinogen A (25 kDa) and ribonuclease A (13.7 kDa) (GE Healthcare).Enzyme assays
All reaction mixtures contained 50 mM Tris-HCl (pH 7.5) and were incubated at 37 °C. The conditions of the reaction mixtures for identification of the prenylated products are described in Table S9 in the ESI.†Kinetic parameters of 6-DMATSMo toward DMAPP were determined with 1 mM of 1a as the prenyl acceptor. Assays containing 229.9 nM 6-DMATSMo and DMAPP at final concentrations of 0.002 to 0.5 mM were incubated for 30 min. For determination of the kinetic parameters of different DMATSs towards 1a, DMAPP at 1 mM and 1a at final concentrations of up to 1 mM were used. Assays with 36.2 nM FgaPT2, 99.2 nM 5-DMATS, 118.5 nM 5-DMATSSc, 121.7 nM 6-DMATSSa, 231.0 nM 6-DMATSSv, 229.9 nM 6-DMATSMo, and 228.8 nM 7-DMATS were incubated for 15 min or in the case of 6-DMATSMo for 30 min. For determination of the kinetic parameters of 1b, the maximal substrate concentration was increased to 2 mM or even 4 mM in the case of 5-DMATSSc. The assays contained 905.8 nM FgaPT2, 396.8 nM 5-DMATS, 473.9 nM 5-DMATSSc, 486.6 nM 6-DMATSSa, 461.9 nM 6-DMATSSv, 459.8 nM 6-DMATSMo, or 1.4 μM 7-DMATS and were incubated for 30 min, or in the case of 5-DMATSSc for 15 min. The assays for determination of the kinetic parameters of 6-DMATSMo for 6a and 6b contained 1 mM DMAPP, 459.8 nM of the recombinant protein, 6a from 0.002 to 0.5 mM or 6b from 0.01 to 1 mM. The reaction mixtures were incubated for 30 min. For the 5-DMATSSc reactions with 6a and 6b, the assays contained 7.1 μM and 473.9 nM of the recombinant protein and 0.002 to 0.5 mM of 6a or 6b, which were incubated for 15 min. The conditions for all other enzyme assays are given in the legends of respective figures or tables.
Assays for isolation of the enzyme products of 6-DMATSMo were prepared in large scales (10 ml) containing 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1 mM L-tryptophan or its analogues, 2 mM DMAPP, and recombinant protein in an adequate quantity (2.3–6.9 μM). Enzyme products of 6-DMATSMo with 5-methyl-D-tryptophan as well as 5-DMATSSc with D-tryptophan and 5-methyl-D-tryptophan were isolated from up-scaled assays (5 ml) containing 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1 mM D-tryptophan or 0.5 mM 5-methyl-D-tryptophan, 2 mM DMAPP and the recombinant protein in an adequate quantity (4.7–11.9 μM 5-DMATSSc or 4.6–11.5 μM 6-DMATSMo). Assays were incubated for 16 h.
All enzyme reactions were terminated by addition of one volume of methanol. Assays of up to 100 μl were centrifuged at 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 20 min before further analysis on HPLC. Isolation assays of 5–10 ml were centrifuged at 4500g for 30 min and the obtained supernatants were concentrated on a rotating vacuum evaporator at 35 °C to a volume of 1 ml for isolation on HPLC.
000g for 20 min before further analysis on HPLC. Isolation assays of 5–10 ml were centrifuged at 4500g for 30 min and the obtained supernatants were concentrated on a rotating vacuum evaporator at 35 °C to a volume of 1 ml for isolation on HPLC.
HPLC and LC-MS analyses
The methods and the column used for HPLC measurements are listed as conditions 1–9 in Table S1 in the ESI.† For all analyses with the exception of condition 4, an Agilent HPLC 1200 (Böblingen, Germany) was used. Reaction mixtures for comparison of the substrate specificities of 6-DMATSSa, 6-DMATSSv, and 6-DMATSMo were analysed under condition 1. For investigation of the substrate preferences of DMATSs towards L- und D-isomers of tryptophan and analogues thereof, the incubation mixtures were analysed with a CHIRALPAK® Zwix(+) column (150 × 3 mm, 3 μm, Chiral Technologies Europe, Daicel Group, Illkirch Cedex, France) and an isocratic run (condition 2, Table S1, ESI†). Condition 2 was also used to determine the kinetic parameters of 5-methyl-L-tryptophan and 5-methyl-D-tryptophan. Incubation mixtures for kinetic studies on tryptophan were analysed under condition 1.To detect prenylation, the assays were analysed on an Agilent HPLC 1260 equipped with a Bruker microTOF QIII mass spectrometer. The applied method is illustrated as condition 3. Chromatograms with UV detection and the extracted ion chromatograms (EIC) are given in Fig. 1 and 4 as well as in Fig. S25–S48 (ESI†).
Reaction mixtures for better separation of the enzyme products with prenylations at different positions were analysed under condition 4 for L- and D-tryptophan or under condition 5 for the L- and D-forms of 5-, 6-, and 7-methyltryptophan as substrates. Enzyme assays with 5-DMATSSc, illustrated in Fig. 3, were measured under condition 6. The enantiomeric substrates were separated isocratically with the same chiral column by using methanol as the elution solvent (condition 7).
For isolation of the prenylated products, the same HPLC equipment under condition 8 or 9 was used. Detection was carried out with a photodiode array detector and illustrated at 277 nm in this paper.
Structure elucidation by NMR analysis
NMR spectra of the enzyme products were recorded on a JEOL ECA-500 MHz (1a-C6, 1b-C6, 1a-C5,C6, and 1a-C5,C7), a JEOL ECX-400 MHz (1b-C5, 1b-C6, 1b-C7, 2a-C6, 3a-C6, 4-C6, 7-C6, and 9a-C6) (JEOL Germany GmbH, Munich, Germany) or a Bruker Avance III HD 500 (Bruker Biospin GmbH, Rheinstetten, Germany) (5-C5, 5-C6, 5-C7, 6-C6, 6-C7, 8-C6, and 1b-C5,C6) spectrometer. All spectra were processed with MestReNova 6.0.2-5475 and 1H chemical shifts were referenced to those of the solvent signals at 3.31 ppm for methanol. The NMR data are given in the ESI† (Tables S5–S8, S10, and Fig. S4–S22, S57–58, S66†).Circular dichroism (CD) spectroscopy of the isolated substrates
For determination of the configuration, CD spectra of the L- and D-forms of tryptophan and its methylated derivatives were taken on a Jasco J-810 CD spectrometer. The samples were dissolved in water and measured in the range of 180–350 nm by using a 1 mm path length quartz cuvette.Accession numbers
The nucleotide sequence of MolI14.36 is available in the GenBank database under accession number AJ628421.2 and the corresponding protein sequence under accession number CAF31565.1.Acknowledgements
We thank Lena Ludwig for synthesis of DMAPP, Stefan Newel and Rixa Kraut for taking NMR and mass spectra, respectively. J. W. was partially financed by the LOEWE program of the State of Hessen (SynMikro to S. M. L.). S. M. L. also acknowledges the Deutsche Forschungsgemeinschaft for funding the Bruker microTOF QIII mass spectrometer. (INST 160/620-1)References
- A. Shimada and H. Ozaki, Life, 2012, 2, 215–228 CrossRef CAS PubMed.
- L. A. Nguyen, H. He and C. Pham-Huy, Int. J. Biomed. Sci., 2006, 2, 85–100 CAS.
- X. Gao, Q. Ma and H. Zhu, Appl. Microbiol. Biotechnol., 2015, 99, 3341–3349 CrossRef CAS PubMed.
- S. D'Aniello, I. Somorjai, J. Garcia-Fernandez, E. Topo and A. D'Aniello, FASEB J., 2011, 25, 1014–1027 CrossRef PubMed.
- F. Cava, H. Lam, M. A. de Pedro and M. K. Waldor, Cell. Mol. Life Sci., 2011, 68, 817–831 CrossRef CAS PubMed.
- R. C. Strauch, E. Svedin, B. Dilkes, C. Chapple and X. Li, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 11726–11731 CrossRef CAS PubMed.
- D. M. Veine, H. Yao, D. R. Stafford, K. S. Fay and D. L. Livant, Clin. Exp. Metastasis, 2014, 31, 379–393 CrossRef CAS PubMed.
- B. D. Welch, A. P. VanDemark, A. Heroux, C. P. Hill and M. S. Kay, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 16828–16833 CrossRef CAS PubMed.
- J. Kumar and V. Sim, Prion, 2014, 8, 119–124 CrossRef CAS PubMed.
- S.-M. Li, Nat. Prod. Rep., 2010, 27, 57–78 RSC.
- J. Winkelblech, A. Fan and S.-M. Li, Appl. Microbiol. Biotechnol., 2015, 99, 7379–7397 CrossRef CAS PubMed.
- B. Wollinsky, L. Ludwig, A. Hamacher, X. Yu, M. U. Kassack and S.-M. Li, Bioorg. Med. Chem. Lett., 2012, 22, 3866–3869 CrossRef CAS PubMed.
- B. Botta, A. Vitali, P. Menendez, D. Misiti and M. G. Delle, Curr. Med. Chem., 2005, 12, 717–739 CrossRef PubMed.
- A. H. Liu, D. Q. Liu, T. J. Liang, X. Q. Yu, M. T. Feng, L. G. Yao, Y. Fang, B. Wang, L. H. Feng, M. X. Zhang and S. C. Mao, Bioorg. Med. Chem. Lett., 2013, 23, 2491–2494 CrossRef CAS PubMed.
- A. Oya, N. Tanaka, T. Kusama, S. Y. Kim, S. Hayashi, M. Kojoma, A. Hishida, N. Kawahara, K. Sakai, T. Gonoi and J. Kobayashi, J. Nat. Prod., 2015, 78, 258–264 CrossRef CAS PubMed.
- T. Sasaki, Y. Igarashi, M. Ogawa and T. Furumai, J. Antibiot., 2002, 55, 1009–1012 CrossRef CAS PubMed.
- I. A. Unsöld and S.-M. Li, Microbiology, 2005, 151, 1499–1505 CrossRef PubMed.
- S. Takahashi, H. Takagi, A. Toyoda, M. Uramoto, T. Nogawa, M. Ueki, Y. Sakaki and H. Osada, J. Bacteriol., 2010, 192, 2839–2851 CrossRef CAS PubMed.
- R. Satou, M. Izumikawa, Y. Katsuyama, M. Matsui, M. Takagi, K. Shin-ya and Y. Ohnishi, J. Antibiot., 2014, 67, 231–236 CrossRef CAS PubMed.
- J. Winkelblech and S.-M. Li, ChemBioChem, 2014, 15, 1030–1039 CrossRef CAS PubMed.
- T. Ozaki, M. Nishiyama and T. Kuzuyama, J. Biol. Chem., 2013, 288, 9946–9956 CrossRef CAS PubMed.
- S. Subramanian, X. Shen, Q. Yuan and Y. Yan, Process Biochem., 2012, 47, 1419–1422 CrossRef CAS.
- C. Wu, C. Du, J. Gubbens, Y. H. Choi and G. P. van Wezel, J. Nat. Prod., 2015, 78, 2355–2363 CrossRef CAS PubMed.
- A. Fan, J. Winkelblech and S.-M. Li, Appl. Microbiol. Biotechnol., 2015, 99, 7399–7415 CrossRef CAS PubMed.
- X. Yu, Y. Liu, X. Xie, X.-D. Zheng and S.-M. Li, J. Biol. Chem., 2012, 287, 1371–1380 CrossRef CAS PubMed.
- J. Winkelblech, M. Liebhold, J. Gunera, X. Xie, P. Kolb and S.-M. Li, Adv. Synth. Catal., 2015, 357, 975–986 CrossRef CAS.
- N. Steffan, I. A. Unsöld and S.-M. Li, ChemBioChem, 2007, 8, 1298–1307 CrossRef CAS PubMed.
- A. Kremer, L. Westrich and S.-M. Li, Microbiology, 2007, 153, 3409–3416 CrossRef CAS PubMed.
- X. Yu, G. Zocher, X. Xie, M. Liebhold, S. Schütz, T. Stehle and S.-M. Li, Chem. Biol., 2013, 20, 1492–1501 CrossRef CAS PubMed.
- J. M. Schuller, G. Zocher, M. Liebhold, X. Xie, M. Stahl, S.-M. Li and T. Stehle, J. Mol. Biol., 2012, 422, 87–99 CrossRef CAS PubMed.
- M. Jost, G. Zocher, S. Tarcz, M. Matuschek, X. Xie, S.-M. Li and T. Stehle, J. Am. Chem. Soc., 2010, 132, 17849–17858 CrossRef CAS PubMed.
- U. Metzger, C. Schall, G. Zocher, I. Unsöld, E. Stec, S.-M. Li, L. Heide and T. Stehle, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 14309–14314 CrossRef CAS PubMed.
- A. Kremer and S.-M. Li, Appl. Microbiol. Biotechnol., 2008, 79, 951–961 CrossRef CAS PubMed.
- K. Miyamoto, F. Ishikawa, S. Nakamura, Y. Hayashi, I. Nakanishi and H. Kakeya, Bioorg. Med. Chem., 2014, 22, 2517–2528 CrossRef CAS PubMed.
- Y. Ding, R. M. Williams and D. H. Sherman, J. Biol. Chem., 2008, 283, 16068–16076 CrossRef CAS PubMed.
- A. Markert, N. Steffan, K. Ploss, S. Hellwig, U. Steiner, C. Drewke, S.-M. Li, W. Boland and E. Leistner, Plant Physiol., 2008, 147, 296–305 CrossRef CAS PubMed.
- A. Fan, H. Chen, R. Wu, H. Xu and S.-M. Li, Appl. Microbiol. Biotechnol., 2014, 98, 10119–10129 CrossRef CAS PubMed.
- H.-L. Ruan, E. Stec and S.-M. Li, Arch. Microbiol., 2009, 191, 791–795 CrossRef CAS PubMed.
- L. Jia, K. Schweikart, J. Tomaszewski, J. G. Page, P. E. Noker, S. A. Buhrow, J. M. Reid, M. M. Ames and D. H. Munn, Food Chem. Toxicol., 2008, 46, 203–211 CrossRef CAS PubMed.
- M. Tanaka, X. Li, H. Hikawa, T. Suzuki, K. Tsutsumi, M. Sato, O. Takikawa, H. Suzuki and Y. Yokoyama, Bioorg. Med. Chem., 2013, 21, 1159–1165 CrossRef CAS PubMed.
- P. T. Di, S. Danese, C. R. De and S. Rutella, Expert Opin. Ther. Pat., 2010, 20, 229–250 CrossRef PubMed.
- K. Saito, C. Y. Chen, M. Masana, J. S. Crowley, S. P. Markey and M. P. Heyes, Biochem. J., 1993, 291(Pt 1), 11–14 CrossRef CAS PubMed.
- B. Heller, D-Phenylalanine Treatment, US Pat., 4355044, 1982 Search PubMed.
- T. Fukushima, A. Sugiura, I. Furuta, S. Iwasa, H. Iizuka, H. Ichiba, M. Onozato, H. Hikawa and Y. Yokoyama, Int. J. Tryptophan Res., 2015, 8, 1–5 CrossRef CAS PubMed.
- A. B. Woodside, Z. Huang and C. D. Poulter, Org. Synth., 1988, 66, 211–215 CrossRef CAS.
- T. Kieser, M. J. Bibb, M. J. Buttner, K. F. Chater and D. A. Hopwood, Practical Streptomyces Genetics, John Innes Foundation, Norwich, UK, 2000 Search PubMed.
- J. Sambrook and D. W. Russell, Molecular Cloning: a Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2001 Search PubMed.
- J. M. Jez, J. L. Ferrer, M. E. Bowman, R. A. Dixon and J. P. Noel, Biochemistry, 2000, 39, 890–902 CrossRef CAS PubMed.
- U. K. Laemmli, Nature, 1970, 227, 680–685 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ob01803c |
| This journal is © The Royal Society of Chemistry 2016 |