Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Targeted SERS nanosensors measure physicochemical gradients and free energy changes in live 3D tumor spheroids†
Lauren E.
Jamieson
a,
Victoria L.
Camus
a,
Pierre O.
Bagnaninchi
b,
Kate M.
Fisher
a,
Grant D.
Stewart
c,
William H.
Nailon
de,
Duncan B.
McLaren
f,
David J.
Harrison
g and
Colin J.
Campbell
*a
aEaStCHEM, School of Chemistry, University of Edinburgh, Edinburgh, EH9 3FJ, UK. E-mail: colin.campbell@ed.ac.uk
bMRC Centre for Regenerative Medicine, University of Edinburgh, EH16 4UU, UK
cAcademic Urology Group, University of Cambridge, Addenbrooke's Hospital, Cambridge, CB2 0QQ, UK
dEdinburgh Radiation Research Collaborative, Department of Oncology Physics, Western General Hospital, Edinburgh EH4 2U, UK
eSchool of Engineering, University of Edinburgh, Edinburgh, EH9 3JL, UK
fEdinburgh Radiation Research Collaborative, Edinburgh Cancer Centre, Western General Hospital, Edinburgh EH4 2U, UK
gSchool of Medicine, University of St Andrews, KY16 9TF, UK
First published on 6th September 2016
Abstract
Use of multicellular tumor spheroids (MTS) to investigate therapies has gained impetus because they have potential to mimic factors including zonation, hypoxia and drug-resistance. However, analysis remains difficult and often destroys 3D integrity. Here we report an optical technique using targeted nanosensors that allows in situ 3D mapping of redox potential gradients whilst retaining MTS morphology and function. The magnitude of the redox potential gradient can be quantified as a free energy difference (ΔG) and used as a measurement of MTS viability. We found that by delivering different doses of radiotherapy to MTS we could correlate loss of ΔG with increasing therapeutic dose. In addition, we found that resistance to drug therapy was indicated by an increase in ΔG. This robust and reproducible technique allows interrogation of an in vitro tumor-model's bioenergetic response to therapy, indicating its potential as a tool for therapy development.
Introduction
The microenvironment of cancer, and hence the generation and distribution of energy by cancer cells and stroma for growth and survival, is complex. Oxygen gradients are common and hypoxia is often a feature of the tumor microenvironment since the blood supply to the mass of rapidly proliferating cells in vivo is poor due to disordered and leaky vasculature that fails to deliver oxygen and nutrients uniformly. In addition, many cancers exhibit the Warburg effect,1 whereby metabolism shifts away from oxidative phosphorylation (which is optimized for ATP generation) towards glycolysis, a less energy efficient metabolism but generating more molecules supportive of rapid cell proliferation. The metabolic dysregulation associated with the Warburg effect has been identified as an “Emerging Hallmark of Cancer” and this altered metabolism may play an important role in the response of tumors to therapy.2,3Conventional 2D monolayer tissue culture techniques overlook microenvironmental effects, but 3D tissue culture techniques such as the formation of multicellular tumor spheroids (MTS) may provide a better model of tumor structure.4–8 MTS develop a radial structure with a core that may be necrotic or quiescent, surrounded by quiescent cells and an outer layer that contains proliferating cells (Fig. 1); this structure has gradients in oxygen, nutrients and pH (Fig. 1) that mimic gradients present in vivo.4,7 However, the analysis of MTS models is more difficult due to their 3D nature and often misses either temporal or spatial resolution (i.e. fast methods typically overlook 3D heterogeneity and methods with good spatial resolution are time intensive and may require fixing and sectioning).9 Here we report a method for the measurement of redox potential and pH with spatial and temporal resolution that may be used to probe MTS metabolic phenotype and response to radiation and drug treatment in live MTS.
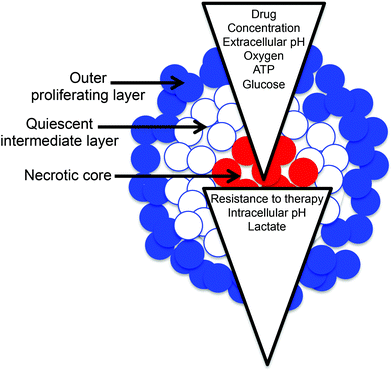 | ||
| Fig. 1 MTS grow into a spherical structure with outer, intermediate and core regions, and can support gradients in many important physiological characteristics. | ||
Cellular redox potential is a measure of the oxidizing/reducing potential of the cellular environment. It is a function of various factors such as oxygen concentration and metabolic pathway (e.g. glycolysis provides more reducing equivalents of NADPH than oxidative phosphorylation, so it may be expected that glycolysis will make a cell more reductive). Similarly pH is a function of hydrogen ion concentration [H+] and changes as a result of metabolism since lactic acid is a major product of glycolysis, but not oxidative phosphorylation (Fig. 2).
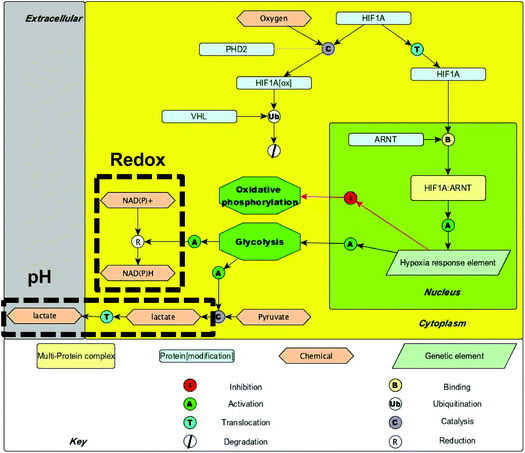 | ||
| Fig. 2 A pathway map showing cellular metabolic processes associated with hypoxia – areas likely to have a large bearing on intracellular redox potential and pH are highlighted. | ||
Currently there is very limited knowledge about the quantitative nature of the redox potential and pH gradients that develop in cancer models. An analytical technique allowing quantitative measurement of gradients could be a valuable tool for screening and monitoring responses to therapies such as radiation and drug candidates.
We have previously shown that surface enhanced Raman scattering (SERS) nanosensors can be used to quantitatively measure redox potential and pH in live cells in monolayer culture.10–16 The SERS nanosensors used are gold nanoshells (NS) coated with redox or pH responsive molecules and they are delivered to the cytosol of cells and report in a ratiometric manner.17,18 Simultaneous measurements of redox and pH are important as, for our reporters (and most cellular redox couples), redox potential is pH dependent and therefore can only be reported accurately if this is taken into account.16Eqn (1) shows the Nernst equation for a two-electron, two-proton coupled reduction where E is the redox potential of the environment, Eθ is the standard potential for the redox couple, n is the number of electrons transferred, F is Faraday's constant, [Ox] is the concentration of the oxidised form, [Red] is the concentration of the reduced form and [H+] is the concentration of protons.19
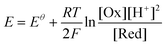 | (1) |
Here we show that simultaneous measurements using redox and pH nanosensors, can be made to calculate a redox potential and probe the response of MTS to drug treatment with both spatial and temporal resolution. This is accomplished by targeting nanosensors to distinct zones of the MTS, and validating their distribution using photothermal optical coherence tomography (PTOCT). Nanosensor targeting allows radial resolution of redox potential using a simple, robust, fiber-based Raman system that is amenable to high throughput automation.
Results
Visualizing and controlling 3D nanosensor distribution
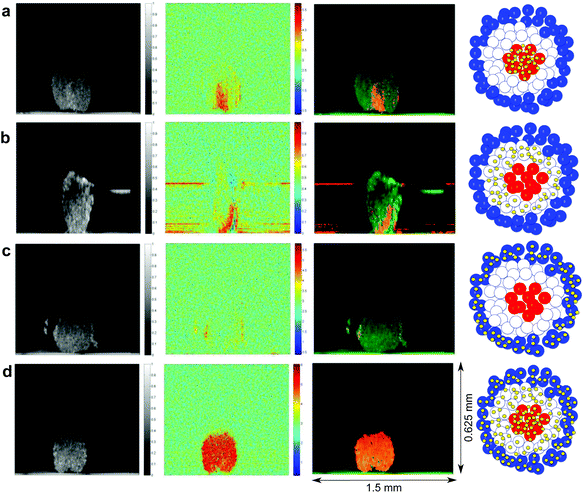
In order to measure SERS signals from nanosensors in specific regions of MTS we developed protocols for targeting nanosensors to the core, intermediate and outer zones. Transmission electron microscopy (TEM) confirmed nanosensor delivery with subcellular resolution (see ESI Fig. 1†), however, TEM fails to provide information on global distribution of NS in whole MTS. To evaluate the efficacy of our targeting, we used photothermal OCT (PTOCT)20 as it allows intact MTS to be imaged with appropriate spatial resolution to show MTS targeting and does not require sectioning or staining. For PTOCT, nanosensors in the MTS were photothermally heated by a modulated laser (785 nm), which created a modulation in the phase of the OCT signal over time. The modulated phase was then Fourier transformed to assign a color for each pixel (indicating the presence/absence of nanosensors) based on the presence or absence of phase modulation (see ESI Fig. 2 and 3†).Photothermal OCT images in Fig. 3 show the localization of nanosensors in MTS grown using the different protocols described in the Experimental section. Briefly, in order to target NS to the core of MTS, we seeded MTS with cells containing NS and then subsequently added cells in suspension that had not been treated with NS. Fig. 3a shows the highest intensity of photothermal signal in the central region of MTS indicating successful targeting. A similar technique was used to deliver NS predominantly to the outer region (proliferating layer) of the MTS and predominantly to the intermediate region (quiescent layer) – in each case MTS were seeded with cells not treated with NS. For delivery to the intermediate region a suspension of cells that had been treated overnight with NS were added on the second day of MTS growth and for delivery to the outer region on the fifth day of MTS growth. Fig. 3b shows that for the intermediate region NS are between the core and the outer region (although not uniformly distributed throughout the MTS) and Fig. 3c shows targeting to the outer zone of the MTS. In a control experiment where MTS were grown for 6 days from cell suspension containing NS, the NS are homogeneously distributed through the MTS (Fig. 3d). By confirming successful NS targeting to the outer, intermediate and core regions of MTS, we can ensure that measurements made in each case come exclusively from the region being probed. As SERS signals are exclusively obtained from NS, NS only report on redox and pH of their immediate environment and thus by confirming successful targeting we confirm that only the targeted region contributes to SERS measurements of redox and pH. As each region is targeted separately, we ensure no mutual interference of SERS spectra acquired from each region.
Determining gradients in MCF7 MTS using SERS
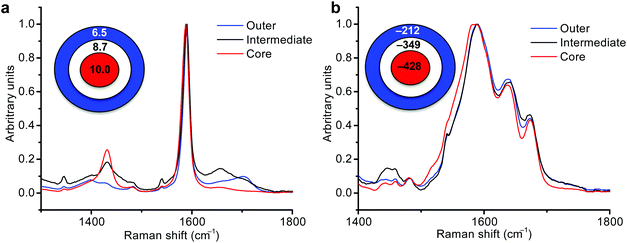
Redox potential and pH are important conduits of metabolism and their measurement in MTS should provide important information on how metabolism is regulated in distinct microenvironments. Having demonstrated that NS can be targeted to distinct zones within MTS we next sought to make measurements using pH and redox nanosensors. SERS measurements were made using a 785 nm laser via a simple fiber optic probe (see ESI Fig. 4†). Since MTS were grown under identical conditions we used the average pH from each condition to correct the average redox measurement in order to give a pH-corrected redox potential in accordance with eqn (1). Fig. 4a and b show examples of spectra from MBA-NS (pH nanosensor) and NQ-NS (redox nanosensor) respectively, measured in the core, intermediate and outer zones of MTS grown from MCF7 (breast cancer) cells.Combining data from technical and biological replicates we calculated the redox potential gradient in MCF7 MTS. Fig. 5a shows that the redox potential, became more reducing going from the outside to the intermediate and then to the core of MTS (see ESI Fig. 5† for pH measurements and ESI Tables 1 and 2† for values obtained, and ESI Fig. 6† for an example of the importance of making pH corrections to redox values). This redox potential gradient confirms the hypothesis that intracellular redox potential becomes more reductive (hypoxic) when moving towards the center of an MTS.
Monitoring MCF7 MTS gradient changes in response to apoptosis
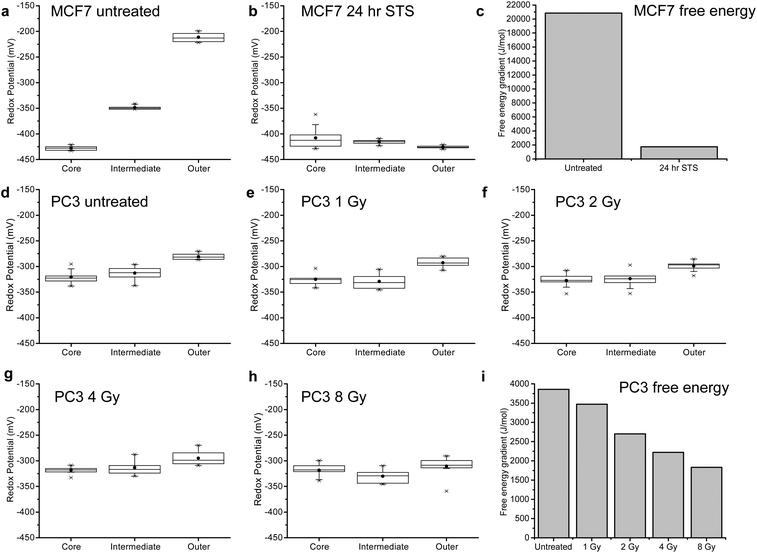
The gradients generated by MTS are an emergent property of a viable system (and should dissipate as a result of death) – in other words, it takes free energy to generate a gradient and in the absence of free energy input the system will move towards equilibrium. We therefore propose that the measured gradient can be used as a measure of MTS viability by converting the potential gradient to a free energy difference associated with viability via ΔG = −nFΔE.Having established the nature of the redox potential distribution throughout an untreated MTS, we sought to examine how the gradient changed as the MTS were treated with the drug staurosporine (STS, a potent tyrosine kinase inhibitor that induces apoptosis).21 As predicted, after 24 hours the gradient is dissipated (Fig. 5b) and the resultant free energy is lower than in the case of the untreated MTS (Fig. 5c). These findings tie in well with morphological studies of MCF7 cells treated with STS in 2D that show nuclear condensation, cell rounding and membrane blebbing (all indicative of apoptosis) (see ESI Fig. 7†). For pH measurements see ESI Fig. 5 and 6† and for details of values see ESI Tables 1 and 2.†
By taking the largest difference in redox potential between MTS regions and converting to free energy we can see that a quantitative decrease in free energy is observed upon STS treatment (Fig. 5c). This supports our hypothesis that there is a correlation between loss of the free energy required to maintain a gradient and loss of viability. ESI Fig. 8† illustrates an independent repeat of redox potential gradient response in MCF7 cells to staurosporine treatment. While the overall trend in free energy change is consistent between repeats, there are subtle changes in the gradients observed. This can be attributed to varying spheroid size between independent data sets and a further extensive study could investigate the subtle effects of spheroid size and age on free energy gradient. MCF7 spheroids had an average diameter of ca. 0.5–1 mm (see ESI Fig. 9†).
Radiation therapy is used to treat nearly 50% of patients with cancer in the UK and there is an ongoing interest in analytical methods that enable the effect of radiation on the viability of live tumor models to be monitored in order to improve treatment planning. For this reason and in order to investigate the extent to which the gradient could be used to measure relatively subtle differences in viability we treated MTS grown from a prostate cancer cell-line (PC3) with radiation of increasing dose from 0 to 8 Gy. MTS were treated with radiation in a single dose and their pH and redox potential measured after 24 hours (see ESI Fig. 10† for pH measurements and ESI Tables 3 and 4† for values obtained). When the pH corrected redox potentials were measured in these MTS (Fig. 5d–h), it is clear that the potential gradient (Fig. 5i) decreases with an increase in dose, confirming that this measure of viability can discriminate between increasing doses of radiation. The increasing effect of radiation can also be seen on the morphology of the MTS in ESI Fig. 11,† where higher doses lead to increased degradation of the MTS. The PC3 MTS images in ESI Fig. 11† also highlight the slightly smaller size of PC3 MTS compared to MCF7 MTS with an average diameter of ca. 0.4–0.5 mm. Again this could explain the difference in redox potential values observed in the untreated spheroids, but could also be a result of the differing cell lines. The technology developed in this paper would allow an extensive study into the influence of cell line, MTS size and age on pH and redox gradients in developing MTS.
Application to cisplatin resistant prostate cancer cell line
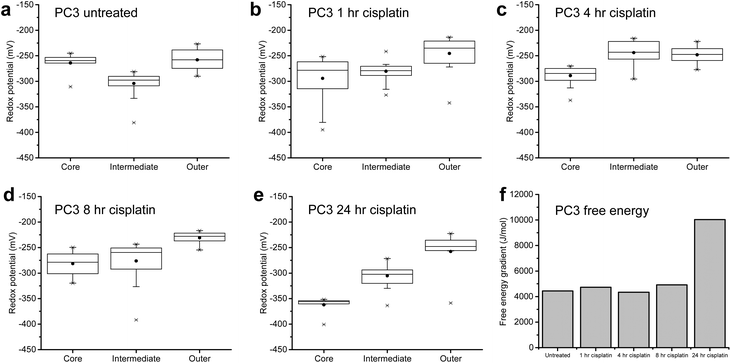
In order to investigate whether MTS respond differently when treated with a drug to which they have developed resistance, we used our technique to examine the response of MTS generated from PC3 cells treated with cisplatin at a concentration at which they are known to be resistant (50 μM).22 (For comparison, ESI Fig. 12 and 13† illustrate the pH and redox gradient changes, respectively, upon treatment of PC3 MTS with staurosporine.) Firstly, these MTS have a different structure with the most reductive part of the MTS being the intermediate layer. Again this highlights potential size variation effect on redox gradients between independent experiments. Upon treatment with cisplatin there was no loss of the initial gradient, suggestive of resistance, and there was an increase in gradient after 24 h cisplatin treatment (Fig. 6) suggestive of a compensatory anti-oxidant response by resistant cells (see ESI Fig. 14† for pH data, ESI Fig. 15 and 16† for an independent repeat experiment and ESI Tables 5 and 6† for values). Again, this finding was in line with morphological measurements of PC3 cells treated with cisplatin in 2D culture (see ESI Fig. 17†), which show the cells’ resistance to treatment. Resistance to cisplatin is known to be mediated in part by free-thiols, and while drug treatment would be expected initially to cause oxidative stress, a robust reductive response seems to have been important to re-establishing and maintaining the MTS viability. This behavior can be seen clearly in Fig. 6f where for PC3 MTS, free energy remained constant on initial treatment with cisplatin followed by an increase suggesting maintenance of MTS viability and a strongly compensating response, consistent with PC3 resistance to cisplatin.Discussion
We have demonstrated that MTS can be grown such that nanosensors are targeted to distinct zones and we confirmed this targeting using photothermal OCT. Targeting of nanosensors to MTS allowed us to use a simple fiber optic Raman probe to measure ‘average’ SERS spectra under different conditions to establish quantitative information about MTS bioenergetics. As well as providing previously unknown quantitative information on these gradients and thus providing potentially vital information for therapy development, we have demonstrated the potential for this assay to be used for monitoring response to therapy. By measuring redox potential gradients, after treatment of MCF7 MTS with the apoptosis inducer STS, we could observe a loss of redox potential gradient that correlates with a loss of MTS viability. In order to quantify MTS viability in terms of potential gradients, we expressed the gradients in terms of free energy with a loss of free energy indicative of loss of MTS viability. By treating PC3 MTS with increasing doses of therapeutic radiation we have demonstrated that the measurement of potential gradients is sensitive to subtle differences in MTS viability with MTS gradients decreasing as the dose of radiation increases. In PC3 MTS (resistant to the drug cisplatin) treatment with cisplatin showed no initial significant change in gradient, followed by an increase in gradient that we suggest is linked to drug resistance.This newly established method for fast detection of redox potential gradients in MTS uses a simple, and potentially portable, fiber optic probe, and provides a new platform for monitoring response to drugs and therapy in this 3D cell model, in a way that allows 3D integrity to be maintained in live cultures. Since the radial distribution comes from nanosensor targeting and not from complex optics, there is great potential for automation and scale-up. While this proof of concept paper includes experimental steps that require transfer of the intact MTS onto a gold coated chip for measurement, we believe that this step could be eliminated in an optimized high-throughput platform. For example, hanging drop plates, which allow MTS to be formed in a 384 drop array,23 could be used in a Raman plate-reader setup where Raman measurements would be taken directly from the MTS growing in the plate since the SERS enhancement is strong enough for spectra to be acquired above potential background contributions from glass or plastic substrates. In this setup it is even possible to take measurements from partially and fully dissociated MTS, which could result from particular drug treatments. As the MTS has been structurally destroyed in this case, it is likely that the gradient would be diminished and, while care must be taken in interpretation if dissociation occurred, an absolute loss of gradient could still be used as an efficacy indicator.
Increasing movement is being made towards the use of 3D cell culture models for drug screening due to their increased physiological relevance, however current techniques for probing response of MTS to drug candidates are limited by z penetration depths in imaging techniques, meaning that MTS often have to be partially digested and analyzed by flow cytometry, or require extensive processing of fixed and sectioned samples.4,9 Therefore this novel SERS method for quick quantitative assessment of the structure and viability of intact live MTS could prove to be a vital tool in monitoring response to drugs and therapy in a 3D culture model and lead to an improved phenotypic assay for drug candidate selection.
Methods
Nanoshell functionalisation
1,8-Diaza-4,5-dithian-1,8-di(2-chloro-[1,4]-naphthoquinone-3-yl)octane (NQ) was synthesized according to the previously reported synthetic route.12 MethylNQ (MeNQ) was synthesized in our group according to the previously reported synthetic route (reported as compound 12 by Thomson et al.14). NS were functionalized with 100 μM NQ (10% EtOH in water), 100 μM MBA solution (10% EtOH in water) or 100 μM MeNQ (1% DMSO in water) overnight. Functionalized NS were washed 2–3 times with water.Nanosensor calibration MBA
Calibration and analysis of spectra was carried out in accordance with Jamieson et al.16 Briefly, ca. 20 μl of poly-L-lysine (PLL) solution (0.1 mg ml−1) was deposited on the surface of a gold ‘chip’ (cut glass slide coated with a thin layer of gold) attached to a larger glass slide. After ca. 2 h the PLL solution was removed and ca. 20 μl of MBA-NS were added to the same area and left to dry. A series of buffers were prepared with pH values between 0 and 14. By attaching a well around the gold ‘chip’, buffers were added to the ‘chip’ in turn and multiple spectra acquired by focusing the laser spot from the fibre optic probe on the area of NS and acquiring each spectrum for 30 s. Intensity of the pH sensitive peaks at ca. 1400 cm−1 and ca. 1700 cm−1 and the peak at ca. 1590 cm−1 were used to form calibration plots from which pH values could be determined for unknown spectra.Nanosensor calibration NQ
SERS spectroelectrochemistry and analysis of spectra was carried out as described by Auchinvole et al.12 Briefly, gold microfabricated electrodes were prepared with a deposition of NS and self assembled monolayer of NQ. The functionalized electrode was connected to a potentiostat (along with a reference and auxiliary electrode) and added to a Petri dish containing phosphate buffer. The potentiostat was used to set redox potential values, and multiple SERS spectra were acquired for each potential value with a 10 s acquisition time. Intensity of the redox sensitive peak at ca. 1635 cm−1 and the peak at ca. 1580 cm−1 were used to form calibration plots from which redox potential values could be determined for unknown spectra.Nanosensor calibration MeNQ
SERS spectroelectrochemistry was carried out as described by Thomson et al.14 Analysis of spectra was performed in MATLAB to subtract a 16 point baseline followed by extraction of peak area from 1500 cm−1 to 1612 cm−1 (peak) and area from 397 cm−1 and 1750 cm−1 (total) and plotting (total area − peak area)/(total area) vs. redox potential.Cell culture and nanoshell delivery to cells
MCF7 human breast cancer cells were cultured in Dulbecco's Modified Eagle's Medium and PC3 prostate cancer cells were cultured in RPMI 1640 media, in both cases supplemented with penicillin/streptomycin (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 units per ml) and 10% heat-inactivated Fetal Calf Serum (FCS). Cells were incubated at 37 °C and 5% CO2 in a humidified incubator.
000 units per ml) and 10% heat-inactivated Fetal Calf Serum (FCS). Cells were incubated at 37 °C and 5% CO2 in a humidified incubator.
For MTS growth a hanging drop technique was used. 20 μl drops of cell suspension were pipetted onto the lid of a petri dish and approximately 10 ml of media was added to the dish. The lid containing hanging drops was placed on the petri dish and MTS grew over a period of 6 days with media added every other day.
For delivery of SERS nanosensors to different regions of MTS different protocols were followed. In all cases, media for cells growing in monolayer culture was exchanged for serum free media for 1–2 h. For homogenous distribution of nanosensor through MTS, cells in monolayer culture were then incubated overnight with MBA-NS, NQ-NS or MeNQ-NS at a concentration of ca. 170 fM before formation of hanging drops the following day. For nanosensor delivery to central regions of MTS the same protocol was followed, however on the second day of MTS growth, 5 μl of media was removed and 10 μl of cell suspension not treated with nanosensors was added. For nanosensor delivery to intermediate and outer regions of MTS, MTS were grown from cells initially not treated with nanosensors. For delivery to intermediate region, 5 μl of cell suspension from cells incubated overnight with MBA-NS, NQ-NS or MeNQ-NS was added on the second day of growth. For delivery to the outer region, 5 μl of cell suspension from cells incubated overnight with MBA-NS, NQ-NS or MeNQ- was added on the fifth day of growth. Spectra were collected on the sixth day of growth.
Cellular SERS measurements
On the sixth day of growth, MTS in media were pipetted onto a gold-coated glass slide. (For PTOCT measurements, MTS were subsequently fixed in 0.5% glutaraldehyde in PBS or 4% paraformaldehyde.) For SERS measurements, the laser from the fibre optic probe was focused on the MTS and a total of 15 SERS spectra were collected under each condition from at least three independent MTS. An acquisition time of between 10 s and 30 s was used for each spectral acquisition. Analysis of spectra was performed using Origin 9 and MATLAB. Baseline subtraction was performed followed by extraction of maximum peak intensity or peak area in the regions of interest used for calibration experiments. These values were used along with calibration data to determine corresponding pH or redox potential. Redox potential measurements were adjusted dependent on pH according to a variation in half cell potential of probe molecules by ca. 60 mV per pH unit as previously reported by Thomson et al.14Staurosporine treatment
Media was removed from hanging drop and replaced with media containing STS at a concentration of 10 μM. MTS were analyzed as previously 24 h after STS treatment.Radiation treatment
MTS were irradiated in a Faxitron Cabinet X-ray System operated at 180 kV. MTS were exposed to 1 Gy, 2 Gy, 4 Gy and 8 Gy of radiation and analyzed as previously 24 h after irradiation.Cisplatin treatment
Media was removed from hanging drop and replaced with media containing cisplatin at a concentration of 50 μM. MTS were analyzed as previously 1, 4, 8 and 24 h after cisplatin treatment.SERS instrumentation and measurements
SERS spectra were recorded with an Ocean Optics QE65 Pro spectrometer with fibre optic probe and 785 nm laser with 350 mW power and Spectra Suite software. Spectra were acquired over a 10–30 s acquisition time. SERS spectra were processed using WIRE™ 2.0, Origin 9 and MATLAB software. Aqueous suspensions of NS (2.2 × 109 particles per ml, 3.65 pM) were purchased from Nanospectra Biosciences. NS had a 150 nm diameter with a 125 nm silica core and 25 nm gold shell and were resonant at 782 nm.Photothermal OCT
The system used in this study was similar to the system used by Pai et al.20 The custom system is built around a commercial OCT engine (Callisto, Thorlabs) which consist of a spectrometer and a superluminescent laser diode (SLD) light source centred at λo = 930 nm with a FWHM bandwidth of 90 nm, providing a resolution of 5 μm in tissue.The light beam was then collimated out of a single mode fibre onto a pair of galvanometers (Thorlabs), to form a custom laser scanning head (LSH) that raster the beam into an inverted microscope (SPi95, Brunel). Two f = 50 mm double achromats in a telecentric configuration were used as the imaging lens. As opposed to conventional optical coherence tomography the system was in a common path configuration where the first reflection from the sample was used as a reference. The common path configuration resulted in increased phase stability rejecting common noise more efficiently. Control and acquisition algorithms were written in labview and matlab. Spectra were collected at a rate of 1200 spectra per second. In depth intensity and phase information along the depth axis, Z, were retrieved by fast Fourier transform of the spectra after resampling in the k-space.
In order to introduce the excitation infrared laser (785 nm), we made use of the illumination path of the inverted microscope. The excitation laser was modulated by an optical shutter operated at 330 mHz frequency, heating the gold nanocages at their absorption peak on and off. Thus producing a phase modulated at the excitation laser modulation frequency due to the local variation in temperature induced by the plasmonics photothermal effect.
The photothermal effect was measured by recording successive B scans (N = 64) at the same location with an acquisition frequency of 3 Hz. A typical photothermal in depth spheroid cross-section dataset consisted of 64 time-lapse images (400 × 512 pixels (x,z)). Considering the small size of the spheroids, images were cropped down to 256 pixel in z to speed up image processing. The time varying phase in each pixel was then Fourier transformed and filtered in the spectral domain with a bandpass filter corresponding to the excitation laser frequency. The amplitude was plotted as a heat map showing the distribution of the gold nanoparticles in the spheroids. In addition, the mean intensity over successive B scan was calculated to produce spheroids images. Finally, an overlay was built by combining the two using the thresholded mean intensity picture as a mask for the photothermal map.
Author contributions
All authors reviewed the manuscript. LEJ wrote the manuscript text, ESI and prepared figures and carried out all experiments on MCF7 cells. VLC carried out all experiments on PC3 cells. POB carried out PTOCT measurements. KMF contributed to data analysis. GDS, WHN and DBM helped conceive ideas. DJH helped conceive ideas and contributed to manuscript text. CJC contributed to the manuscript text and helped conceive ideas.Acknowledgements
CJC is grateful to the Leverhulme trust for support (Project Grant RPG-2012-680). VLC is grateful to the Jamie King Cancer Research Fund for funding. The authors gratefully acknowledge the EaSTCHEM School of Chemistry at the University of Edinburgh, Peter Mullen at the University of St Andrews and Carol Ward at the University of Edinburgh for cell culture training and Fraser Robinson for MATLAB script contribution.Notes and references
- R. A. Cairns, Cancer J., 2015, 21, 56–61 CrossRef CAS PubMed.
- M. Archetti, Cell Proliferation, 2015, 48, 259–269 CrossRef CAS PubMed.
- J. A. Menendez, J. Joven, S. Cufí, B. Corominas-Faja, C. Oliveras-Ferraros, E. Cuyàs, B. Martin-Castillo, E. López-Bonet, T. Alarcón and A. Vazquez-Martin, Cell Cycle, 2013, 12, 1166–1179 CrossRef CAS PubMed.
- L. E. Jamieson, D. J. Harrison and C. J. Campbell, Analyst, 2015, 140, 3910–3920 RSC.
- I. Vega-Naredo, R. Loureiro, K. A. Mesquita, I. A. Barbosa, L. C. Tavares, A. F. Branco, J. R. Erickson, J. Holy, E. L. Perkins, R. A. Carvalho and P. J. Oliveira, Cell Death Differ., 2014, 21, 1560–1574 CrossRef CAS PubMed.
- J. A. Menendez and T. Alarcon, Front. Oncol., 2014, 4, 1–21 Search PubMed.
- R.-Z. Lin, R.-Z. Lin and H.-Y. Chang, Biotechnol. J., 2008, 3, 1172–1184 CrossRef CAS PubMed.
- L.-B. Weiswald, D. Bellet and V. Dangles-Marie, Neoplasia, 2015, 17, 1–15 CrossRef PubMed.
- C. R. Thoma, M. Zimmermann, I. Agarkova, J. M. Kelm and W. Krek, Adv. Drug Delivery Rev., 2014, 69–70, 29–41 CrossRef CAS PubMed.
- V. Mallikarjun, D. J. Clarke and C. J. Campbell, Free Radicals Biol. Med., 2012, 53, 280–288 CrossRef CAS PubMed.
- M. A. Ochsenkuhn, P. R. Jess, H. Stoquert, K. Dholakia and C. J. Campbell, ACS Nano, 2009, 3, 3613–3621 CrossRef CAS PubMed.
- C. A. Auchinvole, P. Richardson, C. McGuinnes, V. Mallikarjun, K. Donaldson, H. McNab and C. J. Campbell, ACS Nano, 2012, 6, 888–896 CrossRef CAS PubMed.
- J. Jiang, C. Auchinvole, K. Fisher and C. J. Campbell, Nanoscale, 2014, 6, 12104–12110 RSC.
- P. I. T. Thomson, V. L. Camus, Y. Hu and C. J. Campbell, Anal. Chem., 2015, 87, 4719–4725 CrossRef CAS PubMed.
- A. Jaworska, L. E. Jamieson, K. Malek, C. J. Campbell, J. Choo, S. Chlopicki and M. Baranska, Analyst, 2015, 140, 2321–2329 RSC.
- L. E. Jamieson, A. Jaworska, J. Jiang, M. Baranska, D. J. Harrison and C. J. Campbell, Analyst, 2015, 140, 2330–2335 RSC.
- S. W. Bishnoi, C. J. Rozell, C. S. Levin, M. K. Gheith, B. R. Johnson, D. H. Johnson and N. J. Halas, Nano Lett., 2006, 6, 1687–1692 CrossRef CAS PubMed.
- J. Kneipp, H. Kneipp, B. Wittig and K. Kneipp, Nano Lett., 2007, 7, 2819–2823 CrossRef CAS PubMed.
- F. Q. Schafer and G. R. Buettner, Free Radicals Biol. Med., 2001, 30, 1191–1212 CrossRef CAS PubMed.
- J.-H. Pai, T. Liu, H.-Y. Hsu, A. B. Wedding, B. Thierry and P. O. Bagnaninchi, RSC Adv., 2014, 4, 27067–27073 RSC.
- L. Y. Xue, S. M. Chiu and N. L. Oleinick, Exp. Cell Res., 2003, 283, 135–145 CrossRef CAS PubMed.
- J. Gumulec, J. Balvan, M. Sztalmachova, M. Raudenska, V. Dvorakova, L. Knopfova, H. Polanska, K. Hudcova, B. Ruttkay-Nedecky, P. Babula, V. Adam, R. Kizek, M. Stiborova and M. Masarik, Int. J. Oncol., 2013, 44, 923–933 Search PubMed.
- Y.-C. Tung, A. Y. Hsiao, S. G. Allen, Y. Torisawa, M. Ho and S. Takayama, Analyst, 2011, 136, 473–478 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: TEMs confirming intracellular nanosensor delivery. Methodology for OCT and photothermal OCT. Experimental set-up for SERS experiments. Redox and pH data for all experiments. Optical images of cells and spheroids after treatments. See DOI: 10.1039/c6nr06031e |
| This journal is © The Royal Society of Chemistry 2016 |