Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Overproduction and identification of butyrolactones SCB1–8 in the antibiotic production superhost Streptomyces M1152†
John D.
Sidda
a,
Vincent
Poon
a,
Lijiang
Song
a,
Weishan
Wang
b,
Keqian
Yang
b and
Christophe
Corre
 *a
*a
aDepartment of Chemistry and School of Life Sciences, University of Warwick, Coventry, CV4 7AL, UK. E-mail: C.Corre@warwick.ac.uk; Fax: +44 (0)2476 524 112; Tel: +44 (0)2476 523 557
bState Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, No. 1 West Beichen Road, Chaoyang District, Beijing 100101, People's Republic of China
First published on 10th May 2016
Abstract
Gamma-butyrolactones (GBLs) are signalling molecules that control antibiotic production in Streptomyces bacteria. The genetically engineered strain S. coelicolor M1152 was found to overproduce GBLs SCB1–3 as well as five novel GBLs named SCB4–8. Incorporation experiments using isotopically-labelled precursors confirmed the chemical structures of SCB1–3 and established those of SCB4–8.
Introduction
Genome mining has revealed that Streptomyces bacteria typically harbour 20–30 machineries or gene clusters dedicated to the production of bioactive natural products. Many of these compounds find use in human or veterinary medicine as well as in agriculture.1 Hundreds of gene clusters proposed to direct the biosynthesis of unknown (cryptic) antibiotic-like molecules are currently available in publicly accessible databases. The characterisation of these cryptic metabolites is often challenged by the presence of transcriptional repressor proteins that prevent the expression of biosynthetic genes when bacteria are grown in laboratory culture conditions.1Since the discovery of the γ-butyrolactone (GBL) A-factor in 1967 and subsequent characterisation of its intracellular target, the transcriptional repressor ArpA, distinct classes of antibiotic production inducers have been discovered.2,3 In particular butenolides and 2-alkyl-4-hydroxymethylfuran-3-carboxylic acids (AHFCAs) have been shown to trigger the expression of antibiotic-like biosynthetic gene clusters by interacting with specific ArpA-like transcriptional repressors.4–6
Early investigations of signalling molecules in S. coelicolor A3(2) reported at least seven GBL-like signalling molecules produced by this strain.7 The chemical structures of S. coelicolor GBLs SCB1–3 were elucidated in the 2000s and the scbA gene, an afsA-like butenolide synthase, was shown to direct their biosynthesis.8–11 SCBs act as antibiotic production inducers and directly control the expression of a cluster of genes, named cpk, responsible for the biosynthesis of the coelimycin polyketide antibiotic in S. coelicolor.12,13
The molecular mode of action of SCBs involves direct binding to the transcriptional repressor ScbR. In the absence of SCBs, ScbR binds to specific operator sequences in particular upstream of the transcriptional activator cpkO and prevents coelimycin production.14 An additional transcriptional repressor, ScbR2, is also responsible for repressing the expression of cpk genes.14 Both scbR and scbR2 genes are adjacent to the cpk gene cluster but on opposite sides (Fig. 1). Genetic inactivation of scbR2 resulted in overproduction of coelimycin and a related yellow pigment.15,16 In addition ScbR2 is known to bind to the scbA–scbR intergenic region and repress expression of scbA.16 Consequently SCB1 was found to be overproduced in the S. coelicolor M145 scbR2 mutant.16
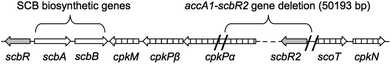 | ||
| Fig. 1 Genetic organisation of the scb/cpk gene cluster in Streptomyces coelicolor highlighting the deleted DNA fragment (∼50 kb) during the construction of the superhost S. coelicolor M1152. | ||
The S. coelicolor superhost strain M1152, derived from S. coelicolor M145, has been genetically optimised for the production of antibiotic-like natural products whose biosynthesis is directed by heterologous metabolic pathways.17 Construction of this strain involved deletion of multiple biosynthetic gene clusters, including ≈50 kb of the coelimycin gene cluster. This deletion included the gene scbR2 but not the GBL-biosynthetic genes scbA and scbB (Fig. 1).
In this study we reveal that the engineered M1152 overproduces GBLs fortuitously. We report the overproduction of known S. coelicolor GBLs SCB1–3 as well as production of five new SCBs, SCB4–8 in M1152.17 The chemical structures of SCB1–8 were determined by a combination of incorporation experiments using stable isotope-labelled precursors and mass spectrometry analyses.
Results and discussion
Identification of SCBs in Streptomyces M1152
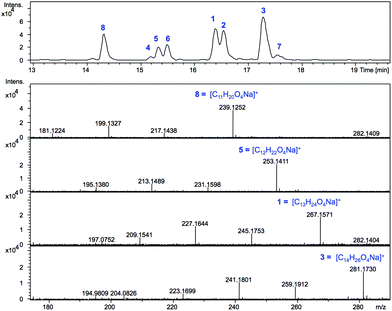
SCBs are produced at nanomolar concentration in the parent strain S. coelicolor M145: ≈320 μg of SCB1 were purified from a 300 L culture in the original study.8 In S. coelicolor M1152 we expected the scbR2 deletion to result in scbAB overexpression and in turn overproduction of SCBs. S. coelicolor M1152 was therefore grown on the minimal culture medium AlaMM and ethyl acetate extracts of the culture supernatant were submitted to liquid chromatography-mass spectrometry, LC-MS, analyses (Fig. 2).Based on the calculated m/z values for the sodium adducts of SCB1 (1) and SCB2 (2), m/z = 267.1567 [C13H24O4 + Na]+, and that of SCB3 (3) m/z = 281.1723 [C14H26O4 + Na]+, positive ion mode chromatograms were extracted for these ions. In addition, m/z values consistent with derivatives carrying shorter or longer alkyl chains (m/z = 239.1, 253.1 and 295.2) were also extracted and revealed the presence of metabolites 4–8. Each of the compounds 1–8 exhibited fragmentation patterns consistent with those previously reported for SCBs,18i.e. daughter ions corresponding to [M − H2O + H]+ and [M − 2H2O + H]+ in addition to [M + H]+ and [M + Na]+ molecular ions (Fig. 2 and S1†).
The high accuracy of the mass spectrometer used in these experiments combined with the isotopic pattern identified for each of these ions allowed masses to be generated within 5 ppm of those corresponding to the predicted molecular formulae for 1–8 (Fig. 2 and Table S1†). The molecular formulae of compounds 1 and 2 matched that of SCB1–2 (C13H24O4) while for compound 3 it was consistent with that expected for SCB3 (C14H26O4). For the previously uncharacterised metabolites, a common C12H22O4 molecular formula was determined for 4–6, while 7 and 8 exhibited formulae of C14H26O4 and C11H20O4 respectively (Table S1†). In addition to fragmentation patterns and molecular formulae, the respective retention times on reverse phase HPLC were also consistent with 1–3 corresponding to SCB1–3. The identity of SCB1 was also confirmed by HPLC purification and NMR analysis (Fig. S11 and S12†).
The structural diversity between 1–8 was proposed to directly derive from the nature of the precursor incorporated into SCBs. The enzyme ScbA is the only butenolide synthase encoded in S. coelicolor M1152 genome and has been shown to be essential for SCB1–3 biosynthesis.10 ScbA is an orthologue of AfsA that has been shown to catalyse the condensation of dihydroxyacetone phosphate (DHAP) with a β-ketothioester intermediate hijacked from fatty acid metabolism to yield a butenolide phosphate intermediate.11 This intermediate is then reduced by the butenolide phosphate reductase BprA in Streptomyces griseus to generate A-factor.11 Located downstream of scbA is scbB, which encodes for a homologue of BprA (76% identity, 84% similarity over 279 amino acids).
The SCB-like molecules 1–8 observed in this culture extract were therefore proposed to each result from the incorporation of different β-ketothioester intermediates. Specific primary metabolites, namely leucine, isoleucine, valine, propionic and butyric acids, are known to act as precursor of starter units in fatty acid biosynthesis and are converted into β-ketothioester intermediates that are subsequently incorporated into butenolides and GBLs such as SCB1 (Scheme 1).
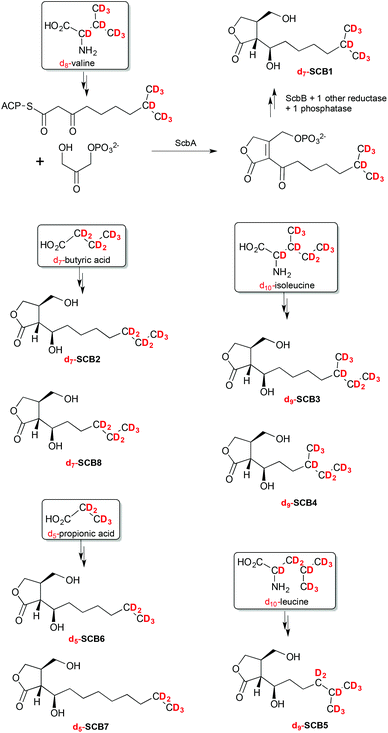 | ||
| Scheme 1 Specific incorporation of deuterated precursors into compounds 1–8 revealing the chemical structure of SCB1–8. | ||
Structural characterisation of SCB1–8 using stable-isotope labelled precursors
To unambiguously determine the nature of the alkyl side chains in each of the SCB-like compounds 1–8, S. coelicolor M1152 was grown in AlaMM media supplemented with 1 mM final concentration of deuterium-labelled d8-DL-valine, d10-L-leucine, d10-L-isoleucine, d5-propionic acid or d7-butyric acid. This method was used previously to probe incorporation of specific deuterium-labelled precursor molecules into the AHFCA signalling molecules.5,19 Metabolites were extracted from each of the supplemented cultures and analysed by LC-MS. As expected, feeding with each labelled precursor altered the mass spectra of specific SCB-like metabolites.Feeding with d8-DL-valine resulted in a proportion of compound 1 (SCB1) to specifically contain seven deuterium atoms, suggesting the intact incorporation of a β-ketothioester primed with isobutyryl-CoA. The molecular formula of 1 [M + Na]+ with m/z = 274.2 was confirmed to be C13H17D7O4Na (Fig. S5†). The m/z values of [M + H]+, [M − H2O + H]+ and [M − 2H2O + H]+ presented a similar pattern where seven deuterium atoms were incorporated (Fig. S5 and Table S2†). In this particular experiment, 1 was the only metabolite amongst 1–8 revealing an intact incorporation of the labelled precursor. This experiment confirmed that the alkyl chain of 1 terminates with an isopropyl group, which agrees with the published structure for SCB1 (Scheme 1). In a similar way the linear nature of the alkyl chain in 2, SCB2, was confirmed by detecting the intact incorporation of seven deuterium atoms from d7-butyric acid (Fig. S6†). Compound 3, revealing the incorporation of 9 deuterium atoms from d10-isoleucine, was confirmed to correspond to the previously reported SCB3 (Fig. S7†).
Feeding with d10-isoleucine also resulted in specific labelling of compound 4. The difference between SCB3 (3) and 4 rises from the fact that their β-ketothioester precursors have gone through a different number of chain extensions: 3 extensions in the case of SCB3 (3) and only 2 for compound 4. Metabolites 5 and 6 were found to specifically incorporate nine deuterium atoms from d10-leucine and five deuterium atoms from d5-propionic acid, respectively (Fig. S3 and S4†). Compound 7 was specifically labelled upon addition of d5-propionic acid and 8 specifically incorporated d7-butyric acid (Fig. S8 and S2†).
The structures of the new natural products 4–8, named SCB4–8 respectively, were therefore as indicated in Scheme 1. All the molecular formulae generated from UHR-LC-MS analyses of the labelled and unlabelled compounds 1–8 are reported in Tables S1 and S2.†
Compounds 4–8 have not been previously reported from natural sources, however 6–8 were previously synthesised and tested on the dissociation of ScbR from its DNA binding site using an in vivo kanamycin reporter assay.9 Compounds 6 and 7 were shown to be 10 times less effective than SCB1 and 8 was 480 times less effective at stimulating dissociation of ScbR.9
Conclusion
In conclusion, we have established that the superhost strain Streptomyces M1152, increasingly used as a heterologous host for the expression of bacterial gene clusters, overproduces SCB1–3 (mg L−1 scale compare to μg L−1 scale in the wild-type strain) and at least five additional SCBs. The chemical nature of SCB1–8 was determined using a combination of incorporation experiments and mass spectrometry.Overproduction of SCB antibiotic production inducers in S. coelicolor M1152 might provide this strain an advantage in triggering the expression of otherwise silent GBL-dependent metabolic pathways from heterologous origin. However, in most cases, the cost of GBL overproduction in terms of precursor supply and energy is more likely to impair the yield of metabolite of interest. In addition, we observed that 1–8 interfered significantly with the purification of novel metabolites co-eluting with SCB1–8. As a result of the present work, construction of an improved superhost strain has been undertaken. Inactivation of the biosynthetic gene scbA in the S. coelicolor M1152 background is expected to particularly improve the production of polyketide natural products as they share the same pool of precursors as those used in SCB biosynthesis.
Acknowledgements
This work was mainly supported by a University Research Fellowship (UF090255) from The Royal Society and by an EPSRC DTA studentship to JDS. The Bruker MaXis mass spectrometer used in this research was obtained with support from AWM and the ERDF. YKQ was supported by National Natural Science Foundation of China 31570031. We are very grateful to Prof. Mervyn Bibb for providing strain Streptomyces M1152.Notes and references
- P. J. Rutledge and G. L. Challis, Nat. Rev. Microbiol., 2015, 13, 509 CrossRef CAS PubMed.
- Y. Ohnishi, S. Kameyama, H. Onaka and S. Horinouchi, Mol. Microbiol., 1999, 34, 102 CrossRef CAS PubMed.
- J. M. Willey and A. A. Gaskell, Chem. Rev., 2011, 111, 174 CrossRef CAS PubMed.
- K. Arakawa, N. Tsuda, A. Taniguchi and H. Kinashi, ChemBioChem, 2012, 13, 1447 CrossRef CAS PubMed.
- C. Corre, L. Song, S. O'Rourke, K. F. Chater and G. L. Challis, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 17510 CrossRef CAS PubMed.
- S. O'Rourke, A. Wietzorrek, K. Fowler, C. Corre, G. L. Challis and K. F. Chater, Mol. Microbiol., 2009, 71, 763 CrossRef PubMed.
- M. Kawabuchi, Y. Hara, T. Nihira and Y. Yamada, FEMS Microbiol. Lett., 1997, 157, 81 CrossRef CAS.
- E. Takano, T. Nihira, Y. Hara, J. J. Jones, C. J. Gershater, Y. Yamada and M. J. Bibb, J. Biol. Chem., 2000, 275, 11010 CrossRef CAS PubMed.
- N.-H. Hsiao, et al. , Chem. Biol., 2009, 16, 951 CrossRef CAS PubMed.
- N.-H. Hsiao, et al. , Microbiol., 2007, 153, 1394 CrossRef CAS PubMed.
- J. Kato, N. Funa, H. Watanabe, Y. Ohnishi and S. Horinouchi, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2378 CrossRef CAS PubMed.
- E. Takano, et al. , Mol. Microbiol., 2005, 56, 465 CrossRef CAS PubMed.
- J.-P. Gomez-Escribano, L. Song, D. J. Fox, V. Yeo, M. J. Bibb and G. L. Challis, Chem. Sci., 2012, 3, 2716 RSC.
- M. Gottelt, S. Kol, J. P. Gomez-Escribano, M. J. Bibb and E. Takano, Microbiology, 2010, 156, 2243 CrossRef PubMed.
- J. Wang, et al. , Mol. Microbiol., 2011, 82, 236 CrossRef CAS PubMed.
- X. Li, J. Wang, S. Li, J. Ji, W. Wang and K. Yang, Sci. Rep., 2015, 5, 14831 CrossRef CAS PubMed.
- J. P. Gomez-Escribano and M. J. Bibb, Microb. Biotechnol., 2011, 4, 207 CrossRef CAS PubMed.
- H.-S. Joo, Y.-H. Yang, C.-S. Lee, J.-H. Kim and B.-G. Kim, Rapid Commun. Mass Spectrom., 2007, 21, 764 CrossRef CAS PubMed.
- C. Corre, S. W. Haynes, N. Malet, L. Song and G. L. Challis, Chem. Commun., 2010, 46, 4079 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures and spectroscopic data. See DOI: 10.1039/c6ob00840b |
| This journal is © The Royal Society of Chemistry 2016 |