Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
High-affinity host–guest chemistry of large-ring cyclodextrins
Khaleel I.
Assaf
*a,
Detlef
Gabel
a,
Wolfgang
Zimmermann
b and
Werner M.
Nau
*a
aDepartment of Life Sciences and Chemistry, Jacobs University Bremen, Campus Ring 1, 28759 Bremen, Germany. E-mail: k.assaf@jacobs-university.de; w.nau@jacobs-university.de
bDepartment of Microbiology and Bioprocess Technology, Institute of Biochemistry, Leipzig University, Johannisallee 21-23, 04103 Leipzig, Germany
First published on 20th July 2016
Abstract
The host–guest chemistry of large-ring cyclodextrins (LRCDs) has been largely unexplored due to the lack of suitable guest molecules that bind with significant affinities to enable potential applications. Herein, we report their complexation with dodecaborate anions (B12X122−), a novel class of guest molecules. The binding constants of the inorganic guests (104–106 M−1) allow their classification as the first tight binders for LRCDs.
Introduction
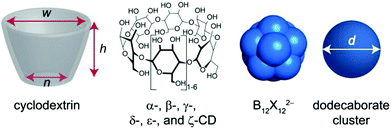
Cyclodextrins (CDs, Fig. 1) are native water-soluble macrocyclic molecules that consist of α(1–4)-linked D-glucopyranose units.1 The smallest homologues, α-, β-, and γ-CD with 6, 7, and 8 units, are cone-shaped with a hydrophobic cavity that is capable of encapsulating small organic guests.2 Larger CD homologues are also available. The first evidence for the existence of large-ring CDs (LRCDs, see δ-, ε-, and ζ-CD in Fig. 1),3 which are composed of 9 or more glucoses, dates back to the work by Freudenberg and Cramer,4 which was confirmed by Pulley and French.5 The structure of LRCDs was found to be different from the annular shape of small CDs. The crystal structures of δ-CD and ε-CD display a distorted elliptic boat-like shape, while even larger rings have more folded conformations.6 Molecular dynamics studies showed that the distorted shape of LRCDs is induced by steric encumbrance caused by large-ring strain.7A number of potential applications have been proposed for LRCDs, including food-industry and drug-formulation-related ones, owing to their non-toxicity, which is a general asset of CDs.3b,c,8 However, even though evidence for the formation of inclusion complexes could be obtained from solubility enhancements of guests with limited water solubility3b,8a–c,e or from crystal structures of the precipitating solids,9 the host–guest chemistry of LRCDs has received little attention. In particular, their affinities to guest molecules proved to be disappointingly low, which has been related to their large cavities and their high flexibility,3b,8b,e,9,10 affinity-limiting features which are also known for the large derivatives of other classes of macrocycles such as cucurbiturils11 and calixarenes.12 The status-quo of host–guest chemistry of LRCDs can be summed up by Table 1, which includes the guests for which low binding affinities or upper limits have been estimated.
| Host | Guest | K a/M−1 | Ref. |
|---|---|---|---|
| a Measured by the solubility method, while the structural evidence for inclusion was obtained by 1H NMR. b Measured by the solubility method. c Measured by electrophoresis. d The highest affinity was obtained for δ-CD. e The highest affinity was obtained for 3,5-dimethoxy benzoate with ζ-CD. f The highest affinity was obtained for δ-CD and no value was reported for ε-CD. g Spectroscopic evidence for complexation was obtained by UV-Vis titration. h Structural evidence for inclusion was obtained by XRD. | |||
| δ-CD | Digitoxin | 1700a | 8e |
| δ-CD | Spironolactone | 820b | 8a |
| δ-, ε-, ζ-CD | 4-tert-Butyl benzoate | <50c,d | 10c |
| δ-, ε-, ζ-CD | Ibuprofen | <30c,d | 10c |
| δ-, ε-, ζ-CD | Benzoate derivatives | 2–10c,e | 10c |
| δ-CD | 1-Adamanatane carboxylate | 4–8c,f | 10c |
| δ-CD | C70 | n.d.g | 10d |
| δ-CD | Cycloundecanone | n.d.h | 9 |
Recently, we have reported the complexation of large dodecaborate cluster dianions (B12X122−, X: H, Cl, Br, and I, Fig. 1) with γ-CD; the binding affinities reached micromolar in aqueous solution.13 The driving force for complexation was traced back to the chaotropic effect, based on the superchaotropic nature of the dodecaborate anions.13 Additionally, the large polarizability of the clusters contributes to the high stability of the formed inclusion complexes.13,14 The binding constant with γ-CD reaches its maximum for B12Br122−, while B12I122− already becomes too large and binds more weakly.13 We reckoned that these globular clusters, and in particular the largest ones, could serve as ideal guests for LRCDs and now present our results on the binding of dodecaborate clusters with δ-, ε-, and ζ-CD, synthesized and purified as described previously.15
Results and discussion
The key-and-lock principle in host–guest chemistry describes a complementarity between the guest size and the cavity size of the host as well as the shape of both. Table 2 lists pertinent structural parameters of common CD homologues, including LRCDs, and dodecaborate anions. The cavity size of CDs spans from 174 to 794 Å3, while the size of the clusters spans from 152 to 520 Å3. The size match between the guest and the host represents a quick estimator for the steric goodness of fit of host–guest complexation. For example, the smallest cluster, B12H122−, is too small to efficiently fill the cavity of large CDs, but matches that of the small CDs, such as α- and β-CD.13 On the other hand, the larger clusters, such as B12Br122− and B12I122−, are too large to fit inside α-CD and β-CD, but they are expected to lock better into the cavities of γ-CD up to ζ-CD.| Host | w/Å | n/Å | h/Å | V cavity/Å3 | Guest | d/Å | V/Å3 |
|---|---|---|---|---|---|---|---|
| a From ref. 1. b Linearly extrapolated values by assuming an annular CD shape, cf.Fig. 5. c From ref. 13. | |||||||
| α-CD | 8.0 | 5.0 | 9.0 | 174a | B12H122− | 8.0 | 152c |
| β-CD | 9.7 | 5.6 | 9.0 | 262a | B12F122− | 9.0 | 176 |
| γ-CD | 10.7 | 7.0 | 9.0 | 427a | B12Cl122− | 10.5 | 333c |
| δ-CD | 12.6b | 7.8b | 9.0 | 541b | B12Br122− | 11.1 | 416c |
| ε-CD | 13.9b | 8.8b | 9.0 | 667b | B12I122− | 11.7 | 520c |
| ζ-CD | 15.3b | 9.7b | 9.0 | 794b | |||
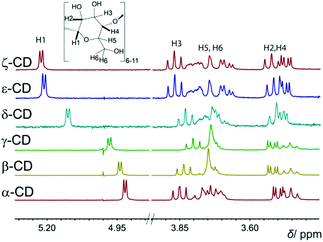
The 1H NMR chemical-shift differences among α-, β-, and γ-CD are very small, amounting, for example, to only 0.05 ppm for the H1 proton (Fig. 2). In contrast, for LRCDs, larger differences are observed (up to 0.3 ppm, see Fig. 2), as expected from their distinct, folded structures.6a,e,f
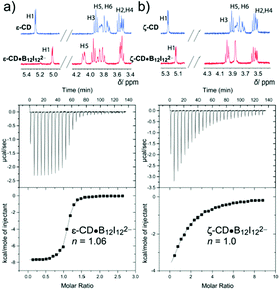
Complexation of the clusters by LRCDs was first probed by complexation-induced 1H NMR shifts. The small clusters, B12H122− and B12F122−, showed either no or heavier changes in the 1H NMR spectra. The encapsulation of the perhalogenated clusters inside the large hosts caused significant down-field shifts, selectively of the inner protons, H3 and H5 (Fig. 3 and 4); this confirmed the formation of inclusion complexes. The magnitude of the chemical shifts signals how deeply the cluster protrudes into the cavity (see 1H NMR in Fig. 3). δ-CD showed 1H NMR shifts, and, therefore, complexation with all perhalogenated clusters. For B12Cl122−, a large shift was observed for H5, which is located inside the cavity near the lower (narrower) rim, while a smaller shift was observed for H3 (0.2 versus 0.1 ppm), in line with a deep inclusion. For the largest cluster (B12I122−), the H5 proton was significantly shifted, but an even larger shift was observed for H3 (0.2 versus 0.4 ppm), indicating that this large cluster cannot protrude as deeply as the smaller perchlorinated one. ε-CD and ζ-CD showed either no or small changes in the 1H NMR upon the addition of B12Cl122− or B12Br122−; only the largest guest, B12I122−, showed sizable 1H NMR shifts even for the two largest investigated homologues (Fig. 4). In general, we concluded that when ΔδH3 > ΔδH5, a partial inclusion of the cluster inside the cavity applies, while full inclusion is signaled by ΔδH5 > ΔδH3. The 1H NMR results are in accordance with expectations from the size complementarity principle.
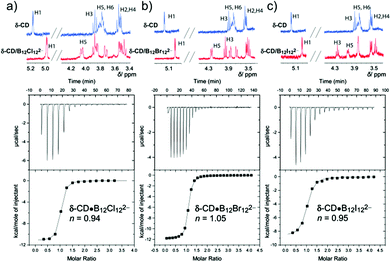 | ||
| Fig. 3 1H NMR spectra (top) and ITC data (bottom) for the complexation of δ-CD with a) B12Cl122−, (b) B12Br122−, and (c) B12I122−. | ||
 | ||
| Fig. 4 1H NMR spectra (top) and ITC data (bottom) for the complexation of B12I122− with (a) ε-CD and (b) ζ-CD. | ||
ITC was used to determine the association constants of the halogenated clusters to LRCDs. The resulting binding affinities are shown in Table 3. Very strong binding affinities to δ-CD were observed, with the highest affinities measured for B12Br122− (2.6 × 106 M−1) and B12Cl122− (2.5 × 106 M−1), followed by B12I122− (6.8 × 105 M−1). These values even exceed the values previously measured for these clusters to γ-CD, and set record benchmarks for LRCDs (compare Table 3 with Table 1). The increase in affinity from γ-CD to δ-CD occurs at the expense of a decreased selectivity, that is, the binding constants for B12Cl122−, B12Br122−, and B12I122− vary by almost a factor of 70 for γ-CD but by less than a factor of 4 for δ-CD. Presumably, the higher flexibility of δ-CD allows for a better induced fit. For example, it is likely that the smaller B12Cl122− cluster is accommodated through an elliptic distortion of the LRCD cavity, which has been experimentally observed in the crystal structure of δ-CD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6a and which has also been found in molecular dynamics simulations of LRCDs.7a,d,f The resulting clamp-like binding site (Fig. 5) offers a tighter cavity with more dispersive contact points, which accounts for the absolute (from 0.014 to 2.5 × 106 M−1) and relative (to B12Br122−) enhancement in binding of B12Cl122− with the larger macrocycle; this offsets simple size complementarity arguments, which would have led to the expectation of a reduced affinity of the smaller guest as the cavity becomes larger. These simple arguments are again sufficient to account for the variation in affinities as the LRCD series expands from δ-CD to ζ-CD. Particularly noteworthy is the fact that B12I122− becomes the strongest binder for ε-CD where also a micromolar affinity is achieved. And even for ζ-CD a sizable binding constant of 8000 M−1 is obtained.
6a and which has also been found in molecular dynamics simulations of LRCDs.7a,d,f The resulting clamp-like binding site (Fig. 5) offers a tighter cavity with more dispersive contact points, which accounts for the absolute (from 0.014 to 2.5 × 106 M−1) and relative (to B12Br122−) enhancement in binding of B12Cl122− with the larger macrocycle; this offsets simple size complementarity arguments, which would have led to the expectation of a reduced affinity of the smaller guest as the cavity becomes larger. These simple arguments are again sufficient to account for the variation in affinities as the LRCD series expands from δ-CD to ζ-CD. Particularly noteworthy is the fact that B12I122− becomes the strongest binder for ε-CD where also a micromolar affinity is achieved. And even for ζ-CD a sizable binding constant of 8000 M−1 is obtained.
Thermodynamic parameters for complexation are shown in Table 4. In general, the binding is an enthalpically driven process with an entropic penalty (ε-CD was an exception), in agreement with our previous report on the binding of same clusters with γ-CD.13 In our previous study with γ-CD, a correlation between enthalpy and guest size was observed: the enthalpy of complexation (ΔH°) increased with increasing the dianion size. This trend discontinues for the LRCDs, presumably due to the different binding modes (Fig. 5). However, large enthalpy values are accompanied by an increased entropic penalty for the LRCDs as well, that is, enthalpy–entropy compensation applies, as is common for CDs.2c,13
| Host | B12Cl122− | B12Br122− | B12I122− | |
|---|---|---|---|---|
a Measured by ITC in H2O at 25 °C and analyzed for a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexation model; errors in ΔH and TΔS are 10% or ±0.8 kcal mol−1, whichever is larger.
b Measured as sodium salts.
c From ref. 13. 1 complexation model; errors in ΔH and TΔS are 10% or ±0.8 kcal mol−1, whichever is larger.
b Measured as sodium salts.
c From ref. 13.
|
||||
| γ-CDc | ΔH° | −14.4 | −21.4 | −25.0 |
| TΔS° | −8.6 | −13.3 | −18.4 | |
| δ-CD | ΔH° | −11.2 | −11.9 | −8.7 |
| TΔS° | −2.5 | −3.1 | −0.8 | |
| ε-CD | ΔH° | −3.6 | −4.6 | −7.7 |
| TΔS° | 2.5 | 2.4 | 0.9 | |
| ζ-CD | ΔH° | −16.9 | −6.4 | −10.4 |
| TΔS° | −12.5 | −1.3 | −5.1 | |
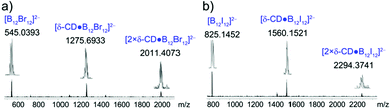
Besides the studies in aqueous solution, we have tested the stability of the formed complexes in the gas phase using mass spectrometry experiments. Fig. 6 shows the mass spectra of δ-CD with B12Br122− and B12I122−. For both clusters, doubly charged ions were observed at m/z 545 and 825, corresponding to the naked anions, B12Br122− and B12I122−, respectively. The 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes were also observed as doubly negatively charged species (δ-CD·B12Br122− at m/z 1276 and δ-CD·B12Br122− at m/z 1560). Moreover, 2
1 complexes were also observed as doubly negatively charged species (δ-CD·B12Br122− at m/z 1276 and δ-CD·B12Br122− at m/z 1560). Moreover, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host–guest complexes were observed in the gas phase, which has also been observed for γ-CD in a crystal structure.13
1 host–guest complexes were observed in the gas phase, which has also been observed for γ-CD in a crystal structure.13
Until now, even association constants on the order of 100 M−1 have been very difficult to achieve for LRCDs (Table 1). For example, the binding constants for 1-adamantane carboxylate, which presents a well-known gold standard in the CD field,16 reaches only ca. 8 M−1 for δ-CD.10c Additionally, most studies on the host–guest complexes with LRCDs have shown no defined stoichiometry.
Recently, we have utilized the B12H122− core as an innovative anchoring group, tethered to a chromophore.17 The hybrid anchor-dyes were optimized for indicator displacement assays and sensing applications.17 The high-affinity binding (106 M−1) for the perhalogenated dodecaborate clusters to LRCDs makes them excellent choices as potential anchoring groups for indicator displacement applications; in particular, it should allow for a convenient screening method to explore the affinity to guest libraries to identify additional strong binders and to advance structure–affinity relationships in a broader context. Monofunctionalized halogenated clusters have recently been synthesized,18 which paves the way in this direction.
Conclusions
In summary, we have conducted a systematic study on the host–guest complexation of LRCDs with dodecaborate clusters. A rational choice of the substituent X (H, F, Cl, Br, and I) allows for a systematic variation of the size and polarizability of the guest, while its shape remains globular (icosahedral). We have found that perhalogenated clusters act as strong binders of LRCDs, with micromolar affinities for δ-CD as well as ε-CD and millimolar affinity for ζ-CD. The size match plays a key role for the stability of these complexes, in which B12Br122− fits well inside γ-CD and δ-CD, while B12I122− binds tightly to the larger homologues. The discovery of dodecaborate anions as tight binders for LRCDs opens the door for potential applications of these unconventional hosts.19Experimental
The LRCDs were synthesized and purified as described previously.15,20 The dodecaborate clusters (Na2B12H12, Na2B12Cl12, Na2B12Br12, and Na2B12I12) were synthesized according to published procedures,21 while K2B12F12 was purchased from Sigma-Aldrich (Germany) and used without further purification. Nuclear Magnetic Resonance (NMR) spectra were recorded with a JEOL ECX 400 MHz NMR spectrometer in D2O. Isothermal titration calorimetry experiments were carried out in water (unbuffered) on a VP-ITC from Microcal, Inc., at 25 °C, pH 6.5–7. The binding equilibria were studied using a cellular host concentration of 50 μM, to which a 10–30 times more concentrated guest solution was titrated. Typically, 27 consecutive injections of 10 μL were used. All solutions were degassed prior to titration. Heats of dilution were determined by titration of the guest solution into water. The first data point was removed from the data set prior to curve fitting (Origin 7.0 software) according to a one-set-of-sites model. The knowledge of the complex stability constant (Ka) and molar reaction enthalpy (ΔH°) enabled the calculation of the standard free energy (ΔG°) and entropy changes (ΔS°) according to ΔG° = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka = ΔH° − TΔS°. Mass spectrometry experiments were performed with a Bruker Micro-TOF MS.
Ka = ΔH° − TΔS°. Mass spectrometry experiments were performed with a Bruker Micro-TOF MS.
Acknowledgements
The authors acknowledge support from the DFG within grant NA-686/8 as part of the priority program SPP 1807 “Control of London Dispersion Interactions in Molecular Chemistry” (W. M. N.).Notes and references
- J. Szejtli, Chem. Rev., 1998, 98, 1743 CrossRef CAS PubMed.
- (a) W. Saenger, Angew. Chem., Int. Ed. Engl., 1980, 19, 344 CrossRef; (b) K. A. Connors, Chem. Rev., 1997, 97, 1325 CrossRef CAS PubMed; (c) M. V. Rekharsky and Y. Inoue, Chem. Rev., 1998, 98, 1875 CrossRef CAS PubMed.
- (a) K. Koizumi, H. Sanbe, Y. Kubota, Y. Terada and T. Takaha, J. Chromatogr., A, 1999, 852, 407 CrossRef CAS; (b) K. L. Larsen, J. Inclusion Phenom. Macrocyclic Chem., 2002, 43, 1 CrossRef CAS; (c) H. Ueda and T. Endo, in Cyclodextrins and Their Complexes, Wiley-VCH, 2006, p. 370 Search PubMed; (d) F. Ellouze, N. Ben Amar and A. Deratani, C. R. Chim., 2011, 14, 967 CrossRef CAS; (e) T. Endo, Trends Glycosci. Glycotechnol., 2011, 23, 79 CrossRef CAS.
- K. Freudenberg and F. Cramer, Z. Naturforsch., 1948, 3b, 464 CAS.
- (a) A. O. Pulley and D. French, Biochem. Biophys. Res. Commun., 1961, 5, 11 CrossRef CAS PubMed; (b) D. French, A. O. Pulley, J. A. Effenber, M. A. Rougvie and M. Abdullah, Arch. Biochem. Biophys., 1965, 111, 153 CrossRef CAS PubMed.
- (a) T. Fujiwara, N. Tanaka and S. Kobayashi, Chem. Lett., 1990, 739 CrossRef CAS; (b) J. Jacob, K. Gessler, D. Hoffmann, H. Sanbe, K. Koizumi, S. M. Smith, T. Takaha and W. Saenger, Angew. Chem., Int. Ed., 1998, 37, 606 CrossRef CAS; (c) W. R. Saenger, J. Jacob, K. Gessler, T. Steiner, D. Hoffmann, H. Sanbe, K. Koizumi, S. M. Smith and T. Takaha, Chem. Rev., 1998, 98, 1787 CrossRef CAS PubMed; (d) K. Gessler, I. Uson, T. Takaha, N. Krauss, S. M. Smith, S. Okada, G. M. Sheldrick and W. Saenger, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 4246 CrossRef CAS PubMed; (e) T. Endo, H. Nagase, H. Ueda, S. Kobayashi and M. Shiro, Anal. Sci., 1999, 15, 613 CrossRef CAS; (f) K. Imamura, O. Nimz, J. Jacob, D. Myles, S. A. Mason, S. Kitamura, T. Aree and W. Saenger, Acta Crystallogr., Sect. B: Struct. Sci., 2001, 57, 833 CAS.
- (a) P. M. Ivanov and C. Jaime, J. Phys. Chem. B, 2004, 108, 6261 CrossRef CAS PubMed; (b) M. G. Gotsev and P. M. Ivanov, Int. J. Quantum Chem., 2007, 107, 1657 CrossRef CAS; (c) I. Maestre, I. Bea, P. M. Ivanov and C. Jaime, Theor. Chem. Acc., 2007, 117, 85 CrossRef CAS; (d) G. Raffaini and F. Ganazzoli, Chem. Phys., 2007, 333, 128 CrossRef CAS; (e) M. G. Gotsev and P. M. Ivanov, J. Phys. Chem. B, 2009, 113, 5752 CrossRef CAS PubMed; (f) P. M. Ivanov, J. Phys. Chem. B, 2010, 114, 2650 CrossRef CAS PubMed; (g) P. Ivanov, J. Mol. Struct., 2012, 1009, 3 CrossRef CAS; (h) P. Ivanov, E. Atanassov and C. Jaime, J. Mol. Struct., 2014, 1056, 238 CrossRef.
- (a) I. Miyazawa, H. Ueda, H. Nagase, T. Endo, S. Kobayashi and T. Nagai, Eur. J. Pharm. Sci., 1995, 3, 153 CrossRef CAS; (b) K. Tomono, A. Mugishima, T. Suzuki, H. Goto, H. Ueda, T. Nagai and J. Watanabe, J. Inclusion Phenom. Macrocyclic Chem., 2002, 44, 267 CrossRef CAS; (c) T. Furuishi, T. Fukamil, H. Nagase, T. Suzuki, T. Endo, H. Ueda and K. Tomono, Pharmazie, 2008, 63, 54 CAS; (d) S. Machida, S. Ogawa, X. H. Shi, T. Takaha, K. Fujii and K. Hayashi, FEBS Lett., 2000, 486, 131 CrossRef CAS PubMed; (e) H. Ueda, A. Wakamiya, T. Endo, H. Nagase, K. Tomono and T. Nagai, Drug Dev. Ind. Pharm., 1999, 25, 951 CrossRef CAS PubMed; (f) D. Wistuba, A. Bogdanski, K. L. Larsen and V. Schurig, Electrophoresis, 2006, 27, 4359 CrossRef CAS PubMed.
- K. Harata, H. Akasaka, T. Endo, H. Nagase and H. Ueda, Chem. Commun., 2002, 1968 RSC.
- (a) H. Akasaka, T. Endo, H. Nagase, H. Ueda and S. Kobayashi, Chem. Pharm. Bull., 2000, 48, 1986 CrossRef CAS PubMed; (b) S. Kitamura, K. Nakatani, T. Takaha and S. Okada, Macromol. Rapid Commun., 1999, 20, 612 CrossRef CAS; (c) K. L. Larsen, T. Endo, H. Ueda and W. Zimmermann, Carbohydr. Res., 1998, 309, 153 CrossRef CAS; (d) T. Furuishi, T. Endo, H. Nagase, H. Ueda and T. Nagai, Chem. Pharm. Bull., 1998, 46, 1658 CrossRef CAS; (e) H. Ueda, J. Inclusion Phenom. Macrocyclic Chem., 2002, 44, 53 CrossRef CAS.
- (a) S. M. Liu, A. D. Shukla, S. Gadde, B. D. Wagner, A. E. Kaifer and L. Isaacs, Angew. Chem., Int. Ed., 2008, 47, 2657 CrossRef CAS PubMed; (b) F. F. Li, M. Feterl, J. M. Warner, A. I. Day, F. R. Keene and J. G. Collins, Dalton Trans., 2013, 42, 8868 RSC; (c) Q. Liu, Q. Li, X. J. Cheng, Y. Y. Xi, B. Xiao, X. Xiao, Q. Tang, Y. Huang, Z. Tao, S. F. Xue, Q. J. Zhu and J. X. Zhang, Chem. Commun., 2015, 51, 9999 RSC; (d) Q. Li, S. C. Qiu, K. Chen, Y. Zhang, R. B. Wang, Y. Huang, Z. Tao, Q. J. Zhu and J. X. Liu, Chem. Commun., 2016, 52, 2589 RSC.
- (a) I. Dumazet, J. B. RegnoufdeVains and R. Lamartine, Synth. Commun., 1997, 27, 2547 CrossRef CAS; (b) D. R. Stewart and C. D. Gutsche, J. Am. Chem. Soc., 1999, 121, 4136 CrossRef CAS.
- K. I. Assaf, M. S. U. F. Pan, T. Georgiev, S. S. K. Rissanen, D. Gabel and W. M. Nau, Angew. Chem., Int. Ed., 2015, 54, 6852 CrossRef CAS PubMed.
- J. Warneke, C. Jenne, J. Bernarding, V. A. Azov and M. Plaumann, Chem. Commun., 2016, 52, 6300 RSC.
- Q. S. Qi, X. Y. She, T. Endo and W. Zimmermann, Tetrahedron, 2004, 60, 799 CrossRef CAS.
- (a) W. C. Cromwell, K. Bystrom and M. R. Eftink, J. Phys. Chem., 1985, 89, 326 CrossRef CAS; (b) J. Voskuhl, M. Waller, S. Bandaru, B. A. Tkachenko, C. Fregonese, B. Wibbeling, P. R. Schreiner and B. J. Ravoo, Org. Biomol. Chem., 2012, 10, 4524 RSC.
- K. I. Assaf, O. Suckova, N. Al Danaf, V. von Glasenapp, D. Gabel and W. M. Nau, Org. Lett., 2016, 18, 932 CrossRef CAS PubMed.
- C. Jenne and C. Kirsch, Dalton Trans., 2015, 44, 13119 RSC.
- (a) A. Bogdanski, D. Wistuba, K. L. Larsen, U. Hartnagel, A. Hirsch and V. Schurig, New J. Chem., 2010, 34, 693 RSC; (b) A. Harada, Y. Takashima and M. Nakahata, Acc. Chem. Res., 2014, 47, 2128 CrossRef CAS PubMed.
- (a) Q. S. Qi, M. N. Mokhtar and W. Zimmermann, J. Inclusion Phenom. Macrocyclic Chem., 2007, 57, 95 CrossRef CAS; (b) T. Endo, N. Ogawa, H. Nagase, H. Sambe, T. Takaha, Y. Terada, W. Zimmermann and H. Ueda, Heterocycles, 2007, 74, 991 CrossRef CAS.
- (a) V. Geis, K. Guttsche, C. Knapp, H. Scherer and R. Uzun, Dalton Trans., 2009, 2687 RSC; (b) I. Tiritiris and T. Schleid, Z. Anorg. Allg. Chem., 2004, 630, 1555 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2016 |