Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Organelle-specific bioprobes based on fluorogens with aggregation-induced emission (AIE) characteristics
Fang
Hu
ab and
Bin
Liu
*ac
aDepartment of Chemical and Biomolecular Engineering, National University of Singapore, 4 Engineering Drive 4, Singapore 117585, Singapore. E-mail: cheliub@nus.edu.sg
bNanoscience and Nanotechnology Institute, National University of Singapore, 2 Engineering Drive 3, Singapore 117581, Singapore
cInstitute of Materials Research and Engineering (IMRE), 2 Fusionopolis Way, Innovis, 136834, Singapore
First published on 15th August 2016
Abstract
Bioprobes based on fluorogens with aggregation-induced emission (AIE) characteristics have been increasingly used in chemosensing and bioimaging due to their high sensitivity, photostability and biocompatibility. In this review, we summarize the design of cellular organelle specific (cytoplasm membrane, mitochondria, lysosomes, lipid droplets and nucleus) AIE bioprobes and their applications in organelle imaging, organelle bioactivity monitoring, and image-guided cancer cell ablation.
1. Introduction
Most eukaryotic cells contain cytoplasm membrane, nucleus, vacuoles, mitochondria, lysosomes, lipid droplets, endoplasmic reticulum, Golgi apparatus and so on. These organelles play critical roles in cellular function. The cell membrane is supported mainly by a lipid bilayer with embedded proteins to ensure the relative stability of the intracellular environment. It is selectively permeable to ions and organic molecules to regulate the movement of substances into and out of cells.1 The mitochondrion is the powerhouse of the cell; it is also involved in processes such as cell differentiation, cell communication and cell apoptosis.2 Lysosomes are cleaners; they sweep out disabled bio-macromolecules and organelles and swallow intruding pathogens.3 Lipid droplets could regulate intracellular lipid storage and metabolism.4 The nucleus is just like the brain of a cell; it maintains gene expression.5 Abnormal biological activities of these organelles are the precise signals for a large number of diseases, including Alzheimer's disease, diabetes, Parkinson's disease, angiocardiopathy, atherosclerosis, cancer and so on.6–9 As a consequence, visualization of organelles and their morphology or function changes has great potential in clinical analysis or medical intervention as it gives valuable information on cell bioactivities at the cellular and/or molecular level.The development of advanced technologies for the visualization of the morphology or biological activities of specific organelles plays an indispensable role in disease diagnosis.10 Amongst numerous imaging modalities, fluorescence imaging is a powerful tool for visualizing and analysing the localization and dynamics of ions and biomolecules in different organelles.11 It utilizes fluorescent probes to label organelles or specific inclusions in organelles. Benefiting from the advanced optical microscopy, the spatial resolution of fluorescence imaging reaches micrometre or even nanometre levels.12 Besides, fluorescence imaging has been recognized as one of the most powerful tools in biological systems due to the availability of fluorescent contrast agents and their real-time operation, non-invasive testing and cost-effective performance.13 A variety of fluorescent materials, including organic dyes,14 organic nanoparticles,15 inorganic quantum dots (QDs),16 fluorescent proteins (FPs),17 conjugated polymers,18 noble metal nanoclusters and so on,19 have been developed and utilized in biological sensory and imaging applications. Among them, organic dyes are most widely used and easily accessible. A simple combination of fluorophores with organelle specific ligands can lead to organelle targeting probes. However, typical organic dyes show the notorious effect of aggregation-caused quenching (ACQ) and have relatively poor photostability, which greatly limits their applications in the biological field. Organic fluorescent nanoparticles based on typical organic dyes are also victims of ACQ. QDs are highly emissive and photostable; surface-modified QDs with target peptides/proteins are effective fluorescent markers, but most of them (e.g., CdSe and PbS) are inherently cytotoxic and often blink.20 FPs have attracted much attention because they can be genetically encoded to the targets of interest, but the use of FPs requires transfection processes and their labelling sometimes can disrupt normal cell function.21
Opposite to ACQ is the aggregation-induced emission (AIE) effect.22 Organic dyes with AIE characteristics are almost non-emissive when they are dissolved in a good solvent but they emit efficiently in aggregate states. Fluorogens with AIE characteristics are termed AIEgens. AIEgens generally have propeller-shaped structures, in which several aromatic rings are linked to a conjugated core through chemical bonds. The aromatic rings in isolated molecules of AIEgens are relatively free to rotate around the core, and the intramolecular rotations consume energy from excited states via non-radiative decay. In the aggregate state, the intramolecular rotations are severely restricted due to physical stacking so that AIEgens show bright emission as the non-radiative pathway is blocked. Tang and co-workers defined the restriction of intramolecular rotation (RIR) as the primary operation mechanism for AIEgens.22 In fact, AIE processes are also associated with restriction of intramolecular vibrations (RIV), J-aggregate formation (JAF), twisted intramolecular charge transfer (TICT), excited-state intramolecular proton transfer (ESIPT), etc.23,24
Benefiting from the strong fluorescence in the solid/aggregate state and brighter emission at higher concentrations, AIEgens have been widely used for chemical and biological sensing and imaging. A number of charged AIE molecules have been used for small molecule (e.g., monosaccharides, biothiols, amino acids, amines, and adenosine triphosphate) and macromolecule (e.g., polysaccharides, DNAs, and lipids) detection based on electrostatic interactions.25,26 To further improve the detection selectivity of these assays, specific light-up probes have been developed for protein,27 enzyme28 and cancer cell detection.29,30 Since AIE probes are brighter in the aggregate state, they could be used at higher concentrations than traditional fluorophores and thus show stronger photobleaching resistance and higher signal reliability compared to most ACQ probes. Further encapsulation of AIEgens into the polymer matrix yielded bio-compatible AIE dots, which show higher brightness than QDs for cancer cell detection,31 cell tracking32 and vascular imaging.33 The recent development of multifunctional AIEgens has further broadened their applications to chemotherapy,34 photodynamic therapy,35 and image-guided cancer cell ablation.36 The therapeutic function was also found to be dependent on their organelle location in specific cells.37
This review summarizes the AIE bioprobes which can specifically target different cellular organelles, including cytoplasm membrane, mitochondria, lysosomes, lipid droplets and nucleus. In each subsection, the introduction of a certain organelle and the design principle of specific-targeting AIE bioprobes are discussed, which is followed by examples of organelle imaging applications. Based on the high quality imaging, visualization of biological activities (such as mitophagy) involved in organelles and image-guided cancer cell ablation (such as chemotherapy and photodynamic therapy) are also included in some subsections. The development of high performance organelle-targeting AIE bioprobes not only helps to visualize cell structures, but also opens up endless opportunities for AIEgens to be applied in future clinical diagnosis and disease therapy.
2. Cytoplasm membrane imaging
The main framework of the cell membrane is a phospholipid bilayer embedded with proteins. It plays critical roles like serving as the frontier of a cell, guarding a stable metabolism in the intracellular environment and regulating the nutrient uptake and waste discharge. The cell membrane is also involved in the process of cell recognition and signal transmission. The design of cytoplasm membrane specific bioprobes is mainly based on the components and metabolic activities of the membrane. Three major principles are widely used. (i) Some inclusions such as proteins are especially overexpressed in the cell membrane. A bioprobe combined with a ligand for membrane proteins is certainly a good candidate for cytoplasm membrane imaging.38 (ii) A lipophilic structure can be easily captured by the phospholipid bilayer of a plasma membrane in an aqueous medium as they have similar hydrophobic properties. For example, the two commercial dyes for cell membrane tracking, 3,3′-dioctadecyloxacarbocyanine perchlorate (DiO)39 and 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI),40 both contain two long alkyl chains. (iii) Bioorthogonal turn-on bioprobe which can react with the membrane is also a good choice for cell surface staining.41 Some outstanding AIE bioprobes for cytoplasm membrane imaging have been developed according to these principles.2.1 Cell membrane component tracking
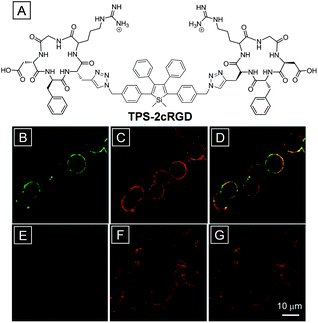
Integrin αvβ3 is overexpressed in tumor cells of different origins; Liu and co-workers developed an AIE bioprobe (TPS-2cRGD) with the aim to specifically light up αvβ3-positive cancer cells. Once stained with TPS-2cRGD at room temperature within 25 min or at 4 °C, the cell membrane with overexpressed integrin αvβ3 was lit up.30 As shown in Fig. 1A, TPS-2cRGD is composed of tetraphenylsilole, an AIEgen, and two cyclic arginine–glycine–aspartic tripeptides (cRGD), ligands targeting integrin αvβ3. The two hydrophilic peptide ligands endow TPS-2cRGD with good water solubility and high integrin αvβ3 affinity. The good water solubility guaranteed a rather low background as the AIE bioprobe was almost non-emissive in the solution state.24,42 The high integrin αvβ3 affinity makes TPS-2cRGD a specific bioprobe for cell imaging. As can be seen from Fig. 1B, high signal-to-noise ratio membrane imaging signals were collected from HT-29 cells, which were induced to have overexpressed integrin αvβ3 on the cellular membrane after incubation with TPS-2cRGD. Co-staining with a commercial membrane tracker gave the visualized location of the cell membranes (Fig. 1D). In sharp contrast, MCF-7 breast cells displayed rather weak fluorescence as they expressed integrin αvβ3 at a low level (Fig. 1E).Furthermore, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays indicate that TPS-2cRGD has a rather good biocompatibility as they show that HT-29 cells remain almost 100% metabolically viable after incubation with the probe. This is the first application of AIEgens for specific fluorescence light-up imaging in live cells and opens up great opportunities for AIEgens to be applied in cellular, subcellular and even in vivo imaging and tumor diagnosis through special component tracking.
2.2 Lipophilic structure
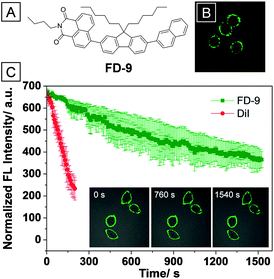
The phospholipid bilayer structure makes the cell membrane prone to capture of organic dyes with hydrophobic chains. For instance, the AIE bioprobe FD-9, which contains two hexyl and one butyl chains (Fig. 2A), has been successfully used to stain and track the cell membrane.43 As an AIEgen, the emission quantum yield (Φem) of FD-9 nanoparticles formed in a THF–water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, v/v) mixture reached as high as 70%. HepG-2 cells showed intense green fluorescence in the membrane region after incubation with FD-9, whereas the emission signals were barely detected in cytoplasm and nucleolus regions, indicating a high signal-to-noise ratio in membrane imaging (Fig. 2B). As mentioned before, DiI is a commercial membrane tracking dye; parallel studies on the photostability of FD-9 nanoparticles and DiI have been conducted in both PBS solution and living cells. As shown in Fig. 2C, continuous scanning of FD-9 and DiI stained HepG-2 cells led to sustained loss of fluorescence signal. However, the signal of FD-9 remained >50% after scanning for 1500 s, while DiI reduced to 35% in 200 s. Because of the AIE property, FD-9 showed much higher photostability than conventional DiI. Benefiting from the high photostability and strong membrane tracking ability, the FD-9 nanoparticles could stain the membrane for as long as 4 days with cell proliferation. Besides, FD-9 showed good biocompatibility since the cellular viability is estimated to be greater than 90% after incubation with 10–50 μM of the nanoparticles for 24 h.
9, v/v) mixture reached as high as 70%. HepG-2 cells showed intense green fluorescence in the membrane region after incubation with FD-9, whereas the emission signals were barely detected in cytoplasm and nucleolus regions, indicating a high signal-to-noise ratio in membrane imaging (Fig. 2B). As mentioned before, DiI is a commercial membrane tracking dye; parallel studies on the photostability of FD-9 nanoparticles and DiI have been conducted in both PBS solution and living cells. As shown in Fig. 2C, continuous scanning of FD-9 and DiI stained HepG-2 cells led to sustained loss of fluorescence signal. However, the signal of FD-9 remained >50% after scanning for 1500 s, while DiI reduced to 35% in 200 s. Because of the AIE property, FD-9 showed much higher photostability than conventional DiI. Benefiting from the high photostability and strong membrane tracking ability, the FD-9 nanoparticles could stain the membrane for as long as 4 days with cell proliferation. Besides, FD-9 showed good biocompatibility since the cellular viability is estimated to be greater than 90% after incubation with 10–50 μM of the nanoparticles for 24 h.
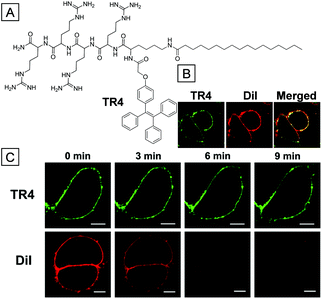
Inspired by the chemical structure of DiO and DiI, Liang and co-workers have developed an amphipathic cell membrane tracker, TR4, through the combination of the AIE moiety (TPE) with a positively charged hydrophilic tetra-peptide sequence and palmitic acid (PA).44 The four positively charged arginine units served as the targeting ligand for the negatively charged cell membrane while the long alkyl chain in PA was used for membrane insertion. Fig. 3B shows that MCF-7 cells treated with the probe were selectively lit up in the membrane region due to restriction of phenyl rotation in TR4. No obvious fluorescence signals were observed in cytoplasm and nucleolus regions. To further verify the membrane tracking ability, MCF-7 cells were treated with TR4 and DiI at the same time. The green TR4 signal was co-localized well with the red DiI signal, suggesting that TR4 is specifically targetable toward the cell membrane. Comparison of photostability was studied by continuous scanning of TR4 and DiI stained MCF-7 cells through confocal laser scanning microscopy (CLSM). Images in Fig. 3C clearly indicate that TR4 is much more photoresistant upon laser irradiation. These results indicate that TR4 is superior to the commercial dye DiI because of its AIE feature. The biocompatibility of TR4 was studied by MTT assays; the cellular viability was calculated to be more than 80% in the presence of TR4 (0.05–50 μM). Additionally, the cell membrane of HUVEC cells was specifically lit up by TR4 through excitation of 700 nm using a two-photon laser. This addresses the short absorption wavelength of TR4 in confocal imaging.
2.3 Bioorthogonal probe
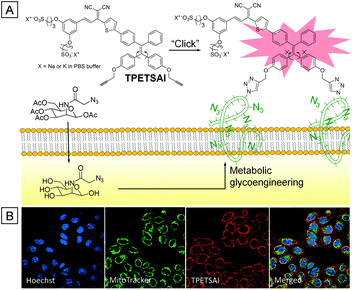
Bioorthogonal chemistry is a powerful tool for bioprobes to be applied in labeling biological substrates. It refers to chemical reactions that can proceed inside biological systems without interruption of living activities.41 Two basic processes are typically used in bioorthogonal modality. Firstly, a substrate with a bioorthogonal functional group is introduced into living systems such as cell organelles, cells, tissues, etc. Secondly, a probe containing the corresponding functional group is subsequently added to react with and label the substrate.45,46 Because AIEgens are generally very emissive when captured by biological substrates but almost non-emissive as molecular species, AIE bioprobes should have great selectivity and signal-to-noise ratio in biological imaging through bioorthogonal reaction. As shown in Fig. 4, Liu and co-workers developed a water-soluble AIE bioorthogonal probe (TPETSAI) that could specifically light up the cytoplasm membrane and subsequently offer photodynamic cancer cell ablation.47 Firstly, azide functionalized peracetylated N-azidoacetylmannosamine was taken up into the cell surface through metabolic glycoengineering. Secondly, alkynyl modified TPETSAI was introduced with catalyst systems (CuSO4, sodium ascorbate and tris(3-hydroxypropyltriazolylmethyl)amine). The membrane labelling was then performed through a bioorthogonal click reaction between azide and alkynyl groups so that the non-emissive water soluble TPETSAI was turned on because of molecular motion restriction. Besides, the electron donor alkoxy groups and electron acceptor dicyanovinyl group narrow the energy gap between the lowest singlet excited state (S1) and the lowest triplet excited state (T1) and thus endow TPETSAI with a reactive oxygen species (ROS) generation ability upon light irradiation. The generated ROS could subsequently damage the stained cancer cells, which is a process of photodynamic therapy.35 The half-maximal inhibitory concentration (IC50) toward HeLa cells was estimated to be 7.3 μm. Furthermore, as a far-red emissive bioprobe, TPETSAI has also been used for multiplexing with mitochondria targeted MitoTracker Green and nucleus targeted Hoechst 33342 (Fig. 4B).3. Mitochondria imaging
The mitochondrion is a semi-autonomous organelle with genetic material and a system as it has its own DNA, which can proceed to have replication and expression independently. It serves as a power workhouse for the oxidation of carbohydrates, fats and amino acids to produce energy currency ATP (adenosine triphosphate).48 In addition to power supply, mitochondria are also involved in many biological processes such as cell differentiation, communication and apoptosis, and have the ability to regulate cell growth and cell cycle. Mitochondrial movement is highly related to their morphology. Two mitochondria moving close to each other may fuse and exchange their contents after fusion. Similarly, fission of mitochondria involves mitochondrial regions’ division to opposite directionality.49 Thus, direct visualization of mitochondrial dynamics and changes through fluorescence microscopy can give valuable information for the understanding of mitochondria involved metabolism and diseases.50 For fluorescence imaging, one of the most important issues is the mitochondrial targeting systems. There is a constant membrane potential of around −180 mV across the outer and inner mitochondrial membranes to pump ions. This negative potential is higher by several orders of magnitude than that in other organelles and offers an opportunity to design appropriate chemical ligands for targeting mitochondria. The triphenylphosphonium (TPP)51 and pyridinium moieties are commonly used to deliver the signal probes to mitochondria because the positively charged ligands are forced to accumulate onto mitochondria by the mitochondrial membrane proton gradient. Sometimes, fluorophores with lipophilic cations, such as cyanine dyes52 and rhodamine,53 can also serve as mitochondrial delivery vehicles following a similar mechanism. Based on these designs, a number of multi-color AIE probes with high photostability and specificity have been developed for mitochondria imaging.3.1 Mitochondria imaging and mitochondrial activity tracking
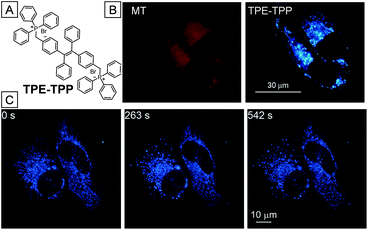
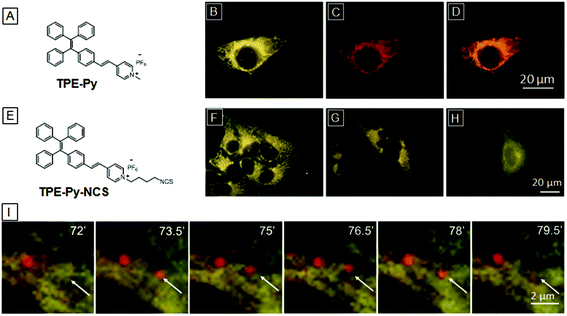
The first AIE bioprobe for mitochondria imaging is a simple integration of TPE and the mitochondrial ligand TPP named TPE-TPP (Fig. 5).54TPE-TPP inherits the AIE property of the TPE moiety and forms strongly emissive nanoparticles with an average size of 144 nm in aqueous solution containing 0.1% DMSO. Firstly, TPE-TPP showed good biocompatibility since MTT assays indicated high cell viability after incubation with the probe. Upon staining of HeLa cells with the nanoparticles, mitochondria were selectively lit up, which was further verified by comparing with the co-stained commercial MitoTracker Red FM (MT). After treatment with carbonyl cyanide m-chlorophenylhydrazone (CCCP), a drug that can trigger a mitochondrial membrane potential decrease, HeLa cells were used to estimate the ability of TPE-TPP and MT for monitoring the change of mitochondria membrane potential. As shown in Fig. 5B, MT has no specificity toward mitochondria of CCCP treated HeLa cells while TPE-TPP could still be used to visualize an elongated network of mitochondria. The high tolerance to low mitochondrial membrane potential makes TPE-TPP a good probe to follow the morphology alterations (Fig. 5C).Though TPE-TPP is an excellent mitochondria imaging probe, the short excitation and emission wavelengths severely reduce its practical application, Tang and co-workers further introduced a pyridinium unit, a suitable electron acceptor and good mitochondria targeting ligand, to the TPE core through a vinyl bond, yielding a yellow emissive mitochondria specific AIE bioprobe, TPE-Py (Fig. 6A).55TPE-Py is weakly emissive in THF but it became a strong yellow emitter at 600 nm when aggregated in aqueous solution. As shown in Fig. 6B, the bright nanoaggregates could clearly visualize the elongated network of mitochondria in HeLa cells, which overlaps well with MitoTracker Red CMXRos and verifies the mitochondrial specificity of TPE-Py. However, the electrostatic and hydrophobic interactions between the bioprobe and mitochondria are not strong enough to retain the probe in mitochondria for monitoring the cell activities. TPE-Py-NCS was subsequently designed as a mitochondrial bioprobe based on TPE-Py which contains an amine-reactive isothiocyanate (NCS) unit. The covalent conjugation between TPE-Py-NCS and mitochondrial proteins makes it suitable for mitochondria tracking.56Fig. 6F and G are CLSM images of living and fixed HeLa cells incubated with TPE-Py-NCS for 15 min, respectively. It is noteworthy that the mitochondria of TPE-Py-NCS treated HeLa cells remain emissive even after washing with acetone, suggesting chemical conjugation between mitochondrial proteins and the probe (Fig. 6H), which makes TPE-Py-NCS suitable for real-time monitoring of the mitophagy process. As shown in Fig. 6I, HeLa cells were treated with TPE-Py-NCS and LysoTracker Red DND-99 (LTR) at the same time. Rapamycin, a mitophagy trigger, was then introduced into the cells. 0–72 min after the addition of rapamycin, no change happens in the yellow fluorescence of mitochondria and the red fluorescence of lysosome. However, a new red emissive spot which overlaps with the yellow mitochondria appears at 73.5 min. This is an indication of acidic autophagosome generation to initiate mitophagy. After 79.5 min, the red spot fades away as the process is completed. This monitoring process is made possible because of the high specificity and good photostability of the designed TPE-Py-NCS bioprobe.
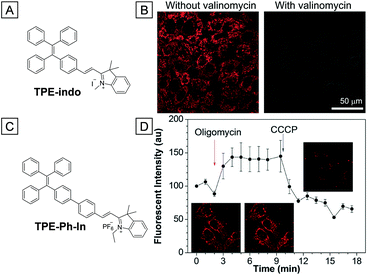
In 2015, two similar red emissive AIE bioprobes both containing the indolium group were reported to target cell mitochondria and monitor the mitochondrial membrane potential.57,58 The indolium moiety endows these probes with red fluorescence and cationic properties, which are more advantageous in mitochondria imaging. Both probes are able to monitor the mitochondrial membrane potential variation through fluorescence intensity changes. As shown in Fig. 7B, the mitochondria of HeLa cells were brightly emissive after incubation with TPE-indo. However, the fluorescence intensity was dramatically decreased after further incubation with valinomycin, which increases the movement of potassium ions through lipid membranes and then decreases the mitochondria potential gradient. Meanwhile, the lit up mitochondria could also be turned off upon addition of H2O2, indicating that TPE-indo could serve as a cell apoptosis probe. For TPE-Ph-In, an apparent enhancement of fluorescence intensity was observed in the presence of oligomycin, a mitochondrial membrane potential enhancer. The fluorescence faded away once the cells were further treated with CCCP (Fig. 7D). Ascribed to its high photostability and sensitivity, TPE-Ph-In was competent to evaluate mouse sperm vitality as the driving force of sperm is highly dependent on mitochondria bioenergy. Energetic sperms were lit up brightly while the unvital ones only showed faint emission.
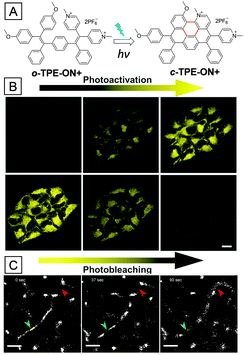
Resolution of conventional confocal microscopy is highly limited due to light diffraction; thus, CLSM images are sometimes not legible enough for tracking mitochondrial activities.59 Fortunately, the development of super-resolution imaging techniques breaks the resolution limitation and shows great advantage for tracking biological activities.12 One of the reliable super-resolution imaging techniques is single-molecule localization microscopy (SLM), which is dependent on emission on/off controllable fluorophores.60 A suitable fluorophore is first in the dark state; then some of the molecules are activated stochastically and subsequently bleached, resulting in precise location recording of the activated molecules. Almost all the fluorophore locations can be recorded by repeating the process many times; piecing together all the information will give a super-resolved image. Tang and co-workers recently developed a photoactivatable AIE bioprobe named o-TPE-ON+, which is applied for mitochondria super-resolution imaging.61 As shown in Fig. 8A, o-TPE-ON+ contains strong electron donating methoxy groups and electron accepting pyridinium groups. The intramolecular donor–acceptor interaction makes it weakly emissive in aqueous media. However, after light irradiation, o-TPE-ON+ was converted to the strongly emissive c-TPE-ON+ through photocyclodehydrogenation. As shown in Fig. 8B, CLSM images were used to monitor the photoactivation and photobleaching processes. After 20% 405 nm laser activation, the fluorescence intensity of mitochondria in HeLa cells gradually increased in 10 s; subsequent continuous scanning with a 100% 514 nm laser for 920 s led to the fluorescence intensity dropping by 90%. The SLM imaging of o-TPE-ON+ treated HeLa cells was then performed with a customized stochastic optical reconstruction microscope (STORM). A full-width at half-maximum of 105 nm was achieved, which is much narrower than that from normal imaging (697 nm). Benefiting from the super-resolution imaging, the dynamic fission and fusion activities of mitochondria were clearly visualized upon treatment with o-TPE-ON+ (Fig. 8C).
Although super-resolution imaging techniques, such as stimulated emission depletion (STED),62 ground-state depletion (GSD),63 and saturated structured-illumination microscopy (SSIM),64 have offered improved lateral and axial resolutions in imaging, they require specific fluorescent probes with excellent photostability. Since AIEgens are strongly emissive in the aggregated state, very stable upon light irradiation and easy to synthesize, they are excellent potential probes for super-resolution imaging.
3.2 Mitochondria-targeted cancer therapy
Sub-cellular organelle specific bioprobes for simultaneous tumor targeting, imaging and treatment are of enormous interest in cancer therapy. The mitochondrion is one of the most important organelles in eukaryotic cells. Delivery of therapy agencies specifically into mitochondria could decrease the therapy dosage as a direct trigger of mitochondrial dysfunctions will induce severe damage to cancer cells.65 Liu and co-workers developed an AIE bioprobe, AIE-mito-TPP, which could target cancer cell mitochondria and monitor the chemotherapy process caused by itself.34 The core of AIE-mito-TPP is salicylaldazine, which undergoes AIE- and ESIPT-emission at the same time. The combination of AIE and ESIPT is of great advantage due to the low background and the large Stokes shift. Another functional moiety is the lipophilic cation TPP group, which aims at specific mitochondrial targeting. Images of HeLa cells co-stained with MitoTracker/LysoTracker showed that AIE-mito-TPP could accumulate on the mitochondria with high efficiency. But for normal NIH-3T3 cell lines, the mitochondria and lysosomes were all lit up without specificity. Furthermore, this probe shows high cytotoxicity toward HeLa cells as it could trigger mitochondrial dysfunctions. Tetramethylrhodamine ethyl ester (TMRE), a mitochondrial membrane potential indicator, was incubated with the AIE-mito-TPP pretreated HeLa cells. The lower fluorescence intensity of AIE-mito-TPP pretreated HeLa cells indicates that the bioprobe can cause mitochondrial membrane potential reduction. The ROS indicator dihydroethidium (DHE) also clearly indicates that AIE-mito-TPP could lead to ROS generation to induce cell apoptosis. Further study showed that AIE-mito-TPP exhibited high cytotoxicity to HeLa cells even at a low concentration (0.5–1.0 μM). However, the NIH-3T3 cell viability was still above 80% at the probe concentration of 8 μM. This therapeutic selectivity is associated with the higher affinity of AIE-mito-TPP to cancerous mitochondria (Fig. 9).66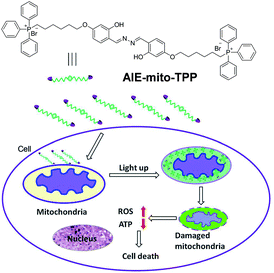 | ||
| Fig. 9 The chemical structure of AIE-mito-TPP and the mechanism for mitochondria targeted imaging and chemotherapy. Copyright 2014, John Wiley and Sons. | ||
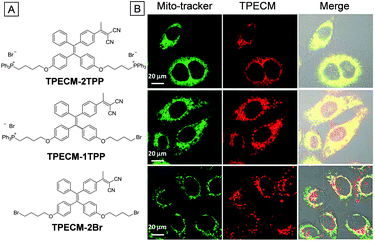
Though chemotherapy is a powerful tool for cancer treatment, drug resistance is difficult to handle. Photodynamic therapy (PDT) is a non-invasive therapy activated by irradiation of photosensitizers (PSs). The combination of PDT and chemotherapy was reported to be effective in weakening the drug resistance with minimized side effects.67TPECM-2TPP, an AIE photosensitizer, was found to show dark cytotoxicity toward cancer cells, achieving image-guided combination of chemotherapy and PDT with mitochondrial specificity.37 As shown in Fig. 10A, TPECM-2TPP is composed of a red emissive AIE core and two mitochondrial ligands TPP. Two control compounds with one TPP group (TPECM-1TPP) and without a TPP group (TPECM-2Br), respectively, were also studied to highlight the advantage of TPECM-2TPP. HeLa cell images showed that TPECM-2TPP could accumulate on its mitochondria effectively. The dark cytotoxicity of TPECM-2TPP to HeLa cells is high with an IC50 of 6.3 μM. However, the IC50 was improved to 0.69 μM under light irradiation, due to the combination of PDT and chemotherapy. For TPECM-1TPP, it also showed good overlap with MitoTracker Green co-staining. But the dark cytotoxicity is rather low, with an IC50 above 10 μM. The lower dark cytotoxicity might be ascribed to the weakened mitochondria targeting affinity although it has the same overlap coefficient with MitoTracker Green as TPECM-2TPP. However, TPECM-2Br is not able to target HeLa cell mitochondria since it has no mitochondrial ligand. It showed low cytotoxicity even under light irradiation. This example illustrates that direct damage of organelles and combination of chemotherapy and PDT can both dramatically improve therapeutic efficiency. Since mitochondria supply almost all the energy for cellular metabolic activities, the targeting to mitochondria and subsequent damage to mitochondria could be one of the most efficient therapeutic solutions to cancer cells.
4. Lysosome imaging
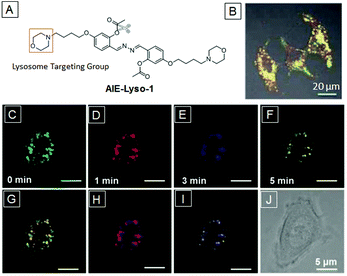
The lysosome is the digestive organelle which contains more than 60 kinds of acid hydrolases in most eukaryotic cells. Lysosomal endocytosis helps to decompose absorbed food vacuoles into nutritional biomacromolecules or to degrade intruding pathogens to safeguard the cells.3 Meanwhile, lysosome autophagy is a process involved in elimination of intracellular disabled biomacromolecules or organelles. Obviously, the dysfunction or enzyme mutation of lysosomes would lead to serious disposal accumulated diseases such as lysosome storage diseases or inclusion-cell disease. Thus, monitoring of lysosome activities is instructive and meaningful for the detection of lysosome related sickness. Fluorescence imaging is able to visualize their morphology, map their distribution and trace their movement in living cells, which represent their ability to perform functional activities.68 There are mainly two design principles for lysosome-specific fluorescent bioprobes. The first is to make the bioprobe a lysosomal enzyme substrate. Initially, the bioprobe is almost non-emissive in aqueous medium. After penetration into cells, the bioprobe becomes strongly emissive upon exposure to lysosomal enzymes such as esterase and protease. The second is to combine a ligand which is specific to the acid lysosomal environment. For example, some weak base groups (N,N-dimethyl amino and morpholine) are only partially protonated in the neutral state to permeate cell membranes and then interact with acid lysosomes in the cytoplasm.The first AIE based lysosome targeting bioprobe is AIE-Lyso-1, which contains lysosome ligand morpholines and esterase substrate site acetoxyl groups.69 The esterase substrate site guarantees that the probe is only emissive in lysosomes and the ligands prevent the active bioprobe from diffusion away from the reactive sites. Initially, hydroxyl groups were blocked by acetyl groups, leading to breakage of hydrogen bonds and free rotation of the N–N bond (both are the off states of ESIPT and AIE, respectively) and hence the quenched fluorescence. Once the probe was incubated with MCF-7 cells, it was captured by the lysosomes directed by morpholine ligands. Subsequently, the protective acetyl groups were hydrolysed by esterase. The generated hydroxyl groups could form intramolecular hydrogen bonds with nitrogen atoms to light up lysosomes due to the activation of both ESIPT and AIE processes. The overlapped yellow color from the green emissive AIE-Lyso-1 and LysoTracker Red co-stained MCF-7 cells further indicate the high specificity of AIE-Lyso-1 to lysosomes (Fig. 11B). Thanks to the high affinity to lysosomes, this probe could also be applied to trace lysosome movements. As shown in Fig. 11C, after the MCF-7 cells were treated with chloroquine, a stimulus for slight lysosome movement, different pseudocolors were used to record the precise location of lysosomes at different times. The merged pictures clearly indicate the subtle movements of lysosomes (Fig. 11G–I). MTT assays showed that cell viabilities were close to 100% under the testing conditions, indicating the low cytotoxicity and good biocompatibility of AIE-Lyso-1.
A similar lysosome specific AIE bioprobe, namely AIE-LysoY, was applied to monitor the autophagy process (Fig. 12).70 The morpholine group serves as the targeting ligand while the “ESIPT + AIE” fluorophore is the fluorescence marker. In the fluorophore, one fluorene is used to replace a phenyl ring in salicylaldazine without disturbance of “ESIPT + AIE” emission character. Meanwhile, the absorption is red-shifted to 390 nm, which is better for imaging due to the expansion of pi-electron conjugation. Fig. 12B shows the images of MCF-7 cells treated with AIE-LysoY, LysoTracker Red and their merged image. The yellow fluorescence from AIE-LysoY overlaps perfectly with the red fluorescence from LysoTracker Red, verifying that AIE-LysoY could selectively light up lysosomes in living cells. In addition, after incubation with AIE-LysoY for 24 h, the viability of HeLa cells remained above 80%, indicating good biocompatibility. Continuous scanning of AIE-LysoY treated MCF-7 cells for 50 times led to only 20% signal loss of emission, while it was above 50% for LysoTracker Red, which verifies the better photostability of AIE-LysoY than commercial LysoTracker Red. The good biocompatibility and high photostability of AIE-LysoY make it suited to monitor the bioactivities of lysosomes.
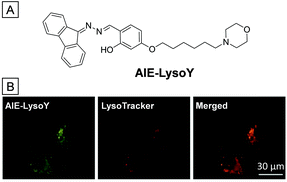 | ||
| Fig. 12 (A) Chemical structure of AIE-LysoY. (B) CLSM images of AIE-LysoY (left), LysoTracker Red (middle) treated MCF-7 cells and their merged photo (right). Copyright 2016, John Wiley and Sons. | ||
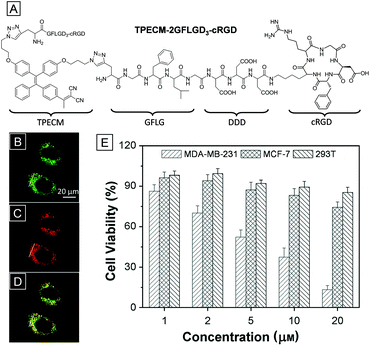
Since lysosomes are one of the most vital organelles in eukaryotic cells, disruption of lysosomes in cancerous cells should be an efficient method for cancer therapy. An AIE photosensitizer combined with a lysosomal protease responsive peptide and a cancer cell targeting ligand was successfully applied in image-guided photodynamic therapy (Fig. 13).36 The cRGD sequence, an αvβ3 integrin ligand mentioned before, guarantees the accumulation of the bioprobe toward αvβ3 integrin overexpressed cancer cells. Meanwhile, the -Gly-Phe-Leu-Gly- (GFLG) peptide sequence, a cleave site for the lysosomal enzyme cathepsin B, ensures that the bioprobe can only be activated in cancer cell lysosomes. TPECM, a red emissive AIE fluorophore, was designed to serve as a switch-on imaging agent and a photosensitizer. At first, the bioprobe was water-soluble and almost non-emissive in aqueous solution. However, after incubation with MDA-MB-231 cells, αvβ3 integrin overexpressed cancer cells, the hydrophilic peptide sequences were cut off by cathepsin B expressed in lysosomes. The probe was converted into a hydrophobic compound and specifically lighted up lysosomes of MDA-MB-231 cells. The high overlap coefficient with LysoTracker Green DND-26 shown in Fig. 13B proves the lysosome targeting function. Furthermore, after incubation with the probe, the viability of MDA-MB-231 cells decreased dramatically upon light irradiation. This is ascribed to the harmful ROS generated through the photosensitization process. However, the viability of some other cell lines with low αvβ3 integrin expression remained high under the same conditions due to poor uptake of the probe. The rational design of the probe makes it a good example of cancer lysosome-targeted imaging and PDT.
5. Lipid droplet imaging
Lipid droplets (LDs), existing mainly in adipose tissues, are lipid rich organelles that serve as a reservoir for neutral lipid storage.71 For a long time since they were discovered in the 17th century, LDs were merely considered as fat depots and inert content of cells. However, the discovery of a series of proteins in LDs made people aware that LDs are highly dynamic organelles which regulate intracellular lipid storage and metabolism.72 They are also highly involved in membrane transfer for supplementation of acyl-glycerols and cholesterol. The abnormality of lipid droplet activities or numbers is a critical signal of various diseases, such as fatty liver diseases, type II diabetes and inflammatory myopathy.73 Monitoring the location and distribution of LDs is useful for early diagnosis of related diseases. Fluorescence imaging shows great advantages in visualizing LDs in cells. The design of LD targeted bioprobes takes into consideration the highly hydrophobic lipid environment, which is less polar than other cellular parts. Generally, TICT fluorophores are weakly emissive in a polar environment, but they could become strongly emissive with blue-shifted emission in a nonpolar environment. Therefore, fluorophores with TICT characteristics, such as Nile red74 and BODIPY,75 are great candidates for LD imaging. Several AIE bioprobes have been proven to be good LD imaging agents due to their high photostability and selectivity.The AIE bioprobe TPE-AmAl, whose emission color alters according to environmental polarity, is an excellent LD imaging agent with high photostability and selectivity (Fig. 14).76 The strong electron-donating groups (dimethylamino) and electron-accepting groups (formyl) endow the dye with TICT and solvatochromism characteristics. TPE-AmAl was faintly emissive when dissolved in THF and became strongly emissive in a THF/water mixture when the water ratio was above 80%, showing typical AIE property. However, the emission color was gradually red-shifted with an increase of the water ratio from 0 to 70%, which is ascribed to the TICT property and increased solvent polarity. When the water ratio is continuously increased from 80% to 99%, the emission color was slightly blue-shifted due to the less polar environment in the formed nanoaggregates. After being incubated with TPE-AmAl, the spherical LDs in HeLa cells were specifically lit up with a low background. However, parallel experiments with Nile red gave poor LD imaging due to poor selectivity (Fig. 14C). The biocompatibility of TPE-AmAl was verified through a MTT assay and the photostability was better than that of Nile red due to the AIE property. Though TPE-AmAl is an orange emitter in aqueous solution, the emission color turns greenish blue when accumulated onto LDs due to a highly hydrophobic and non-polar environment. Subsequently, a red-emissive LD probe of TPE-Ac was synthesized (Fig. 14B), which is suitable for in vivo imaging.77
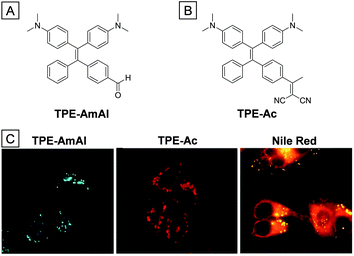 | ||
| Fig. 14 Chemical structures of TPE-AmAl (A) and TPE-Ac (B). (C) Fluorescence images of TPE-AmAl, TPE-Ac and Nile red treated HeLa cells. Copyright 2014 and 2016, Royal Society of Chemistry. | ||
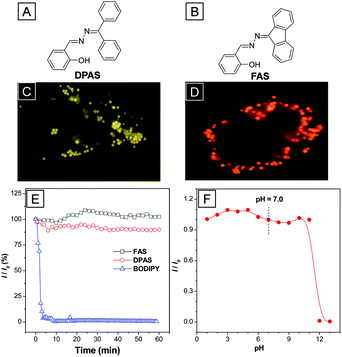
In addition to typical AIE bioprobes, two “ESIPT + AIE” fluorophores named DPAS and FAS with yellow and orange emission, respectively, were also reported to target LDs in living and fixed cells.78DPAS and FAS were both obtained through a dehydration reaction between ketone and hydrazine hydrate. The salicylaldehyde Schiff-base structures endow both AIEgens with the ESIPT property. Thanks to the ESIPT property, the Stokes shifts of DPAS and FAS are as large as ∼200 nm, which are superior to the BODIPY dye based LD stains. As shown in Fig. 15C and D, the LDs in A549 cells were lit up with high resolution after incubation with DPAS or FAS. The overlap ratios of DPAS and FAS with the commercial LD targeted BODIPY dye were as high as 98% and 97%, respectively, suggesting great LD targeting affinity. No distinct inhibitory effect was observed on HeLa cell growth in culture medium with high concentrations of up to 10 μM of FAS and DPAS, indicating excellent biocompatibility of the two probes. Upon continuous light irradiation, the fluorescence intensities of DPAS, FAS and BODIPY were recorded to study their relative photostability (Fig. 15E). The signal loss of BODIPY reached 90% after 5 min irradiation while the fluorescence intensity of DPAS and FAS almost remained the same after 60 min of irradiation. Furthermore, the fluorescence of DPAS and FAS does not change in a wide range of pH from 1.0 to 11.0, indicating their great tolerance to pH variations.
6. Nucleus imaging
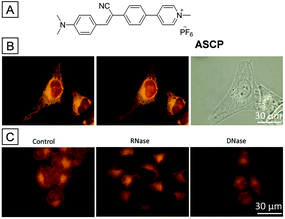
The nucleus, the biggest organelle in the cell, was first discovered in the early 18th century. It serves as the brain of the cell because it contains the most genetic materials and maintains the gene expression. In most eukaryotic cells, the genetic materials are chromosomes composed of long DNA molecules and nucleoproteins such as histones. During the gene expression process, one of the most important procedures is the transcription of rDNA to rRNA which occurs in the nucleolus. After that, the RNA is assembled with proteins to form a precursor of ribosomes, which is then transported to the cell cytoplasm. Fluorescence imaging is a powerful tool to visualize the bioactivities and morphology of the nucleus.79 Since large molecules are forbidden to permeate the nuclear membrane, few fluorogens are reported to stain the nucleus. For nucleus imaging, the bioprobes generally have an ability to target special nuclear inclusions such as nucleoproteins or nucleic acids.80 One of the main design principles is to introduce DNA targeting sites. The probes should be almost non-emissive, but the fluorescence intensities are enhanced dramatically after interaction with DNA. The probes could target different parts of the double helix frame of DNA. These probes are classified as intercalating dyes (ethidium bromide, propidium iodide), minor-groove binders (DAPI, Hoechst dyes) and other nucleic acid stains (acridine orange).81 The other design principle is to stain RNA in the nucleolus. Molecules with positive charge have the potential to target the negatively charged RNA. Besides, molecular beacons with a matched base sequence could offer better selectivity.82ASCP is a newly designed AIEgen bearing the TICT property and could be used as a dual-color agent for mitochondria and nucleus imaging.83 Owing to the intramolecular electron push–pull effect, the emission color of ASCP could be readily tuned by solvent polarity. When dissolved in DMSO, it was faintly red emissive. However, the fluorescence was gradually enhanced and blue-shifted with the increase of the toluene ratio in a DMSO/toluene mixture. Firstly, the cytotoxicity was studied; the cell viability remains high at ASCP concentrations as high as 10 μM, suggesting that ASCP possesses good biocompatibility. After incubation with ASCP, the HeLa cells were almost totally lit up inside and outside the nucleus. Interestingly, fluorescence images indicated that the emission color outside the nucleus was orange while that inside was red. Referenced by MitoTracker Green, the orange emissive parts of HeLa cells were proven to be mitochondria. Thus, ASCP could be used as a dual-color agent for mitochondria and nucleus imaging. Since most components of mitochondria and the nucleus are phospholipids and nucleic acids, the fluorescence properties of ASCP with lipid vesicles and nucleic acids were studied to understand its nucleus targeting ability. The fluorescence of ASCP was enhanced and red-shifted with nucleic acids while the emission maximum was unaltered with different kinds of lipids. This makes it clear that the nucleus targeting ability of ASCP comes from the affinity toward nucleic acids, especially for RNAs. For a better understanding, fixed cells incubated with ASCP were treated with deoxyribonuclease (DNase) and ribonuclease (RNase). Fig. 16C shows that the red fluorescence of RNase treated HeLa cells disappeared while the red fluorescence remained for DNase treated HeLa cells, which further indicated that ASCP stained the nucleus with red fluorescence through targeting the RNAs in the nucleolus. SYTO RNASelect, a commercial probe for nucleolus, performed similar to ASCP in parallel experiments.
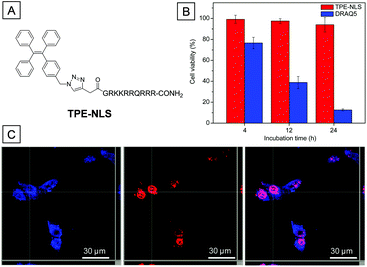
The conjugation of AIEgens with water soluble targeting ligands is one of the basic design principles to realize turn-on AIE bioprobes.42TPE-NLS, a probe composed of a TPE group and a nucleus targeting peptide sequence (GRKKRRQRRR), was successfully applied in nucleus imaging without washing procedures.84 The peptide sequence, derived from trans-activator of transcription (TAT) viral proteins, endows the bioprobe with cell penetration and nucleus uptake abilities. When dissolved in aqueous solution, TPE-NLS was almost non-emissive due to a hydrophilic peptide sequence. However, the fluorescence intensity of TPE-NLS dramatically increased after addition of DNA, histone or nuclear lysate, indicating that TPE-NLS could be turned on through interaction with these nucleus inclusions. As shown in Fig. 17C, the nucleus and cytoplasm of MCF-7 cells were lit up with bright blue fluorescence after incubation with TPE-NLS. DRAQ5 is a commercial red emissive nucleus targeting probe. The overlay images between TPE-NLS and DRAQ5 treated MCF-7 cells showed magenta color in the nuclear location, further verifying the nucleus targeting ability of TPE-NLS. Besides, the biocompatibility of TPE-NLS is much better compared to commercial DRAQ5 at the same concentration (Fig. 17B). According to the design principle of TAT conjugated AIEgens, TPE-NLS was rationally designed to stain the nucleus of MCF-7 cells.
7. Conclusions
In this review, we have summarized the applications of AIEgens in specific organelle imaging with excellent performance. Most of the designs are based on the conjugation between AIEgens and organelle-specific ligands. The ligand can not only be selected from moieties with high affinity to special organelle inclusions but also from groups that are prone to accumulate in organelles due to their special micro-environment. Thanks to the bright emission of AIEgens in the aggregated state, they show even stronger fluorescence after accumulation in special organelles rather than quenched fluorescence for typical ACQ dyes. The bright fluorescence of AIE bioprobes in organelles results in high contrast imaging of targeted organelles. Because of their weak fluorescence as molecular probes and strong emission in the aggregated state, higher concentrations of AIE bioprobes could be used in the imaging process without the concern of fluorescence quenching when they are accumulated in organelles. The higher concentration further endows the AIE bioprobes with good photobleaching resistance, which is beneficial to long-term monitoring of organelle activities. Besides, most of the AIE bioprobes have good biocompatibility and can hardly cause any disturbances to cell activities after their accumulation to organelles. The high photostability and biocompatibility make AIE bioprobes great candidates for tracking the morphology, distribution and movements of organelles in living cells. Direct visualization of organelles could provide a large amount of valuable information about organelles, which can assist the further understanding of the organelle metabolism, organelle related diseases and even the early diagnosis of the involved diseases.When properly designed, AIEgens could also be developed to serve as potential chemical drugs or photosensitizers. The corresponding probes will possess dark or light cytotoxicity toward cancerous cells. The natural fluorescence and cytotoxicity of some AIE bioprobes have been used directly for image-guided cancer cell ablation. Once these probes are further introduced into cancerous organelles, the AIE bioprobes can work more efficiently with significantly reduced IC50 values. These organelle targeting AIE bioprobes have great potential in diagnosis and therapy of tumors.
As compared to commercial probes, organelle-specific AIE bioprobes almost always show better photostability because they are imaged in the aggregated state. Although many AIEgens used for organelle staining have shown absorption at short wavelengths, many of them have multi-photon absorption properties so that they could be excited with multiphoton lasers. We strongly believe that the recent development of AIEgens with multiphoton absorption and NIR emission will drive the applications of AIE organelle stains to a new height.
Acknowledgements
We thank the SMART (R279-000-378-592), the Ministry of Education (R279-000-391-112), Singapore NRF Investigatorship (R279-000-444-281) and the Institute of Materials Research and Engineering of Singapore (IMRE/14-8P1110) for financial support. We thank Ms Purnima Manghnani for proof-reading the manuscript.References
- P. Mueller, D. O. Rudin, H. Ti Tien and W. C. Wescott, Nature, 1962, 194, 979 CrossRef CAS.
- D. R. Green and J. C. Reed, Science, 1998, 281, 1309 CrossRef CAS.
- C. de Duve and R. Wattiaux, Annu. Rev. Physiol., 1966, 28, 435 CrossRef CAS.
- Y. Miyanari, K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata and K. Shimotohno, Nat. Cell Biol., 2007, 9, 1089 CrossRef CAS.
- R. Berezney, J. Cell. Biochem., 1991, 47, 109 CrossRef CAS.
- S. Ohta, Curr. Med. Chem., 2003, 10, 2485 CrossRef CAS.
- E. J. Parkinson-Lawrence, T. Shandala, M. Prodoehl, R. Plew, G. N. Borlace and D. A. Brooks, Physiology, 2010, 25, 102 CrossRef CAS.
- N. Mizushima, B. Levine, A. M. Cuervo and D. J. Klionsky, Nature, 2008, 451, 1069 CrossRef CAS.
- F. Camoes, N. Bonekamp, H. Delille and M. Schrader, J. Inherited Metab. Dis., 2009, 32, 163 CrossRef CAS.
- C. P. Satori, M. M. Henderson, E. A. Krautkramer, V. Kostal, M. M. Distefano and E. A. Arriaga, Chem. Rev., 2013, 113, 2733 CrossRef CAS.
- D. Shin, N. Vigneswaran, A. Gillenwater and R. Richards-Kortum, Future Oncol., 2010, 6, 1143 CrossRef.
- M. Fernández-Suárez and A. Y. Ting, Nat. Rev. Mol. Cell Biol., 2008, 9, 929 CrossRef.
- S. M. Borisov and O. S. Wolfbeis, Chem. Rev., 2008, 108, 423 CrossRef CAS.
- D. W. Domaille, E. L. Que and C. J. Chang, Nat. Chem. Biol., 2008, 4, 168 CrossRef CAS.
- B.-K. An, S.-K. Kwon, S.-D. Jung and S. Y. Park, J. Am. Chem. Soc., 2002, 124, 14410 CrossRef CAS.
- X. Michalet, F. Pinaud, L. Bentolila, J. Tsay, S. Doose, J. Li, G. Sundaresan, A. Wu, S. Gambhir and S. Weiss, Science, 2005, 307, 538 CrossRef CAS.
- N. C. Shaner, P. A. Steinbach and R. Y. Tsien, Nat. Methods, 2005, 2, 905 CrossRef CAS.
- J. Liang, K. Li and B. Liu, Chem. Sci., 2013, 4, 1377 RSC.
- J. Li, J.-J. Zhu and K. Xu, TrAC, Trends Anal. Chem., 2014, 58, 90 CrossRef CAS.
- A. M. Smith, H. Duan, A. M. Mohs and S. Nie, Adv. Drug Delivery Rev., 2008, 60, 1226 CrossRef CAS.
- E. Dellambra, G. Pellegrini, L. Guerra, G. Ferrari, G. Zambruno, F. Mavilio and M. De Luca, Hum. Gene Ther., 2000, 11, 2283 CrossRef CAS.
- Y. Hong, J. W. Lam and B. Z. Tang, Chem. Commun., 2009, 4332 RSC.
- J. Mei, Y. Hong, J. W. Lam, A. Qin, Y. Tang and B. Z. Tang, Adv. Mater., 2014, 26, 5429 CrossRef CAS.
- J. Mei, N. L. C. Leung, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, Chem. Rev., 2015, 115, 11718 CrossRef CAS.
- H. Shi, N. Zhao, D. Ding, J. Liang, B. Z. Tang and B. Liu, Org. Biomol. Chem., 2013, 11, 7289 CAS.
- J.-X. Wang, Q. Chen, N. Bian, F. Yang, J. Sun, A.-D. Qi, C.-G. Yan and B.-H. Han, Org. Biomol. Chem., 2011, 9, 2219 CAS.
- J.-P. Xu, Y. Fang, Z.-G. Song, J. Mei, L. Jia, A. J. Qin, J. Z. Sun, J. Ji and B. Z. Tang, Analyst, 2011, 136, 2315 RSC.
- M. Wang, X. Gu, G. Zhang, D. Zhang and D. Zhu, Anal. Chem., 2009, 81, 4444 CrossRef CAS PubMed.
- Y. Huang, F. Hu, R. Zhao, G. Zhang, H. Yang and D. Zhang, Chem. – Eur. J., 2014, 20, 158 CrossRef CAS.
- H. Shi, J. Liu, J. Geng, B. Z. Tang and B. Liu, J. Am. Chem. Soc., 2012, 134, 9569 CrossRef CAS PubMed.
- K. Li, Y. Jiang, D. Ding, X. Zhang, Y. Liu, J. Hua, S.-S. Feng and B. Liu, Chem. Commun., 2011, 47, 7323 RSC.
- G. Feng, C. Y. Tay, Q. X. Chui, R. Liu, N. Tomczak, J. Liu, B. Z. Tang, D. T. Leong and B. Liu, Biomaterials, 2014, 35, 8669 CrossRef CAS PubMed.
- D. Ding, C. C. Goh, G. Feng, Z. Zhao, J. Liu, R. Liu, N. Tomczak, J. Geng, B. Z. Tang and L. G. Ng, Adv. Mater., 2013, 25, 6083 CrossRef CAS PubMed.
- Q. L. Hu, M. Gao, G. X. Feng and B. Liu, Angew. Chem., Int. Ed., 2014, 53, 14225 CrossRef CAS PubMed.
- F. Hu, Y. Huang, G. Zhang, R. Zhao, H. Yang and D. Zhang, Anal. Chem., 2014, 86, 7987 CrossRef CAS PubMed.
- Y. Yuan, C. J. Zhang, M. Gao, R. Zhang, B. Z. Tang and B. Liu, Angew. Chem., Int. Ed., 2015, 54, 1780 CrossRef CAS PubMed.
- C. J. Zhang, Q. L. Hu, G. X. Feng, R. Y. Zhang, Y. Y. Yuan, X. M. Lu and B. Liu, Chem. Sci., 2015, 6, 4580 RSC.
- B. Wang, C. Zhu, L. Liu, F. Lv, Q. Yang and S. Wang, Polym. Chem., 2013, 4, 5212 RSC.
- Q.-Y. Jiang, L.-H. Lai, J. Shen, Q.-Q. Wang, F.-J. Xu and G.-P. Tang, Biomaterials, 2011, 32, 7253 CrossRef CAS PubMed.
- M. Liu, Z. Li, F. Xu, L. Lai, Q. Wang, G. Tang and W. Yang, Biomaterials, 2012, 33, 2240 CrossRef CAS PubMed.
- E. M. Sletten and C. R. Bertozzi, Angew. Chem., Int. Ed., 2009, 48, 6974 CrossRef CAS PubMed.
- J. Liang, B. Z. Tang and B. Liu, Chem. Soc. Rev., 2015, 44, 2798 RSC.
- Y. Li, Y. Wu, J. Chang, M. Chen, R. Liu and F. Li, Chem. Commun., 2013, 49, 11335 RSC.
- C. Zhang, S. Jin, K. Yang, X. Xue, Z. Li, Y. Jiang, W.-Q. Chen, L. Dai, G. Zou and X.-J. Liang, ACS Appl. Mater. Interfaces, 2014, 6, 8971 CAS.
- Z. Hao, S. Hong, X. Chen and P. R. Chen, Acc. Chem. Res., 2011, 44, 742 CrossRef CAS PubMed.
- A. Borrmann and J. C. van Hest, Chem. Sci., 2014, 5, 2123 RSC.
- Y. Yuan, S. Xu, X. Cheng, X. Cai and B. Liu, Angew. Chem., Int. Ed., 2016, 55, 6457 CrossRef CAS PubMed.
- L. S. Jouaville, P. Pinton, C. Bastianutto, G. A. Rutter and R. Rizzuto, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 13807 CrossRef CAS.
- D.-F. Suen, K. L. Norris and R. J. Youle, Genes Dev., 2008, 22, 1577 CrossRef CAS PubMed.
- Z. Xu and L. Xu, Chem. Commun., 2016, 52, 1094 RSC.
- M. Ross, G. Kelso, F. Blaikie, A. James, H. Cocheme, A. Filipovska, T. Da Ros, T. Hurd, R. Smith and M. Murphy, Biochemistry, 2005, 70, 222 CAS.
- S. Luo, E. Zhang, Y. Su, T. Cheng and C. Shi, Biomaterials, 2011, 32, 7127 CrossRef CAS PubMed.
- Z. Zhao, P.-S. Chan, H. Li, K.-L. Wong, R. N. S. Wong, N.-K. Mak, J. Zhang, H.-L. Tam, W.-Y. Wong and D. W. Kwong, Inorg. Chem., 2011, 51, 812 CrossRef PubMed.
- C. W. T. Leung, Y. N. Hong, S. J. Chen, E. G. Zhao, J. W. Y. Lam and B. Z. Tang, J. Am. Chem. Soc., 2013, 135, 62 CrossRef CAS PubMed.
- N. Zhao, M. Li, Y. L. Yan, J. W. Y. Lam, Y. L. Zhang, Y. S. Zhao, K. S. Wong and B. Z. Tang, J. Mater. Chem. C, 2013, 1, 4640 RSC.
- W. J. Zhang, R. T. K. Kwok, Y. L. Chen, S. J. Chen, E. G. Zhao, C. Y. Y. Yu, J. W. Y. Lam, Q. C. Zheng and B. Z. Tang, Chem. Commun., 2015, 51, 9022 RSC.
- L. Zhang, W. W. Liu, X. H. Huang, G. X. Zhang, X. F. Wang, Z. Wang, D. Q. Zhang and X. Y. Jiang, Analyst, 2015, 140, 5849 RSC.
- N. Zhao, S. J. Chen, Y. N. Hong and B. Z. Tang, Chem. Commun., 2015, 51, 13599 RSC.
- C. B. Müller and J. Enderlein, Phys. Rev. Lett., 2010, 104, 198101 CrossRef PubMed.
- S. Manley, J. M. Gillette, G. H. Patterson, H. Shroff, H. F. Hess, E. Betzig and J. Lippincott-Schwartz, Nat. Methods, 2008, 5, 155 CrossRef CAS PubMed.
- X. Gu, E. Zhao, T. Zhao, M. Kang, C. Gui, J. W. Lam, S. Du, M. M. Loy and B. Z. Tang, Adv. Mater., 2016, 28, 5064 CrossRef CAS PubMed.
- S. W. Hell and J. Wichmann, Opt. Lett., 1994, 19, 780 CrossRef CAS PubMed.
- S. W. Hell and M. Kroug, Appl. Phys. B, 1995, 60, 495 CrossRef.
- M. G. Gustafsson, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 13081 CrossRef CAS PubMed.
- D. Trachootham, J. Alexandre and P. Huang, Nat. Rev. Drug Discovery, 2009, 8, 579 CrossRef CAS PubMed.
- J. S. Modica-Napolitano and J. R. Aprille, Adv. Drug Delivery Rev., 2001, 49, 63 CrossRef CAS PubMed.
- A. Khdair, D. Chen, Y. Patil, L. Ma, Q. P. Dou, M. P. Shekhar and J. Panyam, J. Controlled Release, 2010, 141, 137 CrossRef CAS PubMed.
- E. Van Meel and J. Klumperman, Histochem. Cell Biol., 2008, 129, 253 CrossRef PubMed.
- M. Gao, Q. Hu, G. Feng, B. Z. Tang and B. Liu, J. Mater. Chem. B, 2014, 2, 3438 RSC.
- C. W. T. Leung, Z. M. Wang, E. G. Zhao, Y. N. Hong, S. J. Chen, R. T. K. Kwok, A. C. S. Leung, R. S. Wen, B. S. Li, J. W. Y. Lam and B. Z. Tang, Adv. Healthcare Mater., 2016, 5, 427 CrossRef CAS PubMed.
- D. Brasaemle, T. Barber, N. Wolins, G. Serrero, E. Blanchette-Mackie and C. Londos, J. Lipid Res., 1997, 38, 2249 CAS.
- S. Martin and R. G. Parton, Nat. Rev. Mol. Cell Biol., 2006, 7, 373 CrossRef CAS PubMed.
- K. G. M. M. Alberti, P. Zimmet and J. Shaw, Diabetic Med., 2006, 23, 469 CrossRef CAS PubMed.
- P. Greenspan, E. P. Mayer and S. D. Fowler, J. Cell Biol., 1985, 100, 965 CrossRef CAS PubMed.
- J. Spandl, D. J. White, J. Peychl and C. Thiele, Traffic, 2009, 10, 1579 CrossRef CAS PubMed.
- E. Wang, E. Zhao, Y. Hong, J. W. Lam and B. Z. Tang, J. Mater. Chem. B, 2014, 2, 2013 RSC.
- T. Ryan, W. Chris, W. Jacky and B. ZhongáTang, Chem. Commun., 2016, 52, 5957 RSC.
- Z. Wang, C. Gui, E. Zhao, J. Wang, X. Li, A. Qin, Z. Zhao, Z. Yu and B. Z. Tang, ACS Appl. Mater. Interfaces, 2016, 8, 10193 CAS.
- R. W. Horobin, J. C. Stockert and F. Rashid-Doubell, Histochem. Cell Biol., 2006, 126, 165 CrossRef CAS PubMed.
- P. Collas and P. Aleström, Mol. Mar. Biol. Biotechnol., 1997, 6, 48 CAS.
- Z. Darzynkiewicz, F. Traganos, J. Kapuscinski, L. Staiano-Coico and M. R. Melamed, Cytometry, 1984, 5, 355 CrossRef CAS PubMed.
- W. Tan, K. Wang and T. J. Drake, Curr. Opin. Chem. Biol., 2004, 8, 547 CrossRef CAS PubMed.
- Y. Chris, W. Zhang, R. T. Kwok, C. W. Leung, J. W. Lam and B. Z. Tang, J. Mater. Chem. B, 2016, 4, 2614 RSC.
- J. Liang, G. Feng, R. T. K. Kwok, D. Ding, B. Tang and B. Liu, Sci. China: Chem., 2016, 59, 53 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2016 |