Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
High-content analysis of factors affecting gold nanoparticle uptake by neuronal and microglial cells in culture
A.
Stojiljković
a,
K.
Kuehni-Boghenbor
a,
V.
Gaschen
a,
G.
Schüpbach
b,
M.
Mevissen
c,
C.
Kinnear
d,
A.-M.
Möller
c and
M. H.
Stoffel
 *a
*a
aDivision of Veterinary Anatomy, Vetsuisse Faculty, University of Bern, Bern, Switzerland. E-mail: michael.stoffel@vetsuisse.unibe.ch
bVeterinary Public Health Institute, Vetsuisse Faculty, University of Bern, Bern, Switzerland
cDivision of Veterinary Pharmacology and Toxicology, Vetsuisse Faculty, University of Bern, Bern, Switzerland
dAdolphe Merkle Institute, University of Fribourg, Switzerland
First published on 2nd September 2016
Abstract
Owing to their ubiquitous distribution, expected beneficial effects and suspected adverse effects, nanoparticles are viewed as a double-edged sword, necessitating a better understanding of their interactions with tissues and organisms. Thus, the goals of the present study were to develop and present a method to generate quantitative data on nanoparticle entry into cells in culture and to exemplarily demonstrate the usefulness of this approach by analyzing the impact of size, charge and various proteinaceous coatings on particle internalization. N9 microglial cells and both undifferentiated and differentiated SH-SY5Y neuroblastoma cells were exposed to customized gold nanoparticles. After silver enhancement, the particles were visualized by epipolarization microscopy and analysed by high-content analysis. The value of this approach was substantiated by assessing the impact of various parameters on nanoparticle uptake. Uptake was higher in microglial cells than in neuronal cells. Only microglial cells showed a distinct size preference, preferring particles with a diameter of 80 nm. Positive surface charge had the greatest impact on particle uptake. Coating with bovine serum albumin, fetuin or protein G significantly increased particle internalization in microglial cells but not in neuronal cells. Coating with wheat germ agglutinin increased particle uptake in both N9 and differentiated SH-SY5Y cells but not in undifferentiated SH-SY5Y cells. Furthermore, internalization was shown to be an active process and indicators of caspase-dependent apoptosis revealed that gold nanoparticles did not have any cytotoxic effects. The present study thus demonstrates the suitability of gold nanoparticles and high-content analysis for assessing numerous variables in a stringently quantitative and statistically significant manner. Furthermore, the results presented herein showcase the feasibility of specifically targeting nanoparticles to distinct cell types.
Introduction
Nanoparticle is a general term referring to any particle smaller than 100 nm, irrespective of the origin or composition of the material. Their size alone endows nanoparticles with specific physical properties that are beneficial for many different applications in medicine,1 material science, food technology2 and cosmetics.3 However, the health risks stemming from the high cell-penetrating capacity of nanoparticles, which is a result of their size, are poorly understood; nanoparticles from air pollutants,4 food additives, cosmetics or textiles5 interact with various organ systems in largely unknown ways. Therefore, the values and risks of nanoparticles are subjects of intense debate and controversy, emphasizing the need for a better understanding of the interactions of nanoparticles with tissues and organisms.Many different techniques, including dark field microscopy,6 standard fluorescence and confocal laser scanning microscopy, electron microscopy and inductively coupled plasma-mass spectroscopy (ICP-MS), have been used to address this issue in cell culture and laboratory rodent models.7 However, studying particle–tissue interactions in both qualitative and quantitative terms has remained challenging for numerous reasons.8 First, the size gap between nanoparticles and the investigated biological systems hinders the tracking of nanoparticles in the comparatively large environment. Additionally, labelling nanoparticles with fluorescent or other detectable tags inevitably affects the biological properties of the nanoparticles.9 Finally, quantification of the signal intensity has remained a continuous problem in most systems. A method providing statistically significant data on nanoparticle uptake by living cells would represent a major step towards gaining a better understanding of nanoparticle interactions and fate.
Gold nanoparticles (AuNPs) are a versatile tool and hold great potential in nanomedicine.10 Single AuNPs can be detected at the light microscopic level by taking advantage of silver enhancement in combination with epipolarization. AuNPs are easily synthesized in various sizes and shapes, and they can be coated with a broad spectrum of molecules. AuNPs have unique physical properties, such as surface plasmon resonance.11 Due to these advantageous features, AuNPs are already being used for imaging; cell targeting; drug, vaccine and gene delivery; biosensing; and photothermal cancer therapy.12 As the uptake of AuNPs depends not only on their properties, such as size,13 shape14 and surface coating,15 but also on the cell type and differentiation status, the intricate interactions between AuNPs and cells are not yet fully understood.
The present study was conducted to showcase the potential of AuNPs in combination with high-content analysis as a versatile and meaningful tool to investigate the interactions between nanoparticles and cells in culture. Silver enhanced AuNPs were visualized by epipolarization, and a high-content analysis system was used to collect and quantitatively analyse the data. This approach was used to exemplarily provide statistically sound data regarding the uptake of customized AuNPs by two different cell lines. Murine N9 cells were selected as microglial and thus phagocytic cells, whereas human SH-SY5Y neuroblastoma cells were chosen as non-phagocytic neural cells. AuNPs were modified in terms of size, surface charge and protein coating to track the impact of these factors on their cellular internalization. As expected, uptake by phagocytic cells turned out to be more effective than that by non-phagocytic cells. Moreover, a particle diameter between 40 and 80 nm, a positive surface charge, and a wheat germ agglutinin (WGA) coating were shown to substantially enhance internalization by both cell lines. Based on the level of caspase-dependent apoptosis, cytotoxic effects of AuNPs were dismissed. The use of customized AuNPs in combination with epipolarization and high-content analysis thus proved its worth as a powerful tool to investigate the fate and effect of nanoparticles in cell culture.
Material and methods
Synthesis and characterization of AuNPs
AuNPs with diameters of 15, 40 or 80 nm were synthetized according to Frens.16 Briefly, 5 mg of gold(III) chloride trihydrate (HAuCl4·3H2O) was diluted in 50 ml of ultrapure water and heated to boiling. Thereafter, appropriate amounts of a 1% (w/w) trisodium citrate dihydrate (Na3C6H5O7·2H2O) solution (Table 1) were rapidly added while stirring. The colloidal gold solutions were then boiled for another 5 minutes to complete the reaction and allowed to cool to room temperature.| Abbr. | Size and coating | Origin and customization settings | Inhibitory sugar | |
|---|---|---|---|---|
| Size | 15 nm | 17 nm, bare | Custom-made, 1 ml TriNa-citrate | |
| 40 nm | 39 nm, bare | Custom-made, 0.3 ml TriNa-citrate | ||
| 80 nm | 87 nm, bare | Custom-made, 0.2 ml TriNa-citrate | ||
| Charge | Anionic | 15 nm, highly anionic BSA-c™ | Aurion (Wageningen, NL) | |
| Cationic | 15 nm, poly-L-lysine | BBI Solutions (Cardiff, UK) | ||
| Protein coating | BSA | 15 nm, bovine serum albumin | Aurion (Wageningen, NL) | |
| Fet | 15 nm, fetuin | EY Labs (San Mateo, CA, USA) | ||
| Prot G | 20 nm, protein G | Sigma-Aldrich (St Louis, MO, USA) | ||
| Lectin coating | ConA | 10 nm, concanavalin A lectin | BBI Solutions (Cardiff, UK) | 200 mM α-methylmannoside |
| GSL | 17 nm, Griffonia simplicifolia isolectin B4 | Custom-made, conjugation pH 6.5, lectin amount 0.07 mg ml−1 | 200 mM galactose and 50 mM N-acetyl-galactosamine | |
| WGA | 17 nm, wheat germ agglutinin | Custom-made, conjugation pH 9.5, lectin amount 1 mg ml−1 | 500 mM N-acetyl-galactosamine |
Prior to conjugating the 15 nm AuNPs with lectins (as shown in Table 1), the pH of the colloidal gold solution was adjusted to stabilize the AuNPs against flocculation, as determined in preliminary experiments.17–20 Lectins were dissolved in ultrapure water, microfiltered (0.22 μm), and added to the buffered gold solution. The lectin–gold conjugates were stabilized by adding 5% v/v of 1% polyethylene glycol (PEG) 20 kDa.18 The solutions were slowly stirred for 30 minutes. A colour change to blue indicated flocculation. Solutions that remained pink were considered to be stable and were centrifuged at 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 1 hour at 4 °C (high-speed centrifuge HiCen®21C, rotor A24.24 at 20
000g for 1 hour at 4 °C (high-speed centrifuge HiCen®21C, rotor A24.24 at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, Herolab GmBH, Wiesloch, DE) to remove unbound lectin. The pellet was resuspended in 1% PEG. All of the colloidal gold solutions were stored at 4 °C until use.
000 rpm, Herolab GmBH, Wiesloch, DE) to remove unbound lectin. The pellet was resuspended in 1% PEG. All of the colloidal gold solutions were stored at 4 °C until use.
The size and shape of the AuNPs were assessed by transmission electron microscopy (TEM). Carbon-coated 200-mesh copper grids were floated on a drop of native colloidal gold solution for 5 minutes, allowed to dry for 30 minutes at room temperature and examined using a Philips CM12 transmission electron microscope (FEI, Eindhoven, NL). Micrographs were captured at a magnification of 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× with a MegaView III camera (Olympus, Münster, Germany) using iTEM software (version 5.2; Olympus Soft Imaging Solutions GmbH, Münster, Germany). For each custom-made solution, the projected area and the major and minor axes of 250 particles were determined using the open-source platform Fiji.21 The median of the major to minor axis ratio was calculated, and the projected area was used to compute the mean diameter of a coextensive sphere.
000× with a MegaView III camera (Olympus, Münster, Germany) using iTEM software (version 5.2; Olympus Soft Imaging Solutions GmbH, Münster, Germany). For each custom-made solution, the projected area and the major and minor axes of 250 particles were determined using the open-source platform Fiji.21 The median of the major to minor axis ratio was calculated, and the projected area was used to compute the mean diameter of a coextensive sphere.
The concentration of the custom-made AuNPs was established by absorption spectroscopy using a Jasco spectrophotometer (Jasco V-670 UV-VIS-NIR spectrophotometer, Jasco Inc., Easton, MD, USA) according to Haiss et al.22 The same equipment was used to determine the stability of the synthetized particles by comparing the spectra of the native solution, a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution in water, a 1
1 dilution in water, a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution in PBS, and a 1
1 dilution in PBS, and a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution in PBS 1 hour later. Additionally, commercially available, explicitly monodispersed BSA-conjugated and anionic AuNPs were used as standards to validate the utilized methodology, assuming that the commercial AuNPs remain monodispersed in the presence of ions, as is the case when PBS is used as a diluent. The measured parameters included the absorbance at 450 nm (A450), the position of the surface plasma resonance peak (λspr) and the absorbance at the surface plasma resonance peak (Aspr).
1 dilution in PBS 1 hour later. Additionally, commercially available, explicitly monodispersed BSA-conjugated and anionic AuNPs were used as standards to validate the utilized methodology, assuming that the commercial AuNPs remain monodispersed in the presence of ions, as is the case when PBS is used as a diluent. The measured parameters included the absorbance at 450 nm (A450), the position of the surface plasma resonance peak (λspr) and the absorbance at the surface plasma resonance peak (Aspr).
Dynamic light scattering (DLS) with a NanoBrook 90Plus PALS (Brookhaven Instruments Corporation, Holtsville, NY, USA) was used to corroborate the particle size and to substantiate the lectin conjugation of AuNPs by detecting the change in the hydrodynamic radius. Concomitantly, the NanoBrook instrument was used to measure the zeta potential (z-potential) of all of the synthetized nanoparticles. All of the measurements were repeated five times, and the mean values of the hydrodynamic diameter and the polydispersity index (PI) were determined.
The pH values of the solutions were determined using pH test-strips (Sigma-Aldrich, St Louis, MO, USA).
To verify the specific binding of lectin-conjugated AuNPs, paraffin sections of the bovine gut (2 μm thickness) with known lectin binding sites were used as positive controls. The sections were deparaffinized and rehydrated and then incubated with the corresponding lectin-conjugated AuNPs for 1 hour, followed by silver enhancement for 15 minutes (Silver Enhancement Kit, BBI Solutions, Cardiff, UK). The sections were examined under epipolarization illumination with a Zeiss AxioImager Z1 equipped with a digital high-resolution AxioCam MRm (Carl Zeiss Vision, Munich, DE) using the reflector module Pol ACR P&C for HBO 100. To generate negative controls, lectins were pre-incubated with the corresponding inhibitory sugars (Table 1) for 1 hour prior to incubating the sections.
Cell culture
The cell lines used to monitor the uptake of AuNPs included murine N9 microglial cells (N9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 23) and both undifferentiated (SHu) and differentiated (SHd) human SH-SY5Y neuroblastoma (SH) cells (ATCC, Manassas, VA, USA). N9 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% foetal bovine serum (FBS) and 1% penicillin–streptomycin. SH cells were cultured in GlutaMAX™ DMEM supplemented with 10% FBS, 1% penicillin–streptomycin, 4 mM L-glutamine, and 1 mM sodium pyruvate. All of the products for cell culture were purchased from Gibco® (Life Technologies, Carlsbad, CA, USA). Unless otherwise stated, the cells were incubated at 37 °C with 5% CO2. Only cells up to passage 20 were used for the uptake experiments. To obtain an adequate density, 1 × 104 N9 cells per well were seeded into 96-well μClear™ plates (Greiner Bio-One, Kremsmünster, Austria) and incubated for 24 hours prior to exposure to AuNPs. Similarly, 3 × 104 SHu cells per well were cultured for 48 hours before exposure. For the studies with SHd cells, 1 × 104 SHu cells per well were seeded in the plates, and differentiation was achieved by supplementing the standard medium with 10 μM retinoic acid (RA) (Sigma-Aldrich, St Louis, MO, USA).24,25 After 48 hours, the medium was replaced with fresh RA-supplemented medium, and the cells were incubated for another 48 hours prior to starting the experiments. To confirm successful differentiation, an immunofluorescence experiment was performed using a primary chicken anti-microtubule associated protein 2 (MAP2) antibody (Table 2), and the percentage of FITC-conjugated goat anti-chicken (FITC-conj. gt α-ck)-positive cells was assessed. Three different conditions were compared: SHu and SH cells treated with RA for 2 or 4 days.
23) and both undifferentiated (SHu) and differentiated (SHd) human SH-SY5Y neuroblastoma (SH) cells (ATCC, Manassas, VA, USA). N9 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% foetal bovine serum (FBS) and 1% penicillin–streptomycin. SH cells were cultured in GlutaMAX™ DMEM supplemented with 10% FBS, 1% penicillin–streptomycin, 4 mM L-glutamine, and 1 mM sodium pyruvate. All of the products for cell culture were purchased from Gibco® (Life Technologies, Carlsbad, CA, USA). Unless otherwise stated, the cells were incubated at 37 °C with 5% CO2. Only cells up to passage 20 were used for the uptake experiments. To obtain an adequate density, 1 × 104 N9 cells per well were seeded into 96-well μClear™ plates (Greiner Bio-One, Kremsmünster, Austria) and incubated for 24 hours prior to exposure to AuNPs. Similarly, 3 × 104 SHu cells per well were cultured for 48 hours before exposure. For the studies with SHd cells, 1 × 104 SHu cells per well were seeded in the plates, and differentiation was achieved by supplementing the standard medium with 10 μM retinoic acid (RA) (Sigma-Aldrich, St Louis, MO, USA).24,25 After 48 hours, the medium was replaced with fresh RA-supplemented medium, and the cells were incubated for another 48 hours prior to starting the experiments. To confirm successful differentiation, an immunofluorescence experiment was performed using a primary chicken anti-microtubule associated protein 2 (MAP2) antibody (Table 2), and the percentage of FITC-conjugated goat anti-chicken (FITC-conj. gt α-ck)-positive cells was assessed. Three different conditions were compared: SHu and SH cells treated with RA for 2 or 4 days.
| Uptake studies | Apoptosis | Differentiation | ||||
|---|---|---|---|---|---|---|
| Cell line | N9 | SHu/SHd | N9 | SHu | SHu/SHd | |
| a Merck Millipore, Billerica, MA, USA. b Sigma-Aldrich, St Louis, MO, USA. c Bio-Rad Laboratories Inc., Hercules, CA, USA. d Milli-Q EMD Millipore, Billerica, MA, USA. e Jackson ImmunoResearch Laboratories Inc., West Grove, PA, USA. f Abgent, San Diego, CA, USA. g Abcam, Cambridge, UK. h Life Technologies, Carlsbad, CA, USA. i BBI Solutions, Cardiff, UK. | ||||||
| Fixation 20′ | 2.5% GAa + 1 μg ml−1 DAPIb | + | + | − | + | + |
| 1% PFAa | − | − | + | − | − | |
| Wash 2× | PBSa | + | + | + | + | + |
| Permeabilization | 0.02% Triton X-100c, 5′ | + | + | + | + | + |
| Wash 2× | PBS | − | − | + | + | + |
| Ultrapure waterd | + | + | − | − | − | |
| Blocking | 3 mg ml−1 normal gt serume, 30′ | − | − | + | + | + |
| Primary antibody, 60′ | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 CASP3 400 CASP3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) f f |
− | − | + | + | − |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 MAP2g 1000 MAP2g |
− | − | − | − | + | |
| Wash 2× | PBS | − | − | + | + | + |
| Secondary antibody, 30′ | 7.5 μg ml−1 AF®647-conj. gt α-rbe | − | − | + | + | − |
| 2.5 μg ml−1 FITC-conj. gt α-ckg | − | − | − | − | + | |
| Wash 2× | PBS | − | − | + | + | + |
| Nuclei and cell contour staining, 15′ | 1 μg ml−1 DAPI + 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 TRITC-phah 200 TRITC-phah |
− | − | + | − | − |
| Wash 2× | PBS | − | − | + | − | − |
| Silver enhancementi | 7′ for 15 nm, 5′ for 40 nm, 3′ for 80 nm | + | + | − | − | − |
| Wash 3× | Ultrapure water | + | + | − | − | − |
| All of the described procedures were performed at room temperature. The cells were imaged immediately after the last step. | ||||||
Incubation with AuNPs
After removing the culture medium, 50 μl of HEPES-buffered serum-free DMEM (SF-DMEM) (Sigma-Aldrich, St Louis, MO, USA) containing 1 × 109 AuNPs per ml was added, and the cells were incubated for 1 hour at 37 °C. After incubation, the cells were fixed with glutaraldehyde (GA), permeabilized and the AuNPs underwent silver enhancement as summarized in Table 2. Controls included omitting the silver enhancement step and performing silver enhancement of control wells that had not been incubated with AuNPs. For the low-temperature uptake studies (AuNP uptake at 4 °C), all of the parameters were identical as for AuNP uptake at 37 °C except for the temperature. Prior to incubation with AuNPs, the cells were cooled to 4 °C for 10 minutes to reach the desired temperature. Each AuNP uptake experiment was run at least in triplicate, with 5 wells and 30 randomly selected frames per well for each type of AuNP (for more details, see Table 4).Apoptosis
To assess the potential cytotoxic effects of the different AuNPs, plates were prepared as for the uptake studies described above. However, an apoptosis analysis was performed after two different AuNP exposure times, i.e., 1 or 6 hours. Thereafter, immunofluorescence experiments were performed to quantify the percentage of cells that had undergone caspase-dependent apoptosis by staining with a polyclonal rabbit anti-cleaved caspase 3 (CASP3) antibody. Negative controls in the absence of AuNPs were incubated with either regular culture medium or SF-DMEM. Positive controls were incubated with 100 nM staurosporine for 1 or 6 hours.26–28 N9 cells were fixed with 1% paraformaldehyde (PFA) and stained with an Alexa Fluor®647-conjugated goat anti-rabbit secondary antibody (AF®647-conj. gt α-rb). Cell nuclei and cell boundaries were highlighted with DAPI and rhodamine-phalloidin (TRITC-pha), respectively. Apoptosis was assessed in three independent experiments, in which 60 frames were taken from 5 wells for each type of AuNP. The same experiments were performed on GA-fixed SHu cells. Please see Table 2 for additional details.Epifluorescence and epipolarization microscopy
Multiwell plates were imaged for high-content screening in epi-illumination mode with an INCell Analyzer 2000 (INCA; General Electric Healthcare Europe GmbH, Glattbrugg, CH). Imaging was performed with a Nikon 60× 0.70NA objective. The following filter sets were used: 350_50× & 455_50m (DAPI), 490_20× & 525_36m (FITC), 543_22× & 605_64m (GA-autofluorescence and TRITC), and 645_30× & 705_72m (Alexa Fluor® 647). As for the samples fixed with GA, we took advantage of autofluorescence to reveal the cell contours. Silver-enhanced AuNPs were detected with two crossed polarizing filters (Zeiss, Jena, DE). All of the images were captured with the laser autofocus function and a focus offset of 1.5 μm above the bottom of the well. This ensured that the recorded signals originated from an optical plane lying within the cytoplasm, thus excluding AuNPs that might have adhered to the apical plasmalemma. The adequacy of the chosen offset was corroborated by analysing cell height with an Axio Imager Z1 (Carl Zeiss Vision Swiss AG, Feldbach, CH). Samples were imaged in epifluorescence and epipolarization modes using an ApoTome, and 3D images and profiles were generated from Z-stacks. For this purpose, N9 cells were seeded in 12-well plates (TPP, Trasadingen, CH) containing a cover slip. The cells were grown to a final density of 1 × 105 cells per cm2 and then incubated with cationic AuNPs for 1 hour under similar conditions to those reported above in the “Incubation with AuNPs” section. The cells were fixed with 1% PFA and stained with DAPI and TRITC-pha, as previously described. Cover slips were then mounted with Dako Fluorescence Mounting Medium (Dako Denmark A/S, Glostrup, DK).Image analysis
The high-content analysis was completed with INCell Investigator 1.6.2 software (General Electric Healthcare Europe GmbH, Glattbrugg, CH). The segmentation of nuclei, cells and nanoparticles was always performed using the object segmentation function provided by the software. Overlapping nuclei were separated by a two-step nuclear segmentation. Briefly, the first nuclear segmentation was based on the watershed nuclear separation function; the second segmentation relied on the whole nuclear signal and used the first segmentation as a reference for the clump-breaking function. This allowed the vast majority of clumped nuclei to be separated. Nuclei touching an image border were excluded from the analysis because the corresponding cytoplasm was considered to be incompletely imaged. The signal from a given cell cluster was assigned to individual cells using the clump-breaking function, which in turn relied on the second nuclear segmentation. For the assessment of different nuclear and cellular parameters, only cell signals overlapping with the nuclear signal were analysed (Fig. 1). For the AuNPs, only objects showing at least 90% overlap with the cell signal were included in the mean count per cell per frame.For both the differentiation and apoptosis assessments, the same nuclear and cellular segmentations described above were used. For these experiments, the uptake of AuNPs was not recorded. For the MAP2 and CASP3 signals, object segmentation was also used. Cells were considered positive when these signals showed at least 70% overlap with the cellular signal. This value was chosen because the clump-breaking function does not always precisely match the true cell contour, thus reducing the signal overlap to less than 100%.
Intracellular AuNP localization
The internalization and subcellular localization of AuNPs were assessed by TEM. N9 cells were cultured in 12-well plates, as before, and were then exposed to cationized AuNPs. Prior to fixation, the cells were washed twice with PBS and then fixed with 1.5% GA in PBS, pH 7.4, for 30 minutes. The cells were washed once with PBS and three times with 0.1 M cacodylate buffer, pH 7.4. They were then processed for TEM according to standard protocols.29 Sections were examined with the Philips CM12 TEM as reported above, at magnifications ranging from 4400× to 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×.
000×.
Statistics
Statistical analysis was performed with the software NCSS 9 (NCSS, LLC. Kaysville, Utah, USA). The AuNPs were grouped in four different categories (bare; charged; coated with BSA, fetuin or prot G; and coated with lectin), and the number of AuNPs was evaluated for the H0 Hypothesis, which states that “all the medians are equal within the group”, by Kruskal–Wallis one-way ANOVA. Differences with p-values <0.05 were considered significant. Because the H0 Hypothesis was never confirmed, further analyses were performed to identify AuNPs that differed significantly from the others within the same group using a Kruskal–Wallis multiple comparison test (Dunn's Test) with the Bonferroni correction.Results
Nanoparticle characterization
The results from the size measurements of custom-made AuNPs using three different methods are reported in Table 3. This compilation also includes the shape variation (Fig. 2) as corroborated by TEM, the z-potential and the pH values of the native solutions. Except for the largest AuNPs, the shape was always spherical. The AuNPs with an approximate diameter of 80 nm were considered oval, as the median value of the major to minor axis ratio was significantly larger than that for the other custom-made AuNPs.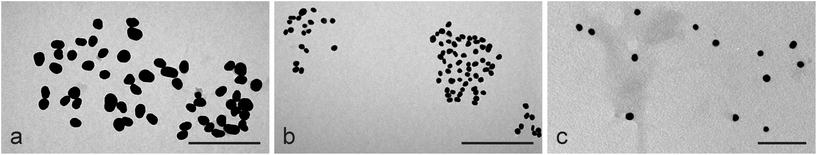 | ||
| Fig. 2 AuNPs in TEM images. (a) Oval-shaped 80 nm AuNPs. (b) Spherical 40 nm AuNPs. (c) Spherical 15 nm AuNPs. Scale bars are 500 nm in (a) and (b) and 100 nm in (c). | ||
| AuNP | TEM | DLS | UV-VIS-spectroscopy | pH | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MA | ma | MA/ma | Ø (nm) | MHD Ø (nm) | PI | ζ (mV) | CS Ø (nm) | CC (NP ml−1) | ||||
| Q1 | Q2 | Q3 | ||||||||||
| a GSL and WGA AuNPs were measured by TEM after conjugation, but no change in TEM Ø was observed. | ||||||||||||
| 80 nm | 88.3 ± 13.7 | 63.2 ± 8.7 | 1.25 | 1.35 | 1.51 | 77.4 ± 8.9 | 35.9 ± 0.6 | 0.326 ± 0.02 | −37.12 ± 7.57 | 72.85 | 11.6 × 109 | 5.5 |
| 40 nm | 42.5 ± 6.1 | 34.9 ± 4.1 | 1.11 | 1.19 | 1.30 | 38.5 ± 4.5 | 37.4 ± 1.0 | 0.255 ± 0.05 | −32.78 ± 6.59 | 50.65 | 37.8 × 109 | 5.5 |
| 15 nm | 16.5 ± 1.9 | 14.2 ± 1.8 | 1.10 | 1.15 | 1.22 | 15.3 ± 1.8 | 24.7 ± 1.4 | 0.212 ± 0.03 | −37.46 ± 8.87 | 14.87 | 1.14 × 1012 | 7.5 |
| GSL | 131.1 ± 7.8 | 0.224 ± 0.04 | −12.12 ± 6.28 | 51.82 | 5.7 × 1012 | 7.0 | ||||||
| WGA | 271.1 ± 50.2 | 0.285 ± 0.11 | −9.49 ± 3.20 | 31.88 | 1.11 × 1014 | 7.0 | ||||||
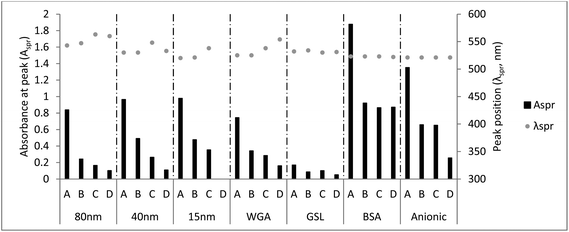
The values for Aspr and λspr, the two parameters used to assess the stability of the customized AuNPs, are shown in Fig. 3. The stability of the commercially available AuNPs was corroborated by spectroscopy, as the Aspr values remained constant irrespective of the diluent. The custom-made GSL-coated AuNPs were similarly stable. For the other custom-made particles, the Aspr decreased when PBS was used instead of water. A prolonged time in PBS led to a further decrease in the Aspr. When diluted in PBS, all of the customized AuNPs, except the GSL-coated AuNPs, had a higher λspr, and visually, the solution turned blueish, both suggesting aggregation.
On tissue sections, both GSL- and WGA-conjugated AuNPs exhibited specific staining patterns, which were abolished after pre-incubation with inhibitory sugars.
Cell differentiation
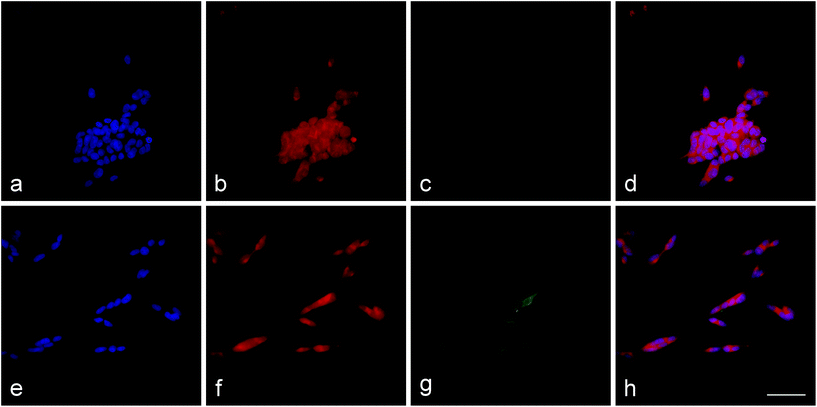
After four days of RA exposure, the cells changed from cell clusters to individual and homogeneously distributed cells with elongated neurites (Fig. 4). As determined by high-content analysis, the proportion of MAP2-positive cells increased from 2% prior to RA treatment to 10% after 2 days of treatment and to 19% after 4 days of treatment.Uptake
The uptake of AuNPs as a function of their respective properties is summarized in Table 4. Because the number of frames remained constant, whereas the proliferation rates differed between the three cell types, the total number of cells analysed per condition was not uniform.Only N9 showed a distinct size preference, preferring AuNPs with a diameter of 80 nm.
When the diameter was constant (15 nm), surface charge (bare, cationic or anionic) had the highest impact on the uptake of AuNPs. The uptake of cationic AuNPs by N9 cells (Fig. 5a) increased by 25-fold compared with that of bare 15 nm AuNPs. The uptake of cationic AuNPs was increased by 17-fold in SHu cells and by 32-fold in SHd cells (Fig. 5b) compared with that of bare AuNPs. A negative surface charge increased the uptake of AuNPs by N9 cells, but no significant effect was observed in either SHu or SHd cells.
Coating 15 nm AuNPs with either BSA, fetuin or prot G increased particle uptake by N9 cells by at least 3-fold compared with that of bare 15 nm AuNPs. The uptake of AuNPs by SH cells was not significantly affected by the tested coatings. The overall trend, however, was similar, with the positive effect of prot G always being stronger than that of BSA or fetuin.
For the three lectin-coated AuNPs, only WGA resulted in a marked increase in particle uptake in both N9 and SHd cells compared with native 15 nm AuNPs (Fig. 5c and d). For both cell types, WGA showed the strongest effect in the coating experiments. Only SHu cells did not show a preferential uptake of WGA-coated AuNPs.
A conspicuous difference in uptake efficiency was noted between the N9 cells and both types of SH cells. The microglial N9 cells always showed a higher (2- to 17-fold) uptake compared with the SH cells (Table 5). The uptake of AuNPs by SHu cells was almost always slightly higher than that by SHd cells, except for the cationic AuNPs and WGA-coated AuNPs, for which the uptake was higher by SHd cells.
| Ratio | 15 nm | 40 nm | 80 nm | Cationic | Anionic | BSA | Fet | Prot G | ConA | GSL | WGA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| N9/SHu | 2.3 | 5.3 | 8.4 | 3.2 | 16.7 | 8.0 | 8.9 | 7.6 | 2.1 | 2.0 | 11.3 |
| SHu/SHd | 1.6 | 1.2 | 1.1 | 0.9 | 1.5 | 1.5 | 1.5 | 1.5 | 1.9 | 1.8 | 0.9 |
Cell loss during fixation was substantially higher after the experiments conducted at 4 °C than after those at 37 °C. Therefore, the number of analysed cells incubated at 4 °C was significantly lower but was still adequate to allow statistical analysis; approximately 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 N9 cells, 800 Shu cells and 1600 SHd cells were analysed. Particle uptake was significantly lower at 4 °C than at 37 °C for all the tested AuNPs (Fig. 6).
000 N9 cells, 800 Shu cells and 1600 SHd cells were analysed. Particle uptake was significantly lower at 4 °C than at 37 °C for all the tested AuNPs (Fig. 6).
Apoptosis
The percentage of cleaved CASP3-positive cells varied between 0 and 5% of all of the cells counted. No significant difference was observed between the negative controls (without exposure to AuNPs) and the cells exposed to different AuNPs. This result was identical for both exposure times (1 or 6 hours). Control cells exposed to 100 nM staurosporine showed a marked increase in CASP3 positivity, with a maximum of 70% positive N9 cells after 6 hours and 55% positive SHu cells after only 1 hour.Intracellular AuNP localization
TEM was used to corroborate the internalization and the intracellular localization of AuNPs. N9 cells were analysed after the uptake of cationic 15 nm AuNPs.Cationic AuNPs were always localized to vesicles and were never observed unbound within the cytoplasm. Vesicles contained either single AuNPs or small AuNP clusters with up to 10 particles (Fig. 7a). AuNPs were also localized to the cell membrane. AuNPs were never observed in the nucleus. These results were corroborated by z-stack fluorescence images using the ApoTome. In these images, the AuNPs were never localized to the nucleus, and the AuNPs close to the nucleus were always surrounded by a red halo, indicating that they were close to the cytoskeleton, which was stained with phalloidin (Fig. 7b).
Discussion
The present study relied on the optical detection of customized unlabelled and labelled AuNPs by epipolarization microscopy. The visualization system was combined with a high-content analysis system to monitor and quantify the uptake of nanoparticles by neural and microglial cells in culture. This novel approach provides an elegant and valuable means to investigate the effect of various parameters, such as size, surface charge and proteinaceous coatings, on the uptake of nanoparticles by cells in culture in a fully quantitative and statistically significant manner.AuNPs are too small to be visualized immediately by light microscopy.12 However, this issue can be solved by silver enhancement,30,31 which magnifies discrete AuNPs of any size to enable their detection with an epipolarization system.32,33 Silver enhancement of colloidal gold is a well-established technique which has been used extensively in immunoelectron microscopy, and it has been shown that every single gold nanoparticle serves as a nucleation site for the deposition of metallic silver.34–40 An appropriate duration of silver enhancement, yielding the smallest detectable particle size of 200–250 nm, was determined in preliminary experiments. As TEM revealed that most intracellular AuNPs were clearly separated (data not shown), signal confluence due to silver enhancement was not considered to be an issue. This was corroborated by the fact that AuNPs as detected by epipolarization microscopy were evenly distributed and of equal size (Fig. 7b) thus precluding the occurrence of substantial aggregates. Silver enhancement thus provided a method for detecting and counting single AuNPs with a crossed polarizing filter set integrated into the high-content analysis system. In addition to providing numerical data, this method is highly specific, as the background after silver enhancement without prior exposure to AuNPs was less than 1.5 AuNPs per N9 cell provided that PBS was omitted after silver enhancement. Although low, this background was always subtracted from the experimental particle counts. Silver enhancement requires washing steps with double distilled water, which could potentially induce cell damage. To prevent osmotic swelling, glutaraldehyde was used as a fixative.41 The accompanying autofluorescence served as a means to allow cell segmentation without the need of an additional cytoplasmic staining step. Unfortunately, silver enhancement can only be performed on fixed samples, thus precluding live cell imaging. However, this limitation may be circumvented by fixing the cells after various incubation times and by conducting pulse-chase experiments.
The detection of AuNPs was restricted to an optical section of 1.5 μm near the bottom of the well to avoid including particles that adhered to the upper cell membrane. The assumption that only intracellular particles were evaluated was clearly validated by the decreased particle uptake at 4 °C. Obviously, these data do not yield total particle counts per cell but allow relative changes to be detected under standardized conditions. Thus, they provide a means to single out the impact of different variables on nanoparticle uptake by using the plane at 1.5 μm from the bottom as a point of reference. Regarding data interpretation, high-content analysis provides a method to track and validate each figure to the very spot in the micrograph from where it originated and to perform retrospective data analyses of additional parameters, such as nuclear morphology, cell distribution, neurite length and branching patterns. Another concern involved the possible toxic effects of nanoparticles. However, the analysis of cleaved caspase-3, an effector caspase involved in the early stages of caspase-dependent apoptosis,42–44 did not indicate any cytotoxicity after six hours of incubation, thus providing evidence that AuNPs are not detrimental to the cell types used in this study. This finding is in accordance with the pertinent literature.7,13,45–48 General limitations related to in vitro techniques also apply to the cell culture system used in this study. Notwithstanding, the approach presented herein provides a powerful and rapid tool for generating extensive data for statistical analysis, indicating the suitability of the method for both the rapid screening and retroactive analysis of numerous variables.
To ascertain that counts were restricted to the intracellular compartment, only AuNPs located within 1.5 μm of the bottom of the well were recorded. Since our approach might have been limited by a nonhomogeneous intracellular distribution of the AuNPs,6 the z-stack analysis was performed to exclude this possibility.
Characterization of AuNPs
But for the 80 nm AuNPs, the custom-made AuNPs were monodispersed and of the intended diameter. The low PI provided evidence of a highly homogenous size distribution thus precluding the presence of aggregates. Size was corroborated by TEM, DLS and UV-VIS-spectroscopy;22 all these methods yielded fully congruent measurements for all the AuNPs, except for the DLS measurements of the 80 nm particles. Repeated measurements with subsequent computation as based on the intensity-weighted Brookhaven algorithm consistently yielded an “effective diameter” near 40 nm, a result which initially eluded any plausible explanation. In hindsight, a close look at the raw data, i.e. the original intensity measurements (data not shown), revealed two particle populations, a minor one with an average diameter of about 6 nm and a major one with an average diameter close to 80 nm. According to the guidelines of the National Institute of Standards and Technology (NIST),49 an aqueous solution is considered monodisperse when the polydispersity index (PI) does not exceed 0.1. Under practical conditions, a value between 0.1 and 0.4 is interpreted as reflecting a moderate polydispersity.50 The bimodal size distribution of the 80 nm AuNP solution resulted in the observed PI of 0.326. The occurrence of two distinct particle subpopulations is consistent with the established fact that lowering the relative amount of citrate in order to produce particles larger than 40 nm entails a bimodal size distribution.51 The growth of AuNPs starts with nanoclusters (≈5 nm in size)52 and a relative lack of citrate – which is a prerequisite for the production of larger particles – does not allow all the particles to reach the full size. In accordance with the recommendations of the NIST,53 diameter was computed as a mean intensity-weighted size. DLS measurements, however, are very susceptible to polydispersity and, thus, only provide meaningful values with monodisperse solutions. The presence of two distinct particle subpopulations thus explains the erratic calculation of the mean “effective diameter” of the 80 nm AuNPs from the DLS measurements as being less than 40 nm. On the other hand, the raw data also revealed that the diameter of the majority (65%) of the particles was indeed ranging between 60 and 80 nm, i.e. fully congruent with the other methods used. Likewise, close examination of all the raw data disclosed a low percentage of similar small particles in the 40 nm AuNP solution but their concentration was too low to affect the computed mean size as based on the DLS measurements.Close scrutiny of the electron micrographs of the 80 nm AuNPs disclosed a low amount of small AuNPs in particular areas. Several reasons may account for the fact that the small particles initially escaped detection. At the magnification used to measure the larger particles, the 6 nm particles passed unnoticed as their cross-sectional surface is about a 175 times smaller than that of the 80 nm particles. Additionally, contrast of small particles is considerably lower and their distribution was inhomogeneous likely as a result of partial segregation. With respect to assessing particle uptake in culture, however, we do not expect a bias due to the presence of a minor proportion of small AuNPs in the 80 nm solution. Rather, keeping the silver enhancement to a minimum does preclude the detection of the 6 nm AuNPs, thus allowing the large particles to be visualized selectively.
Diluting the bare AuNPs with water or PBS abolished the stabilizing effect of the citrate and produced a colour shift towards blue as well as a drop in the absorbance at the peak. Bare AuNPs may thus have undergone transient aggregation prior to being added to the culture medium. This was not the case for the gold conjugates, which is in accordance with the literature,12,14,46,54 and neither the light nor the electron microscopy findings were suggestive of aggregate internalization. The successful conjugation of AuNPs with GSL and WGA was corroborated by DLS and by lectin immunohistochemistry on sections of gut, which was used as a positive control tissue. DLS revealed a substantial increase in the hydrodynamic diameter of both GSL- and WGA-coated particles. As the PI remained unchanged, this expansion cannot be attributed to aggregate formation. The decrease in the z-potential from −35 mV for citrate-stabilized particles to −10 mV for coated particles further validated the conjugation step. Unfortunately, the protein coat cannot be visualized by TEM.13 Accordingly, the concentration of conjugated particles could not be estimated by the Haiss method.55 Therefore, the particle concentration was computed assuming that all of the particles were pelleted during centrifugation.
Uptake
The incubation time was set at one hour because preliminary experiments revealed that the uptake of AuNPs reached a maximum after this period (data not shown), with phagocytic N9 cells consistently internalizing more particles than SH cells. This finding was expected, as these cells belong to the macrophage system.56 In addition, the uptake of native AuNPs was strongly dependent on particle size. Whereas the non-phagocytic SH cells favoured small particles between 15 and 40 nm in diameter, the N9 microglial cells preferentially internalized larger particles with a diameter of 80 nm. This finding is in contrast with the findings of other authors,6,14,45,48,57 who reported a preferential uptake of particles with a diameter between 30 and 50 nm. As these studies were conducted with other cell lines, this difference may simply reflect cell-specific preferences. Notwithstanding, the uptake of particles is limited by size alone, with the upper limit being specific for a given cell type. Thus, macrophages, such as N9 cells, remain able to take up large nanoparticles, which are not easily handled by neuronal cells. The decline in uptake with increasing size was further corroborated by experiments using particles of 230 nm in diameter (data not shown). Cell-specific selectivity and the corresponding peaks in particle uptake as a function of particle diameter rule out a possible bias due to sedimentation. Potential sedimentation effects were further curtailed by reducing the volume of the culture medium to 50 μl per well.58,59Surface charge is the single most important factor that affected particle uptake, irrespective of cell type. Our results showing a dramatic increase in the uptake of cationized particles are in complete accordance with previous reports.60 Obviously, a positive surface charge mediates adhesion to the negatively charged cell surface, thus facilitating particle uptake.17 Unexpectedly, a negative surface charge promoted particle uptake in N9 cells but did not in either SH cell type. This seemingly contradictory finding may be attributed to the fact that the negative charge was imparted to the gold particles by coating them with a highly anionic form of BSA. BSA, however, has been shown to promote receptor-mediated endocytosis.45,61 This is in agreement with our own findings on the stimulatory effect of BSA-coating on particle uptake, and the net uptake of anionic BSA-coated AuNPs may thus simply reflect the balance of the two opposing effects. To resolve this issue, other ways to create a negative charge would be needed.60
With respect to the lectin coating, WGA substantially increased particle uptake in both N9 and SHd cells, clearly demonstrating that particle uptake can be modulated by surface coating.15,46,47,57,62–65 Interestingly, WGA did not promote particle uptake in SHu cells. Previous investigators have pointed to the fact that a higher differentiation status and decreasing mitotic activity result in lower numbers of intracellular nanoparticles.66,67 As for the WGA-coated AuNPs, this general principle is overruled. A plausible explanation involves differences in the composition of membrane-bound sugar residues. As the glycosylation status is dependent on cell differentiation,68,69N-acetylglucosamine sugar residues, which are recognized by WGA, appear to be overexpressed on the surface of differentiated SH cells as compared to SHu.70 Beyond emphasizing the effect of surface coatings on particle uptake, this crucial finding clearly illustrates the potential for the specific targeting of nanoparticles to particular cell types. Conversely, other substances may be used to prevent the uptake of nanoparticles by certain cells.
Binding to the cell membrane has been shown to be a crucial step in endocytosis.15,71,72 The notion that active endocytosis is the main uptake mechanism for AuNPs was further supported by our incubation experiments at 4 °C. Lowering the temperature dramatically reduced internalization, irrespective of particle size, surface charge or coating. This implies that energy-dependent uptake mechanisms occur in both N9 and SH cells. The localization of AuNPs to membrane-bound vesicles, as evidenced by TEM, further corroborates this hypothesis.13,54,73 Therefore, we did not consider direct membrane permeation, as reported by Geiser et al.74 to play a significant role in our setting. Studies using the selective staining of subcellular structures or blockers for specific internalization steps will be necessary to completely elucidate the endocytic pathway.75
Conclusions
The present study demonstrates the utility and versatility of gold nanoparticles to examine the impact of various parameters on nanoparticle uptake in vitro. In combination with high-content analysis, this approach provides a method to assess numerous variables in a stringently quantitative and statistically significant manner. Furthermore, the results presented herein highlight the feasibility of specifically targeting nanoparticles to distinct cell types on principle.Acknowledgements
The technical support of Ernesto Sala with setting up the epipolarization system and the assistance of Angélique Ducray with establishing the cell culture system are gratefully acknowledged. We are deeply indebted to Prof. Paul Bowen, École polytechnique fédérale de Lausanne, Switzerland, who patiently and thoroughly introduced us to the many subtleties and pitfalls of DLS and who provided the clues for reconciling the divergent measurements of the 80 nm AuNPs. This study was performed with the support of the interfaculty Microscopy Imaging Center (MIC) of the University of Bern.References
- M. L. Etheridge, et al. , Nanomedicine, 2013, 9, 1–14 CAS.
- N. Sozer and J. L. Kokini, Trends Biotechnol., 2009, 27, 82–89 CrossRef CAS PubMed.
- A. Weir, et al. , Environ. Sci. Technol., 2012, 46, 2242–2250 CrossRef CAS PubMed.
- P. Gehr, et al., in Comprehensive Physiology, ed. R. Terjung, John Wiley & Sons, Inc, Hoboken, NJ, USA, 2011 Search PubMed.
- A. Sawhney, et al. , Text. Res. J., 2008, 78, 731–739 CrossRef CAS.
- S.-H. Wang, et al. , J. Nanobiotechnol., 2010, 8, 33 CrossRef CAS PubMed.
- R. Lévy, et al. , Nano Rev., 2010, 1, 4889 Search PubMed.
- D. Vanhecke, et al. , Nanomedicine, 2014, 9, 1885–1900 CrossRef CAS PubMed.
- A. E. Nel, et al. , Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- R. Arvizo, R. Bhattacharya and P. Mukherjee, Expert Opin. Drug Delivery, 2010, 7, 753–763 CrossRef CAS PubMed.
- X. Huang and M. A. El-Sayed, J. Adv. Res., 2010, 1, 13–28 CrossRef.
- E. Boisselier and D. Astruc, Chem. Soc. Rev., 2009, 38, 1759 RSC.
- L. Shang, K. Nienhaus and G. Nienhaus, J. Nanobiotechnol., 2014, 12, 5 CrossRef PubMed.
- B. D. Chithrani, A. A. Ghazani and W. C. W. Chan, Nano Lett., 2006, 6, 662–668 CrossRef CAS PubMed.
- P. Nativo, I. A. Prior and M. Brust, ACS Nano, 2008, 2, 1639–1644 CrossRef CAS PubMed.
- G. Frens, et al. , Nat. Phys. Sci., 1973, 241, 20–22 CrossRef CAS.
- S. L. Goodman, et al. , J. Microsc., 1981, 123, 201–213 CrossRef CAS PubMed.
- M. Horisberger and J. Rosset, J. Histochem. Cytochem., 1977, 25, 295–305 CrossRef CAS PubMed.
- N. Benhamou, in Colloidal gold. Principles, methods, and applications, ed. M. A. Hayat, Academic Press, San Diego, 1989, vol. 1, pp. 95–143 Search PubMed.
- G. Egea, in Lectins and glycobiology, ed. H.-J. Gabius and S. Gabius, Springer Laboratory, [S.l.], 1993, pp. 215–233 Search PubMed.
- J. Schindelin, et al. , Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- W. Haiss, et al. , Anal. Chem., 2007, 79, 4215–4221 CrossRef CAS PubMed.
- M. Righi, et al. , Eur. J. Immunol., 1989, 19, 1443–1448 CrossRef CAS PubMed.
- L. Agholme, et al. , J. Alzheimer's Dis., 2010, 20, 1069–1082 CAS.
- M. Encinas, et al. , J. Neurochem., 2000, 75, 991–1003 CrossRef CAS PubMed.
- C. A. Belmokhtar, J. Hillion and E. Ségal-Bendirdjian, Oncogene, 2001, 20, 3354–3362 CrossRef CAS PubMed.
- E. López and I. Ferrer, Mol. Brain Res., 2000, 85, 61–67 CrossRef.
- J.-Y. Ha, et al. , Neurosci. Lett., 2014, 561, 101–106 CrossRef CAS PubMed.
- G. Schätz, et al. , Cells Tissues Organs, 2013, 198, 457–469 CrossRef PubMed.
- C. S. Holgate, et al. , J. Histochem. Cytochem., 1983, 31, 938–944 CrossRef CAS PubMed.
- L. Scopsi, in Colloidal gold. Principles, methods, and applications, ed. M. A. Hayat, Academic Press, San Diego, 1989, vol. 1, pp. 251–295 Search PubMed.
- M. De Waele, et al. , J. Histochem. Cytochem., 1988, 36, 679–683 CrossRef CAS PubMed.
- G. W. Hacker, in Colloidal gold. Principles, methods, and applications, ed. M. A. Hayat, Academic Press, San Diego, 1989, vol. 1, pp. 297–321 Search PubMed.
- W. Baschong and Y. D. Stierhof, Microsc. Res. Tech., 1998, 42, 66–79 CrossRef CAS PubMed.
- B. M. Humbel, et al. , Microsc. Res. Tech., 1998, 42, 43–58 CrossRef CAS PubMed.
- J. M. Robinson and D. D. Vandre, J. Histochem. Cytochem., 1997, 45, 631–642 CrossRef CAS PubMed.
- P. M. Lackie, Histochem. Cell Biol., 1996, 106, 9–17 CrossRef CAS PubMed.
- H. Sawada and H. Esaki, J. Electron Microsc., 1994, 43, 361–366 CAS.
- P. Herter, et al. , J. Microsc., 1993, 171, 107–115 CrossRef CAS PubMed.
- C. Oliver, Methods in Molecular Biology, 2010, 588, 311–316 CAS.
- E. L. Carstensen, et al. , J. Cell Biol., 1971, 50, 529–532 CrossRef CAS PubMed.
- U. Ziegler, News Physiol. Sci., 2004, 19, 124–128 CAS.
- A. M. Gown and M. C. Willingham, J. Histochem. Cytochem., 2002, 50, 449–454 CrossRef CAS PubMed.
- S. A. Love, et al. , Annu. Rev. Anal. Chem., 2012, 5, 181–205 CrossRef CAS PubMed.
- A. Arnida, A. Malugin and H. Ghandehari, J. Appl. Toxicol., 2010, 30, 212–217 CAS.
- A. Albanese and W. C. Chan, ACS Nano, 2011, 5, 5478–5489 CrossRef CAS PubMed.
- A. M. Alkilany and C. J. Murphy, J. Nanopart. Res., 2010, 12, 2313–2333 CrossRef CAS PubMed.
- R. Shukla, et al. , Langmuir, 2005, 21, 10644–10654 CrossRef CAS PubMed.
- V. A. Hackley and J. D. Clogston, in Characterization of Nanoparticles Intended for Drug Delivery, ed. S. E. McNeil, Humana Press, New York, NY, USA, 2011, vol. 697, pp. 35–52 Search PubMed.
- U. Nobbmann, Dispersity or Polydispdersity is a key parameter for GPC SEC, available at: http://www.materials-talks.com/blog/2014/10/23/polydispersity-what-does-it-mean-for-dls-and-chromatography/.
- D. Kumar, et al. , Int. J. Nanosci., 2012, 11, 1250023 CrossRef.
- B.-K. Pong, et al. , J. Phys. Chem. C, 2007, 111, 6281–6287 CAS.
- V. A. Hackley and J. D. Clogston, Measuring the Size of Nanoparticles in Aqueous Media Using Batch-Mode Dynamic Light Scattering. NIST - NCL Joint Assay Protocol, PCC-1 Version 1.1, National Institute of Standards and Technology NIST, Nanotechnology Characterization Lab, Frederick, 2010 Search PubMed.
- C. Brandenberger, et al. , Small, 2010, 6, 1669–1678 CrossRef CAS PubMed.
- T. Hendel, et al. , Anal. Chem., 2014, 86, 11115–11124 CrossRef CAS PubMed.
- M. L. Block, L. Zecca and J.-S. Hong, Nat. Rev. Neurosci., 2007, 8, 57–69 CrossRef CAS PubMed.
- L. Au, et al. , ACS Nano, 2010, 4, 35–42 CrossRef CAS PubMed.
- E. C. Cho, Q. Zhang and Y. Xia, Nat. Nanotechnol., 2011, 6, 385–391 CrossRef CAS PubMed.
- P. M. Hinderliter, et al. , Part. Fibre Toxicol., 2010, 7, 36 CrossRef CAS PubMed.
- D. Hühn, et al. , ACS Nano, 2013, 7, 3253–3263 CrossRef PubMed.
- A. M. Alkilany, et al. , Small, 2009, 5, 701–708 CrossRef CAS PubMed.
- C. H. J. Choi, et al. , Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 1235–1240 CrossRef CAS PubMed.
- K. Niikura, et al. , ACS Nano, 2013, 7, 3926–3938 CrossRef CAS PubMed.
- S. S. Minami, et al. , J. Neuroinflammation, 2012, 9, 22 CAS.
- O. Harush-Frenkel, et al. , Biochem. Biophys. Res. Commun., 2007, 353, 26–32 CrossRef CAS PubMed.
- J. A. Kim, et al. , Nat. Nanotechnol., 2012, 7, 62–68 CrossRef CAS PubMed.
- T. Serdiuk, et al. , J. Biophotonics, 2013, 6, 291–297 CrossRef CAS PubMed.
- C. G. Gahmberg, M. Ekblom and L. C. Andersson, Proc. Natl. Acad. Sci. U. S. A., 1984, 81, 6752–6756 CrossRef CAS.
- T. Ota, et al. , Cancer Res., 1987, 47, 3863–3867 CAS.
- N. Kitamura, et al. , J. Neurosci. Res., 2004, 75, 384–390 CrossRef CAS PubMed.
- B. D. Chithrani and W. C. W. Chan, Nano Lett., 2007, 7, 1542–1550 CrossRef CAS PubMed.
- S. D. Conner and S. L. Schmid, Nature, 2003, 422, 37–44 CrossRef CAS PubMed.
- B. M. Rothen-Rutishauser, et al. , Environ. Sci. Technol., 2006, 40, 4353–4359 CrossRef CAS PubMed.
- M. Geiser, et al. , Environ. Health Perspect., 2005, 113, 1555–1560 CrossRef PubMed.
- D. Vercauteren, et al. , Mol. Ther., 2010, 18, 561–569 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2016 |