 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Total chemical synthesis of lassomycin and lassomycin-amide†
S.
Lear
a,
T.
Munshi
b,
A. S.
Hudson
a,
C.
Hatton
a,
J.
Clardy
c,
J. A.
Mosely
a,
T. J.
Bull
b,
C. S.
Sit
*c and
S. L.
Cobb
*a
aDepartment of Chemistry, Durham University, South Road, Durham, DH1 3LE, UK. E-mail: s.l.cobb@durham.ac.uk
bSt. George's University of London, London, SW17 0RE, UK
cDepartment of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, 240 Longwood Ave, Boston, MA 02115, USA. E-mail: Clarissa_Sit@hms.harvard.edu
First published on 12th April 2016
Abstract
Herein we report a practical synthetic route to the lasso peptide lassomycin (1) and C-terminal variant lassomycin-amide (2). The biological evaluation of peptides 1 and 2 against Mycobacterium tuberculosis revealed that neither had any activity against this bacterium. This lack of biological activity has led us to propose that naturally occurring lassomycin may actually exhibit a standard lasso peptide threaded conformation rather than the previously reported unthreaded structure.
Introduction
The pressing issue of antimicrobial resistance continues to drive the search for novel antibiotic scaffolds.1 Emerging threats such as extensively drug-resistant tuberculosis require the use of innovative approaches in the search for potential leads.2,3 Lassomycin (1), a natural product belonging to the ‘lasso’ peptide class, was recently isolated from an Actinomycete (Lentzea kentuckyensis sp.) in a screen of a library of previously uncultured soil bacteria.4 The lasso peptide was found to possess potent and specific bactericidal activity against mycobacteria, including drug-resistant forms of Mycobacterium tuberculosis. The specificity of activity arises due to the fact that lassomycin targets the ClpC1 ATPase, an essential enzyme in mycobacteria. ClpC1 ATPase is a key component of protein degradation together with the ClpP1P2 proteolytic complex.5The previously reported solution-state NMR structure of lassomycin is shown in Fig. 1. The peptide does not adopt the characteristic knot conformation reported for other homologous lasso peptides: here the C-terminus packs tightly against the N-terminal ring instead of passing through the macrolactam.4 In light of this discovery, and the potential of lasso peptides as novel drug scaffolds in general,6–8 we set out to chemically synthesize the natural product.
 | ||
| Fig. 1 Amino acid sequence and proposed solution-state NMR structure of lassomycin (1).4,9 The N- and C-terminus are labelled. | ||
Herein, we report the first chemical synthesis of the recently identified lasso-peptide lassomycin (1) and a C-terminal amide derivative, lassomycin-amide (2). Synthetic peptides 1 and 2 were also evaluated for activity against M. tuberculosis.
Results and discussion
Two key structural features that need to be incorporated during a chemical synthesis of lassomycin (1) are the isopeptide bond formed between the N-terminus and the carboxylate side chain of Asp-8, and the methylated C-terminus (Fig. 1). We envisaged two possible synthetic approaches to access lassomycin (1) and these are outlined in Scheme 1. Route A involved the preparation of a linear peptide and the formation of the lactam ring on resin using an allyl ester orthogonal protecting group strategy for the side chain of Asp-8. The C-terminal methyl ester would be installed using 2-chlorotrityl chloride resin and the approach reported by Turner et al. (Scheme 1, Route A).10 The alternative approach (Scheme 1, Route B), involved assembling the linear peptide on resin via side chain anchoring. In this approach a di-peptide fragment would be loaded initially onto the resin allowing the C-terminal methyl ester functionality to be installed at this point. Lactam ring formation would be carried out in solution. | ||
| Scheme 1 Proposed synthesis routes to lassomycin (1): (A) on resin cyclization and (B) combined solution/on-resin approach (amino acid side chain protecting groups are omitted for clarity). | ||
Initially, Route A (Scheme 1) was investigated and the linear peptide 7 was successfully assembled via standard Fmoc solid phase peptide synthesis (SPPS). Next, we attempted the on-resin deprotection of the Asp-8 side chain. In our hands all attempts to remove the allyl ester and generate peptide 6 were unsuccessful, despite the use of stoichiometric amounts of tetrakis(triphenylphosphine)palladium(0) catalyst and triphenylsilane, and trialling a number of reaction solvents. The lack of success in allyl ester removal was assumed to arise from a difficulty in coordinating the allyl moiety to the catalyst, presumably due to steric hindrance around the metal centre. This effect will depend upon peptide sequence length and folding of the peptide, and as such is likely to be highly sequence dependent. In light of these difficulties alternative orthogonal protecting groups were considered (i.e. highly acid sensitive,11 photolabile12 or silicon-based13 protection), however the requirement for more specialized deprotection conditions, or the lack of available commercial building blocks, led us to investigate Route B (Scheme 1).
The synthesis of the required orthogonally protected di-peptide (10) for Route B was carried out in solution. Removal of the t-butyl side chain from 10 under standard acidic conditions followed by attachment to Rink amide resin gave the methyl ester protected C-terminal fragment of lassomycin (Scheme 2). The peptide chain was then extended on resin using Fmoc SPPS to give a complete linear version of lassomycin (4). Cleavage from the resin afforded the linear peptide (3) which was ready for lactam ring formation.
While peptide macrocyclizations are often best performed under high dilution to minimize unwanted oligomer formation,14 the ‘pseudo-high dilution’ conditions described by Malesevic et al. were instead utilized as a more convenient alternative in order to minimize solvent usage.15 11.3 mL each of solutions of peptide 3 (0.01 M) and HATU (0.03 M) in DMF were added to a reaction flask containing DIPEA and HOAt over a period of ∼20 h at a rate of 0.01 mL min−1 (syringe pump controlled). Slow addition was used in order to keep the effective reaction concentration low, while requiring only a relatively small final solvent volume (34 mL). Removal of the solvent under vacuum and HPLC purification of the crude reaction product yielded lassomycin (1) in high purity, as confirmed by analytical HPLC and MALDI-TOF MS (Fig. 2).
MALDI LIFT-TOF/TOF16 MS/MS was used to confirm the correct sequence for the linear tail portion of lassomycin (ESI†). However, fragmentation was not observed for the cyclic region, but this matched the data reported by Gavrish et al. from MS/MS sequencing of the isolated natural lassomycin.4 A complete set of fragment ions were observed for the uncyclized peptide.
Ion mobility (IM) mass spectrometry was also carried out on the synthetic peptide. A number of peaks with distinct collisional cross sections were resolved belonging to either lassomycin or a trace amount of the C-terminal acid hydrolysis product (13) (ESI Fig. 7†). Only a single peak was observed for each species suggesting that only one conformation is present, as opposed to distinct populations of threaded and unthreaded peptide (or that the collisional cross section for threaded/unthreaded ions is identical).
It was also possible to access lassomycin-amide (2) a C-terminal amide version of the original lassomycin (1) peptide. Synthesis of the required linear peptide (14) was carried out on Rink amide resin using Fmoc SPPS, and cleavage from the resin afforded peptide 15 (Scheme 3). Formation of the lactam ring in solution and HPLC purification to yield lassomycin-amide (2) were carried out as previously described for the synthesis of 1. Purity and structure were confirmed by analytical HPLC (ESI Fig. 2†), MALDI-TOF MS (ESI Fig. 2†) and tandem mass spectrometry (ESI Fig. 1†).
Peptides 1 and 2, as well as truncated analogue lassomycin (1–9) (16) representing the peptide without the tail were tested against Mycobacterium tuberculosis H37Rv in MIC assays utilizing both solid-agar based and liquid-media based methods. None of the peptides showed inhibitory activity when tested, up to concentrations of 100 μg mL−1. These results were surprising in particular for peptide 1, given the previously reported biological activity profile.4 Given the unlikely nature of spontaneous threading during the synthesis of 1 (and 2) it is logical to assume that the peptides reported herein are un-threaded. This, in combination with the biological data, leads us to speculate that the un-threaded structure previously reported may not be correct.
The structural discrepancy from the original report4 could possibly be due to instability of lassomycin in its threaded conformation. Several examples have been documented of lasso peptides unthreading upon heat treatment. Caulosegnin I unthreads completely after 4 hours at 95 °C, while caulosegnin III unthreads and decomposes at elevated temperatures.17 Similarly, astexin-1 unthreads when heated to 50 °C for 4 hours18 and astexin-2 unthreads and decomposes after heating at 95 °C for 2 hours.19 In contrast, caulosegnin II and astexin-3 show minimal to no unthreading when heated at 95 °C for prolonged periods of time.17,19
In order to further elucidate the effect of temperature on the conformational state of lassomycin, structural characterization of synthetic peptide 1 was carried out using two-dimensional NMR. A comparison of 1H–13C HSQC spectra for 1 at two different temperatures is shown in Fig. 3. The NMR data gathered is also compared in Fig. 3 with the naturally isolated labelled [13C,15N]lassomycin.4 No noticeable changes in chemical shift values are observed between the spectrum of 1 at 25 °C and at 40 °C. Furthermore, clear differences in the 1H–13C HSQC spectrum can be seen between the synthetic peptide (1) and the naturally isolated lassomycin at 40 °C (Fig. 3). These results strongly suggest that the conformation of 1 does not change appreciably upon heating, and they highlight that there are significant differences between the conformation of synthetically prepared lassomycin (1) and that of naturally isolated lassomycin.4
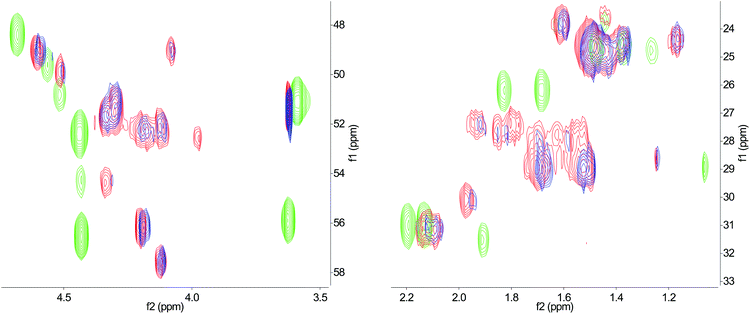 | ||
| Fig. 3 Overlay of expanded regions of 1H–13C HSQC (heteronuclear single-quantum correlation) NMR spectra of naturally isolated [13C, 15N]lassomycin at 40 °C (green), and synthetic lassomycin (1) at 25 °C (blue) and 40 °C (red). Clear differences in chemical shift values at 40 °C are observed between 1 and the natural isolate. In contrast, shift values for the synthetic peptide (1) do not change noticeably over the temperature range surveyed. (Full spectra given in the ESI.†) | ||
In light of this evidence, we propose that synthetic lassomycin (1) either exists in the unconstrained unthreaded form, or as a threaded lasso, but that there is no interconversion between these structures at temperatures up to 40 °C. Given the clear differences observed in the NMR between the synthetic and naturally isolated lassomycin, it seems likely that one peptide is threaded while the other is not. This idea of two distinct conformations being adopted by the synthetic and natural peptides is supported further by the biological data obtained where clear differences in activity are seen.
We propose that (i) neither peptide has undergone unthreading (or threading) upon heating during NMR data collection, or (ii) one peptide has undergone threading from an initial unthreaded state whilst the other has undergone unthreading. For case (ii) the peptide undergoing unthreading cannot be the naturally isolated lassomycin, as this would imply that the synthetic peptide (1) has undergone threading and that it is threaded at the point of NMR data collection, thus paradoxically implying (given the NMR study of temperature dependence already discussed) that a threaded lassomycin cannot unthread. Neither scenario therefore supports the hypothesis that the naturally isolated lassomycin has undergone unthreading upon heating to 40 °C. Furthermore, given the unlikely nature of scenario (ii), taken together with the previously discussed likelihood that synthetic lassomycin is in fact unthreaded, we propose that the previously published naturally isolated lassomycin actually exists as a threaded lasso that does not unthread upon heating to 40 °C.
Conclusions
We have developed a practical synthetic route to an unthreaded version of the lasso peptide lassomycin (1). Lassomycin-amide (2), a C-terminal amide derivative of 1 was also prepared. The biological evaluation of peptides 1 and 2 against Mycobacterium tuberculosis revealed that neither had any activity. A lack of biological activity and our NMR analysis of synthetic lassomycin (1) suggest that naturally occurring lassomycin exhibits a standard lasso peptide threaded conformation rather than the reported unthreaded structure. Further conformational studies to gain additional insight in this area are ongoing.Experimental
All reagents were purchased from Sigma-Aldrich unless otherwise specified. Peptide synthesis grade DMF was purchased from AGTC Bioproducts (Hessle, UK) and amino acid derivatives were purchased from CEM, Novabiochem (Merck) or AGTC. PyBOP® was purchased from Apollo Scientific (Stockport, UK). All resins were purchased from Novabiochem. Peptide molecular weight calculations and mass assignments were carried out using Pep-Calc.com.20Peptide synthesis
Automated SPPS was carried out at 0.10 mmol scale on a CEM Liberty1 single-channel microwave peptide synthesizer equipped with a Discover microwave unit. All reactions were carried out at room temperature in DMF using 2 × 1 h couplings for all residues except Arg, which was triple coupled (3 × 1 h). Fmoc-protected amino acids were used (5 equiv.), with PyBOP® (5 equiv.) as the activator in the presence of DIPEA (10 equiv., 2 M solution in NMP). Amino acid side chain functionality was protected as follows: Fmoc-His(Trt)-OH, Fmoc-Ser(OtBu)-OH and Fmoc-Thr(OtBu)-OH. The Fmoc group was removed by two successive treatments with 20% (v/v) piperidine solution in DMF (5 + 10 min). An additional 10 min deprotection was carried out for the penultimate residue (Leu-2). Bubbling with nitrogen gas was used to ensure efficient agitation of the reaction mixture during each step. Preswelling of dry resin was carried out in DMF for a minimum of 1 h.Cleavage from solid support
Peptide-resin was shrunk in diethyl ether and treated with 2.85 mL TFA, 0.15 mL deionized water and 0.15 mL TIPS for 4 h at room temperature. The resin was then removed by filtration and the filtrate concentrated in vacuo before precipitation using ether and decanting of the liquid (followed by subsequent ether washes). The resulting solid peptide was dissolved in deionized water containing 0.1% TFA and lyophilized.Solution-phase cyclization
The cyclization was carried out using the pseudo-high dilution conditions described by Malesevic et al.15 Fully deprotected peptide (113 μmol) and HATU (339 μmol) were dissolved separately in 11.3 mL each of DMF which had been dried over molecular sieves. Both solutions were added simultaneously using two syringe pumps at a rate of 0.01 mL min−1 each to a stirred solution of HATU (11.3 μmol) and DIPEA (678 μmol) in dry DMF (11.3 mL) over a period of ∼20 h. After the addition was complete, the reaction was stirred for a further 30 min and the solvent was removed under vaccum. The resulting oil was kept at −20 °C until needed for purification.High-performance liquid chromatography
The crude cyclization reaction product was dissolved in 2.5 mL deionized water/acetonitrile, filtered, centrifuged and injected in 0.5 mL amounts (per run) onto a Speck and Burke Analytical C18 Column (5.0 μm, 10.0 × 250 mm) attached to a PerkinElmer Series 200 LC Pump and 785A UV/Vis Detector. A linear gradient of 10–36% B (solvent A = 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95
95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1 MeCN/H2O/TFA; B = 95
0.1 MeCN/H2O/TFA; B = 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1 MeCN/H2O/TFA) over 40 min, followed by 36–100% B over 20 min, with a flow rate of 2 mL min−1 was used for separations. Absorbance data were collected at 220 nm. Selected fractions were lyophilized and a mass assigned using MALDI-TOF MS or ESI-MS, and peak fractions of interest were pooled and lyophilized.
0.1 MeCN/H2O/TFA) over 40 min, followed by 36–100% B over 20 min, with a flow rate of 2 mL min−1 was used for separations. Absorbance data were collected at 220 nm. Selected fractions were lyophilized and a mass assigned using MALDI-TOF MS or ESI-MS, and peak fractions of interest were pooled and lyophilized.
Nuclear magnetic resonance (NMR) spectroscopy
NMR spectra were collected using a Varian VNMRS 700 MHz. Multiplicities: s = singlet, d = doublet, dd = doublet of doublets, m = multiplet, t = triplet. Chemical shifts are reported in δ units and are referenced to residual solvent peaks: CHCl3 (1H 7.26 ppm, 13C 77.0 ppm) and DMSO (1H 2.50 ppm, 13C 39.5 ppm).Analytical liquid chromatography mass spectrometry (LCMS)
Analytical LCMS was carried out using an Acquity UPLC system (Waters Ltd, UK) equipped with a photodiode array detector. Samples were injected onto an Acquity UPLC BEH C18 column (1.7 μm, 2.1 × 50 mm) and a gradient of 5–95% B (solvent A = H2O, 0.1% formic acid; B = MeCN) was run over 3.8 min with a flow rate of 0.6 mL min−1. The flow of solvent from the UPLC system was introduced into the electrospray ion source of an Aquity TQD or QToF Premier mass spectrometer, and positive ions were measured.Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS)
MALDI-TOF mass data was collected using an Autoflex II ToF/ToF mass spectrometer (Bruker Daltonik GmBH) equipped with a 337 nm nitrogen laser. Peptides were dissolved in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 deionized water/MeCN for MS analysis. Sample solution (1 mg mL−1) was mixed with matrix solution (α-cyano-4-hydroxycinnamic acid, ∼50 mg mL−1) in a ratio of 1
1 deionized water/MeCN for MS analysis. Sample solution (1 mg mL−1) was mixed with matrix solution (α-cyano-4-hydroxycinnamic acid, ∼50 mg mL−1) in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, and 1 μL of the resulting solution spotted onto a metal target and placed into the MALDI ion source. MS data was processed using FlexAnalysis 2.0 (Bruker Daltonik GmBH). MALDI MS/MS was performed using LIFT technology, which enables detection of product ions that result from elevating the laser power.
9, and 1 μL of the resulting solution spotted onto a metal target and placed into the MALDI ion source. MS data was processed using FlexAnalysis 2.0 (Bruker Daltonik GmBH). MALDI MS/MS was performed using LIFT technology, which enables detection of product ions that result from elevating the laser power.
Ion-mobility (IM) mass spectrometry
Ion-mobility mass spectrometry was carried out using a Synapt G2S mass spectrometer equipped with an Acquity UPLC (Waters Ltd, UK). 1 μL of sample was injected onto a BEH C18 column (1.7 μm, 2.1 × 100 mm) and a gradient of 5–95% B (solvent A = H2O, 0.1% formic acid; B = MeCN, 0.1% formic acid) was run over 6.0 min with a flow rate of 0.4 mL min−1. A photodiode array detector provided absorbance data from 200 nm to 500 nm. The solvent flow was introduced into the electrospray ion source, and full scan MS carried out for positive ions. IM separation was performed with nitrogen as the gas. MS and photodiode array absorbance data was processed using MassLynx 4.1 (Waters Ltd, UK). IM results are represented as a plot of m/z against drift time in Bins (mobiliogram).MIC testing of compounds against mycobacteria
The MICs of the compounds were determined using standard liquid-broth and solid-agar based methods. Briefly, serial dilutions of each compound were made between 100 mg L−1 and 0.78 mg L−1 with MB7H9/OADC (BD Biosciences) media in 96-well U-bottom plates. Mycobacterium tuberculosis H37Rv was grown to log phase (OD600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) = 1.0) and 104 cells were added to all wells. Solid-agar based method was carried out as previously described21 using a 48-well plate format. Compounds (see above) were tested in triplicate by adding serial dilutions into separate wells overlaid with MB7H11/OADC (BD Biosciences) agar based media. Mycobacterium tuberculosis culture (5 μL containing 104 cells) was then inoculated into the centre of each well on top of the solidified media. Plates were sealed and incubated at 37 °C in 5% CO2 for 10 days. Each plate also contained standardised serial dilution controls of first line TB drugs isoniazid (INH [Sigma, UK]; concentration range 8, 4, 2, 1, 0.5, 0.25, 0.125, 0.0625 μg mL−1) and rifampicin (RIF [Sigma, UK]; concentration range 16, 8, 4, 2, 1, 0.5, 0.25, 0.125 μg mL−1), in addition to negative (media only) controls. The MICs were determined as the first dilution to show complete growth inhibition as determined by a total lack of colony formation within the test period. It was not possible to obtain an authentic sample of naturally isolated lassomycin to compare in this assay because the Lentzea kentuckyensis subspecies from which lassomycin was originally prepared is the property of NovoBiotic Pharmaceuticals and was unavailable.
= 1.0) and 104 cells were added to all wells. Solid-agar based method was carried out as previously described21 using a 48-well plate format. Compounds (see above) were tested in triplicate by adding serial dilutions into separate wells overlaid with MB7H11/OADC (BD Biosciences) agar based media. Mycobacterium tuberculosis culture (5 μL containing 104 cells) was then inoculated into the centre of each well on top of the solidified media. Plates were sealed and incubated at 37 °C in 5% CO2 for 10 days. Each plate also contained standardised serial dilution controls of first line TB drugs isoniazid (INH [Sigma, UK]; concentration range 8, 4, 2, 1, 0.5, 0.25, 0.125, 0.0625 μg mL−1) and rifampicin (RIF [Sigma, UK]; concentration range 16, 8, 4, 2, 1, 0.5, 0.25, 0.125 μg mL−1), in addition to negative (media only) controls. The MICs were determined as the first dilution to show complete growth inhibition as determined by a total lack of colony formation within the test period. It was not possible to obtain an authentic sample of naturally isolated lassomycin to compare in this assay because the Lentzea kentuckyensis subspecies from which lassomycin was originally prepared is the property of NovoBiotic Pharmaceuticals and was unavailable.
Compounds
Abbreviations
| AM | Aminomethyl |
| ATP | Adenosine triphosphate |
| DCM | Dichloromethane |
| DIPEA | N,N-Diisopropylethylamine |
| DMF | N,N-Dimethylformamide |
| DMSO | Dimethyl sulfoxide |
| ES | Electrospray |
| ESI | Electrospray ionization |
| Fmoc | Fluorenylmethyloxycarbonyl |
| HATU | 1-[Bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate |
| HOAt | 3H-[1,2,3]-Triazolo[4,5-b]pyridin-3-ol |
| HPLC | High-performance liquid chromatography |
| IM | Ion-mobility |
| LC | Liquid chromatography |
| LIFT | LIFT |
| MALDI | Matrix-assisted laser desorption/ionization |
| MeCN | Acetonitrile |
| MIC | Minimum inhibitory concentration |
| MS | Mass spectrometry |
| NMM | N-Methylmorpholine |
| NMR | Nuclear magnetic resonance |
| Pbf | 2,2,4,6,7-Pentamethyldihydrobenzofuran-5-sulfonyl |
| PyBOP® | (Benzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate |
| rt | Room temperature |
| SPPS | Solid-phase peptide synthesis |
| tBu | tert-Butyl |
| TFA | Trifluoroacetic acid |
| TIPS | Triisopropylsilane |
| TLC | Thin-layer chromatography |
| TOF | Time of flight |
| Trt | Triphenylmethyl |
| UPLC | Ultra-performance liquid chromatography |
| UV | Ultraviolet |
| HRMS | High-resolution mass spectrometry |
Acknowledgements
We gratefully acknowledge financial support from the EPSRC (Durham University EPSRC DTG, SL). CSS was funded by an Alberta Innovates Health Solutions Fellowship and a Banting Postdoctoral Fellowship. In addition, we thank Dr Juan A. Aguilar Malavia (Durham University) for the collection and processing of 2D NMR data.Notes and references
- M. A. Fischbach and C. T. Walsh, Science, 2009, 325, 1089 CrossRef CAS PubMed.
- S. E. Dorman and R. E. Chaisson, Nat. Med., 2007, 13, 295 CrossRef CAS PubMed.
- T. Kaeberlein, K. Lewis and S. S. Epstein, Science, 2002, 296, 1127 CrossRef CAS PubMed.
- E. Gavrish, C. S. Sit, S. Cao, O. Kandror, A. Spoering, A. Peoples, L. Ling, A. Fetterman, D. Hughes, A. Bissell, H. Torrey, T. Akopian, A. Mueller, S. Epstein, A. Goldberg, J. Clardy and K. Lewis, Chem. Biol., 2014, 21, 509 CrossRef CAS PubMed.
- T. Akopian, O. Kandror, R. M. Raju, M. Unnikrishnan, E. J. Rubin and A. L. Goldberg, EMBO J., 2012, 31, 1529 CrossRef CAS PubMed.
- M. O. Maksimov, S. J. Pan and A. J. Link, Nat. Prod. Rep., 2012, 29, 996 RSC.
- M. Zimmermann, J. D. Hegemann, X. Xie and M. A. Marahiel, Chem. Biol., 2013, 20, 558 CrossRef CAS PubMed.
- H. Nar, A. Schmid, C. Puder and O. Potterat, ChemMedChem, 2010, 5, 1689 CrossRef CAS PubMed.
- The PyMOL Molecular Graphics System, Version 1.2r1, Schrödinger, LLC Search PubMed.
- R. A. Turner, R. J. Weber and R. S. Lokey, Org. Lett., 2010, 12, 1852 CrossRef CAS PubMed.
- (a) S. M. Ocampo, F. Albericio, I. Fernández, M. Vilaseca and R. Eritja, Org. Lett., 2005, 7, 4349 CrossRef CAS PubMed; (b) M. Mergler, F. Dick, B. Sax, P. Weiler and T. Vorherr, J. Pept. Sci., 2003, 9, 36 CrossRef CAS PubMed; (c) C. Yue, J. Thierry and P. Potier, Tetrahedron Lett., 1993, 34, 323 CrossRef CAS.
- (a) A. G. Russell, M.-E. Ragoussi, R. Ramalho, C. W. Wharton, D. Carteau, D. M. Bassani and J. S. Snaith, J. Org. Chem., 2010, 75, 4648 CrossRef CAS PubMed; (b) R. S. Givens, J. F. W. Weber, P. G. Conrad, G. Orosz, S. L. Donahue and S. A. Thayer, J. Am. Chem. Soc., 2000, 122, 2687 CrossRef CAS.
- (a) B. Mothia, A. N. Appleyard, S. Wadman and A. B. Tabor, Org. Lett., 2011, 13, 4216 CrossRef CAS PubMed; (b) C. K. Marlowe, Bioorg. Med. Chem. Lett., 1993, 3, 437 CrossRef CAS.
- C. J. White and A. K. Yudin, Nat. Chem., 2011, 3, 509 CrossRef CAS PubMed.
- M. Malesevic, U. Strijowski, D. Bächle and N. Sewald, J. Biotechnol., 2004, 112, 73 CrossRef CAS PubMed.
- D. Suckau, A. Resemann, M. Schuerenberg, P. Hufnagel, J. Franzen and A. Holle, Anal. Bioanal. Chem., 2003, 376, 952 CrossRef CAS PubMed.
- J. D. Hegemann, M. Zimmermann, X. Xie and M. A. Marahiel, J. Am. Chem. Soc., 2013, 135, 210 CrossRef CAS PubMed.
- M. Zimmermann, J. D. Hegemann, X. Xie and M. A. Marahiel, Chem. Biol., 2013, 20, 558 CrossRef CAS PubMed.
- M. O. Maksimov and A. J. Link, J. Am. Chem. Soc., 2013, 135, 12038 CrossRef CAS PubMed.
- S. Lear and S. L. Cobb, J. Comput. Aided Mol. Des., 2016, 30, 271 CrossRef CAS PubMed.
- D. Evangelopoulos and S. Bhakta, Methods Mol. Biol., 2010, 642, 193 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: NMR, HPLC and MS analysis for all compounds and peptides prepared. See DOI: 10.1039/c6ob00631k |
| This journal is © The Royal Society of Chemistry 2016 |



