Open Access Article
Open Access ArticleMPQ-cytometry: a magnetism-based method for quantification of nanoparticle–cell interactions†
V. O.
Shipunova
a,
M. P.
Nikitin
*abc,
P. I.
Nikitin
*b and
S. M.
Deyev
a
aShemyakin–Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 16/10 Miklukho-Maklaya Street, Moscow, 117997, Russia
bProkhorov General Physics Institute, Russian Academy of Sciences, 38 Vavilov Str., Moscow, 119991, Russia. E-mail: petr.nikitin@nsc.gpi.ru
cMoscow Institute of Physics and Technology (State University), 9 Institutskiy per., Dolgoprudny, Moscow Region, 141700, Russia. E-mail: max.nikitin@phystech.edu
First published on 24th May 2016
Abstract
Precise quantification of interactions between nanoparticles and living cells is among the imperative tasks for research in nanobiotechnology, nanotoxicology and biomedicine. To meet the challenge, a rapid method called MPQ-cytometry is developed, which measures the integral non-linear response produced by magnetically labeled nanoparticles in a cell sample with an original magnetic particle quantification (MPQ) technique. MPQ-cytometry provides a sensitivity limit 0.33 ng of nanoparticles and is devoid of a background signal present in many label-based assays. Each measurement takes only a few seconds, and no complicated sample preparation or data processing is required. The capabilities of the method have been demonstrated by quantification of interactions of iron oxide nanoparticles with eukaryotic cells. The total amount of targeted nanoparticles that specifically recognized the HER2/neu oncomarker on the human cancer cell surface was successfully measured, the specificity of interaction permitting the detection of HER2/neu positive cells in a cell mixture. Moreover, it has been shown that MPQ-cytometry analysis of a HER2/neu-specific iron oxide nanoparticle interaction with six cell lines of different tissue origins quantitatively reflects the HER2/neu status of the cells. High correlation of MPQ-cytometry data with those obtained by three other commonly used in molecular and cell biology methods supports consideration of this method as a prospective alternative for both quantifying cell-bound nanoparticles and estimating the expression level of cell surface antigens. The proposed method does not require expensive sophisticated equipment or highly skilled personnel and it can be easily applied for rapid diagnostics, especially under field conditions.
Introduction
Considerable attention is currently being directed toward investigating the biomedical functionality of a broad spectrum of nanoparticles, particularly their interactions with living cells.1–6 This interest stems from the rapid development of nanotechnology in fundamental life sciences as well as in applied biomedicine. Nanomaterials, which exhibit many valuable structural and physicochemical features unavailable to macro-objects or molecules, are expected to provide new ways and tools for solving challenging problems in the biological sciences.Reliable quantitative analyses of interactions of nanoscale entities with living cells are a key for the development of efficient nanomaterial-based agents and understanding the scope of their biomedical potential. In particular, quantification of nanoparticles in certain tissues and cells is essential for theranostic applications such as hyperthermia, targeted drug delivery or visualization of specific cells. Moreover, the accurate dose estimation is also necessary from pharmacological and toxicological standpoints, namely, for the safe use of nanoscale materials.
“Nanoparticle–cell” interactions are commonly investigated through incorporation of different labels, particularly fluorescent or radioactive ones, into a nanoparticle and subsequent detection by various methods. Different microscopy techniques (such as the spinning disc confocal,7 confocal laser scanning,8,9 fluorescence,10 X-ray fluorescence,11 and electron microscopy12–14) are used for detection of nanoparticles associated with cells, as well as for the structural examination of the nanoscale entities.15–17 However, microscopy (or its combination with other methods, e.g., magnetophoresis18,19) is semi-quantitative and/or requires complicated and time-consuming processing of the obtained 2D or 3D graphical information, which is not always acceptable for routine studies.
Other optical methods, such as flow cytometry,1,4,8,20 fluorescence spectroscopy21 and ultraviolet-visible (UV/VIS) spectrophotometry,22,23 are widely used for investigating the interactions of unmodified or fluorescently labeled nanoparticles with cells. However, fluorescence-based approaches have intrinsic calibration difficulties that may only allow relative rather than absolute measurements.24 Besides, the cells’ autofluorescence often significantly hampers the detection of small amounts of nanoparticles associated with cells, especially when investigating poorly fluorescent nanoparticles or performing the assay in whole blood. In fact, for whole blood cytometry magnetic measurements of cell parameters have been proposed25 as an attractive alternative to light scattering. UV/VIS-spectrophotometry is generally less sensitive than fluorescence-based methods (e.g., detection limit of 3 μg ml−1 of iron oxide nanoparticles can be achieved with Ferrozine staining23), but it may be well suited for studying particles with a high extinction coefficient such as gold nanoparticles. However, this method may be even more susceptible to the calibration and background-related problems discussed above.
In addition to the optical methods, various analytical techniques are often used, including the inductively coupled plasma mass spectrometry (ICP-MS)5,26 or atomic emission and optical emission spectrometry (ICP-AES/OES),27 measurement of the magnetic susceptibility of a sample,18,28 and other nuclear magnetic resonance and ferromagnetic resonance-based methods.6,19,26,29,30 These approaches feature a low detection limit (e.g., up to units of ppb in ICP-MS), however, some of them require complex sample preparation (prolonged acid digestion) and sample destruction. Besides, the absolute quantification of the nanoparticle content may be hampered if the tested cells contain free quantifiable atoms (ICP-MS, ICP-AES/OES). In the case of NMR-based methods it is challenging to perform calibrations for complex samples to identify the absolute amount of nanoparticles,26,30 because the result depends on the spatial distribution of the particles within the sample. Moreover, these techniques require expensive and bulky equipment, and it is difficult to employ them for the rapid analysis of a large series of samples.
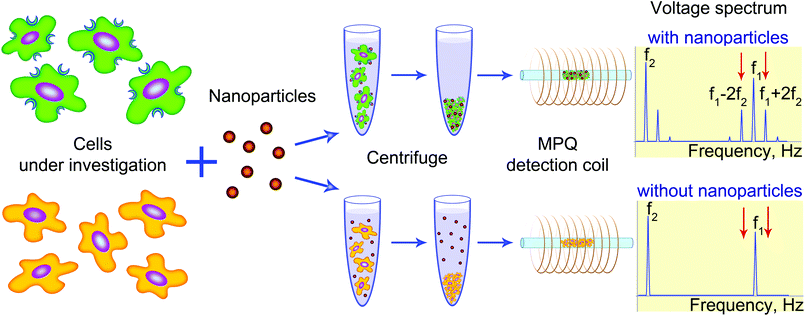
Here, we propose a novel method called by us as MPQ-cytometry for quantitative assessment of nanoparticle–cell interactions. MPQ-cytometry is based on incorporation of a magnetic label into nanoparticles with further counting such labels with the method of magnetic particle quantification (MPQ).
The MPQ detection principle was described by us in detail previously.31,32 Briefly, the sample is exposed to an alternating magnetic field at two frequencies f1 and f2, and the response is recorded at combinatorial frequencies (f = n·f1 ± m·f2, n and m are integers, one of them can be zero). At these frequencies, only non-linear magnetics (e.g., superparamagnetic nanoparticles) contribute to the signal, so there is no background noise from dia- and paramagnetics that always surround the magnetic labels. Therefore, this principle provides a good signal to noise ratio and is well suitable for potential applications oriented on various biological samples such as cells, tissues, or other opaque complex media.33 The MPQ allows measurements of very small relative variations up to 10−8 of magnetic susceptibility and its sensitivity is similar to those of radioactivity sensing techniques with major advantages in safety.34 The MPQ also offers a linear dynamic range of 7 orders of magnitude.35
The proposed method of MPQ-cytometry for detection of cell-bound nanoparticles is schematically illustrated in Fig. 1. It does not require complex sample preparation so that live undigested cells separated from unbound nanoparticles can be used for the measurements. Moreover, MPQ-cytometry overcomes drawbacks of many labeling methods due to magnetic signal stability in comparison with, for example, fluorescence or short-decay radioactive labels. There is no need for cryogenic setups or additional expensive equipment, and the MPQ-cytometry portable device (that can be engineered as shown in Fig. S1† with linear dimensions of 18 × 10 × 4 cm) offers real-time measurement at room temperature with a detection limit in the sub-nanogram level.
Results
For this study, we chose a model cell system that included six cell lines with different levels of surface HER2/neu receptor expression and synthesized superparamagnetic iron oxide nanoparticles (SPIONs) capable of specific binding to this receptor. HER2/neu is a member of the EGFR (epidermal growth factor receptor) family and is overexpressed to a variable degree in approximately 30% of human breast adenocarcinomas, as well as in many other types of human malignancies.36–39 HER2/neu overexpression often correlates with resistance to chemotherapy, a high tumor metastatic potential and also predicts a high risk of disease recurrence and reduced overall survival. However, this receptor is presented to a lesser extent on many types of normal health cells. Thus, accurate and quantitative determination of the HER2/neu tumor status has important clinical implications and the proposed test cell system is a convenient model for estimating the expression of different levels of cell surface-presented molecules.The full-length humanized antibody Trastuzumab (Herceptin®) conjugated to SPIONs was chosen as a targeting molecule for specific cell labeling. This antibody recognizes the extracellular domain of the HER2/neu receptor and is widely used in clinical practice to treat HER2/neu-overexpressing breast tumors.
SPIONs: synthesis, characterization and conjugation to proteins
SPIONs were synthesized by co-precipitating of Fe(II) and Fe(III) salts and stabilized with carboxymethyl-dextran (for the detailed synthesis process and TEM characterization, see the “Methods” section and Fig. S2,† respectively).For the specific cell labeling, we conjugated SPIONs with Trastuzumab (SPION–Trastuzumab) using the EDC/sulfo-NHS coupling method. Investigations of the obtained conjugates by the dynamic light scattering showed their hydrodynamic diameter to be 105 ± 31 nm (see size distribution in Fig. S3†). The ζ-potential of these conjugates was −8.9 ± 0.9 mV. The conjugates of SPIONs with non-specific Human IgG (SPION–HumanIgG) and a non-specific protein, bovine serum albumin (SPION–BSA), were also generated as controls. The hydrodynamic diameters and ζ-potentials of SPION–Trastuzumab, SPION–HumanIgG, SPION–BSA and unconjugated SPIONs are summarized in Table S1.†
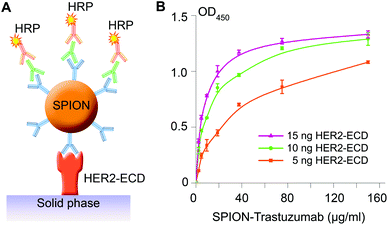
At first, the functional activity of the SPION conjugates, namely, their ability to specifically bind to the HER2/neu receptor, was investigated in a cell-free mode using a standard ELISA format. The SPION–Trastuzumab conjugates were incubated in 96-well plates with 5, 10 and 15 ng of the HER2/neu receptor extracellular domain (HER2–ECD) presorbed overnight and then were sequentially stained with goat anti-human and rabbit anti-goat HRP conjugated antibodies (see Fig. 2A). The dependence of the binding of SPION–Trastuzumab on the amount of HER2–ECD is shown in Fig. 2B.
The SPIONs conjugated to polyclonal human antibodies (SPION–HumanIgG) were used as a control to assess the binding specificity of the obtained conjugates. The dependence of the absorbance at λ = 450 nm on the concentrations of both SPION–Trastuzumab and SPION–HumanIgG conjugates for 5 ng of presorbed HER2–ECD is presented in Fig. S4† and demonstrates the high specificity of the interaction.
Quantification of SPIONs bound to cells by MPQ-cytometry
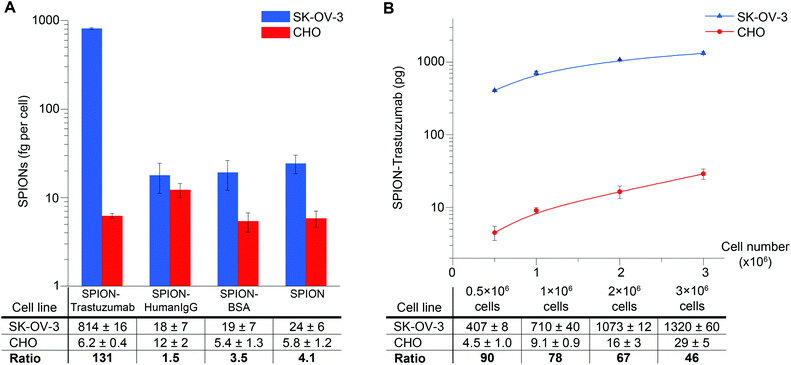
We used the obtained conjugates to quantitatively analyze HER2/neu expression on the cell surface. The quantification of SPIONs bound to cells by the proposed MPQ-cytometry was carried out as schematically illustrated in Fig. 1. Half a million of SK-OV-3 and CHO cells harvested from a culture dish were incubated with SPION–Trastuzumab, and the unbound particles were separated by centrifugation. The pellet of SPION-labeled cells with 30 μl of PBS was then placed on the detection coil of the original device to measure the quantity of the nonlinear magnetic material in the sample. The response signal was calibrated by the known weight of the SPIONs; the calibration curve is presented in Fig. S5.† The detection limit of the synthesized SPIONs achieved with the MPQ technique was found to be 0.33 ng of nanoparticles determined by the 2σ criterion as the minimal SPION quantity, for which the specific signal exceeded the double standard deviation of the signal for buffer without magnetic material. The absolute measured amount of SPIONs in a cell sample was divided by the cell number and further presented as femtograms per cell.The quantity of SPION–Trastuzumab bound to cells was dependent on the concentration of the conjugates during incubation with retention of the interaction specificity within the tested concentration range (Fig. S6†). For subsequent experiments, the concentration of 36 μg ml−1 was chosen as the minimal one required to achieve the maximum interaction specificity with the cells. The maximum ratio of SPION–Trastuzumab bound to SK-OV-3 cells versus CHO cells was more than 130, which evidenced not only the high sensitivity but also the high specificity of the proposed technique for quantifying the HER2/neu expression.
The interaction specificity at the chosen concentration was also verified by additional controls including SPIONs conjugated to non-specific polyclonal Human IgG, SPIONs conjugated to a non-specific protein (BSA), and unconjugated SPIONs (Fig. 3A). The non-specific particles (SPION–HumanIgG, SPION–BSA and unconjugated SPIONs) demonstrated very low binding to SK-OV-3 cells, as did SPION–Trastuzumab to CHO cells without HER2/neu expression.
We further studied the dependence of the quantity of SPION–Trastuzumab bound to the cells on the number of cells. According to the results presented in Fig. 3B, the SPION–Trastuzumab conjugates retain their specificity, and the MPQ technique can be used for quantification of nanoparticles bound to cells up to as high as 3 × 106 cells in the sample.
Detection of HER2/neu-overexpressing cells in a complex mixture
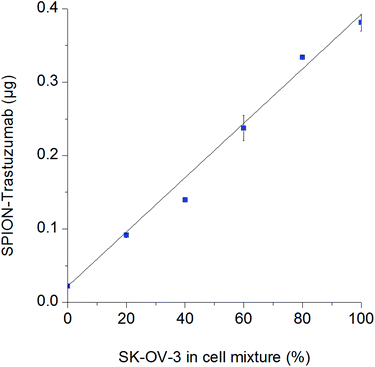
The described method for detection of nanoparticles that specifically bind to the cell surface due to receptor-mediated processes can be applied to determine the number of cells expressing a particular surface antigen in a complex mixture. We realized this concept with the SPION–Trastuzumab conjugates, which were demonstrated to be highly specific during interaction with the cells. SK-OV-3 and CHO cells were mixed in different proportions with the fixed total cell number and incubated with SPION–Trastuzumab as described above. The resulting quantity of SPION–Trastuzumab in the samples proved to depend linearly on the number of SK-OV-3 cells in the mixture (Fig. 4).MPQ-cytometry vs. optical microscopy, flow cytometry and fluorescence spectroscopy
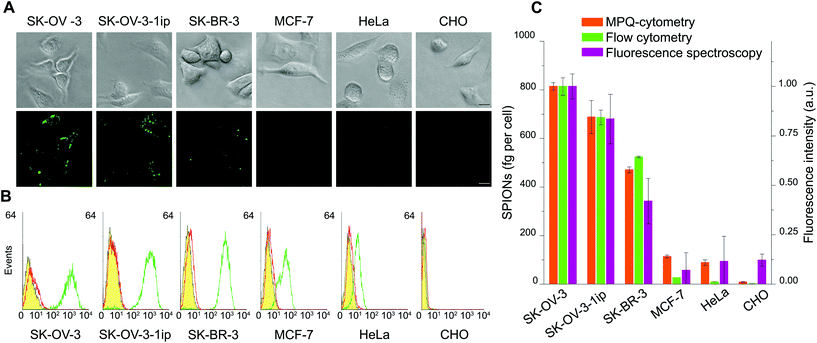
In order to verify the proposed MPQ-cytometry method for both nanoparticle quantification and investigation of cell surface antigen expression, we compared it with three methods commonly used in molecular and cell biology. To this end, we chose six cell lines with different HER2/neu expression levels as follows: three of the lines overexpressed HER2/neu (SK-OV-3, SK-OV-3-1ip and SK-BR-3), two lines featured normal HER2/neu expression levels (HeLa and MCF-7), and the other line did not express HER2/neu (CHO). Firstly, we stained these cells with SPIONs conjugated with Trastuzumab and labeled by using fluorescein isothiocyanate (SPION–Trastuzumab-FITC) and with plain SPION–Trastuzumab conjugates. Then we analyzed the interaction of these conjugates by optical microscopy and MPQ-cytometry, respectively (see optical images in Fig. 5A and orange columns in Fig. 5C for the SPION quantity). The respective control experiments with the cells stained with SPION–HumanIgG-FITC and SPION–HumanIgG conjugates are presented in Fig. S7.† Fluorescence intensities of nanoparticle–cell complexes visualized by optical microscopy appear to be in good correlation with the quantitative data of MPQ cytometry.Next, for demonstration of the capabilities of the proposed method for estimating the expression of membrane-associated proteins, we compared MPQ-cytometry with two quantitative methods, namely, flow cytometry and fluorescence spectroscopy. The immunofluorescence staining of the cell lines described above was performed using Trastuzumab-FITC and Human IgG-FITC as the control antibodies both in suspension and on a culture plate surface for flow cytometry (see Fig. 5B) and fluorescence spectroscopy analyses, respectively. The quantity of SPION–Trastuzumab conjugates bound to the cells (estimated as femtograms per cell) was compared with the fluorescence intensity of the Trastuzumab-FITC labeled cells acquired by flow cytometry and fluorescence spectroscopy (Fig. 5C).
One can see that the MPQ-cytometry data are in good agreement with those provided by the qualitative optical microscopy and two quantitative fluorescence-based methods. We should note that MPQ-cytometry exhibits slightly higher sensitivity while detecting the cells with normal HER2/neu expression levels (here, MCF-7 and HeLa) compared to flow cytometry, but these data are virtually indistinguishable with those provided by the commonly used method of fluorescence spectroscopy. Thus, MPQ-cytometry represents a reliable alternative method for the estimation of cell surface antigen expression.
Discussion
The proposed MPQ-cytometry method offers highly sensitive, room-temperature and rapid quantification of nanoparticle–cell interactions. We have demonstrated the measurement of the integral amount of magnetic nanoparticles that specifically recognize the HER2/neu oncomarker on the surface of cancer cells. By using MPQ-cytometry with SPION–Trastuzumab conjugates, HER2/neu positive cells have been detected in a mixture with HER2/neu negative cells, making the developed method attractive for life science applications as well as biomedical diagnostics.We consider MPQ-cytometry as an effective tool for scientists whose research area involves nanoparticles. The proposed method may facilitate investigation of interaction processes (e.g., uptake and degradation) between nanoparticles and cells. The capability of MPQ-cytometry to quantify targeted cell-bound nanoparticles potentially allows the investigation of specific and non-specific cell binding for selection of optimal particles for nanocarrier-based drug and gene delivery, cell tracking, magnetic force-driven tissue engineering and other applications in biomedicine, biosensing and theranostics. Importantly, thanks to the fact that, along with nano- and microparticles, other biological objects such as proteins,40 bacteria41 and viruses42 can be magnetically labeled, the proposed method can also be applied to study interactions of these nanosized objects with cells.
Concerning the potential of MPQ-cytometry for clinical applications, we believe that the developed method can be used for diagnostics of various membrane-associated disease markers. MPQ-cytometry was shown to be capable of distinguishing subtle differences in the cell expression level of a HER2/neu oncomarker, which is normally present in many healthy tissues. In clinical practice, the HER2/neu tumor status is commonly determined semi-quantitatively by immunohistochemistry at a scale from 0 to 3+ according to the intensity of cell membrane staining. In controversial cases (staining of 2+), patients are sent for an additional more expensive analysis, for example, flow cytometry. Being the gold standard in cell biology, flow cytometry in clinical practice is available to a limited number of diagnostic laboratories due to economic considerations, need for highly skilled personnel and regular maintenance. The herein demonstrated high correlation between the quantitative MPQ-cytometry data on HER2/neu expression and those acquired with flow cytometry allows us to consider the proposed method as a reliable substitute and/or supplement to flow cytometry for medical purposes. We believe that MPQ-cytometry can be potentially used for both in vitro (e.g., blood or biopsy characterization) and ex vivo (e.g., postoperative tumor status determination) diagnostics in cases when the use of expensive and non-portable equipment is impossible, for example under the field conditions or in developing countries.
We do not see any obstacles to expanding MPQ-cytometry to investigation of the expression levels of other cell membrane-associated molecules such as clusters of differentiation and cell adhesion proteins, and consider this method helpful for both fundamental cell biology routine studies and precise medical diagnostics.
In contrast to optical methods, including those based on fluorescence, MPQ-cytometry is not affected by the optical properties of samples. Therefore, it allows highly sensitive quantification of the cell-bound nanoparticles in opaque samples and detection of nanostructures with a complex architecture regardless of many factors that impact fluorescence such as label quenching or oxidation, susceptibility to pH or temperature, sample autofluorescence, nanoparticle modification after cellular internalization and many others. Moreover, MPQ-cytometry does not require complicated sample preparation or expensive setups. Besides, it allows rapid analysis of a large number of similar samples as one measurement takes only a few seconds.
Major limitations and drawbacks of MPQ-cytometry compared with the other techniques in cell biology are its inability to: (i) detect expression of intracellular antigens and (ii) implement at this stage simultaneous multiparametric processing of samples like it is done by some fluorescence-based methods (e.g., flow cytometry, which uses multiple fluorescence channels in different spectral regions). Nevertheless, we should note that multiplex detection with MPQ-cytometry is technically feasible when using nanoparticles with different magnetization curves.43Table 1 below shows a side-by-side comparison of the advantages and shortcomings of MPQ-cytometry and flow cytometry, which are among the most commonly used methods in cell biology.
| Flow cytometry | MPQ-cytometry |
|---|---|
| − Expensive and available only in specialized centers | + Affordable, can be used in diagnostic laboratories in the developing countries |
| − Bulky devices intended for use under stationary conditions require regular maintenance and highly trained personnel | + Hand-held devices (no moving parts) without expensive consumables |
| − Signal affected by cell autofluorescence | + Signal is not affected by linear dia- and paramagnetic materials such as tissues, blood, plastic, and glass, thus providing a low background signal |
| − Complicate analyses in colored or opaque media | + Detection even in optically opaque media |
| − Instable labels | + Signal is stable over time |
| + Multiparametric diagnostics in different fluorescence channels | ± Potential expansion to additional number of parameters |
| + Possibility of intracellular antigen detection | − Detection of intracellular antigens is impossible |
Conclusion
The method of MPQ-cytometry proposed in this work for quantification of nanoparticle–cell interactions and detection of cell surface antigens is a powerful and convenient tool for fundamental life science research that involves nanoparticles. This method, when used with target-specific nanoparticles, provides quantitative information about the expression of cell surface antigens, so it can serve as a reliable alternative to some other label-based methods in molecular and cell biology such as flow cytometry. Moreover, since the related devices are portable, do not contain any moving parts and do not require expensive consumables and highly skilled personnel, MPQ-cytometry is promising as a diagnostic platform for determining the expression of various cell membrane-associated markers of various diseases even under field conditions.Methods
Synthesis of carboxymethyl-dextran-coated superparamagnetic iron oxide nanoparticles
Carboxymethyl-dextran-coated superparamagnetic iron oxide nanoparticles (SPIONs) were synthesized by mixing of FeCl3·6H2O (5.9 g) and FeCl2·4H2O (2.15 g) in 100 ml of degassed MilliQ water with subsequent addition of 12.5 ml of 30% NH4OH. The solution was heated to 85 °C and refluxed with vigorous agitation and N2 bubbling for 2 h. The resulting magnetic particles were then magnetically separated and washed sequentially with 2 M HNO3 and thrice with MilliQ water. Next, the aggregates were magnetically removed, and the particles in the supernatant were coated with carboxymethyl-dextran. For the coating reaction, a carboxymethyl-dextran solution at 300 g l−1 in MilliQ water was added to the particles to obtain the final concentration of 50 g l−1 followed by a 4 h incubation at 80 °C. Then, the resulting nanoparticles were washed from the non-bound carboxymethyl-dextran with MilliQ water by triple centrifugation at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800g for 1–3 h.
800g for 1–3 h.
Conjugation of proteins to SPIONs
Covalent coupling of the protein molecules to the surface of SPIONs was performed using a sodium salt of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC, Sigma, Germany) and a sodium salt of N-hydroxysulfosuccinimide (sulfo-NHS, Sigma, Germany) as crosslinking agents for the formation of amide bonds between the carboxyl groups on the particle surface and protein amino groups. The SPIONs (0.3 mg) suspended in 60 μl of 0.1 M 2-(N-morpholino) ethanesulfonic acid buffer, pH 5.5 were activated with 10 mg EDC and 3 mg sulfo-NHS for 20 min at 20 °C. The excess unreacted reagents were removed by centrifugation for 12 min at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g. Proteins at a concentration of 0.5 g l−1 together with two-fold (wt/wt) excess of BSA were added to the particles in borate buffer (0.4 M H3BO3, 70 mM Na2B4O7, pH 8.0). To prevent aggregation, the conjugates were periodically sonicated in an ultrasonic bath. The reaction was carried out at least overnight. The particles were washed from the non-bound proteins by triple centrifugation at 12
000g. Proteins at a concentration of 0.5 g l−1 together with two-fold (wt/wt) excess of BSA were added to the particles in borate buffer (0.4 M H3BO3, 70 mM Na2B4O7, pH 8.0). To prevent aggregation, the conjugates were periodically sonicated in an ultrasonic bath. The reaction was carried out at least overnight. The particles were washed from the non-bound proteins by triple centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 12 min.
000g for 12 min.
Fluorescein isothiocyanate (FITC) labeled SPION conjugates were prepared by mixing of 50 μl of 3 g l−1 SPIONs in borate buffer with 5 μl of 0.6 g l−1 FITC in dimethyl sulfoxide. The reaction was carried out for 2 h with subsequent centrifugation from unbound FITC molecules at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 12 min.
000g for 12 min.
DLS and zeta potential measurements
The hydrodynamic radius and ζ-potential of the nanoparticles were determined using a Zetasizer Nano ZS (Malvern Instruments Ltd, UK) analyzer in phosphate buffered solution (PBS, pH 7.4) at 25 °C. All measurements were performed in triplicate.Immune-enzyme analysis
The extracellular domain of the HER2/neu receptor was presorbed on 96-well ELISA plates in carbonate–bicarbonate buffer (4 mM Na2CO3, 50 mM NaHCO3, pH 9.2) overnight at 4 °C. The unbound antigen was washed twice with PBS and incubated for 1 h in 5% fat-free milk in PBS at room temperature (to block nonspecific adsorption sites). Thereafter, the particles under investigation were added into the wells in 3% fat-free milk in PBS and incubated for 1 h, and the wells were then washed three times with PBS. After that, the wells were incubated sequentially with 12 μg ml−1 goat anti-human antibodies (Jackson ImmunoResearch Labs Inc., USA) and 0.16 μg ml−1 rabbit anti-goat antibodies conjugated with horseradish peroxidase (Jackson Immuno Research Labs Inc., USA). Antibodies were diluted in PBS with 1% BSA, and both steps were carried out for 1 h at room temperature. After each step, the unbound antibodies were removed by washing three times with PBS with 0.05% Tween-20. Then, the wells were incubated with 0.04% (wt/vol) o-phenylenediamine and 0.01% (vol/vol) hydrogen peroxide solution in citrate buffer (40 mM citric acid, 40 mM Na2HPO4·2H2O, pH 5.0). The color development reaction was stopped with a 0.5 M solution of H2SO4, and the absorbance of the wells was measured at the wavelengths of 450 nm (working) and 630 nm (reference). All samples were analyzed in duplicate.Cell lines
Human breast adenocarcinoma SK-BR-3 and MCF-7 cells, human ovarian adenocarcinoma SK-OV-3 and SK-OV-3-1ip cells, human cervical adenocarcinoma HeLa cells, and Chinese hamster ovary (CHO) cells were cultured in RPMI-1640 medium (HyClone, USA) supplemented with 10% heat inactivated fetal bovine serum (HyClone, USA) and 2 mM L-glutamine (PanEco, Russia) at 37 °C under a humidified atmosphere with 5% CO2. The cells were harvested with 2 mM EDTA solution in PBS without the use of trypsin (to prevent enzymatic cleavage of cell-surface receptors).Analysis of SPIONs binding to the cells
The binding of SPION conjugates to the cells was quantified using the MPQ detection principle31,32 as described above. The cells harvested from culture flasks were washed twice with PBS and resuspended in 500 μl of 3% fat-free milk in PBS and cooled to 4 °C. The SPION conjugates were then added to the cells. The cells were incubated with SPIONs for 50 min on a rotator at 4 °C, and then the unbound particles were separated by centrifugation in 3% milk at 100g. The suspensions of SPION-labeled cells in 30 μl PBS were placed in a plastic tube inside the measuring coil of the MPQ-cytometry device. The measured magnetic signal was divided by the number of cells and calibrated according to the quantity of particles.Fluorescence microscopy
The cell suspensions were seeded into flat-bottomed 96-well plates at a concentration of 2.5 × 104 cells per ml and cultured overnight. Before the experiment, the cells were washed once with PBS, cooled to 4 °C and then conjugates of SPIONs in 3% fat-free milk were added for 45 min at 4 °C. After incubation, the cells were washed three times with PBS and analyzed using an inverted epifluorescence microscope (Axiovert 200, Carl Zeiss, Germany).Flow cytometry
The harvested cells were washed twice with PBS, resuspended in 500 μl of 3% fat-free milk at a concentration of 106 cells per ml and cooled to 4 °C. Then, FITC-labeled antibodies (prepared by rapid mixing of 50 μl of 10 g l−1 antibodies in PBS with 5 μl of 0.1 g l−1 FITC in DMSO with subsequent dialysis with Zeba Spin Desalting Columns, 7 K MWCO) were added to the cells at a final concentration of 20 μg ml−1. Staining was performed for 50 min on a rotator at 4 °C, after which the cells were washed three times with 3% fat-free milk, resuspended in 500 μl PBS with 1% BSA, and analyzed by flow cytometry on a FACS Calibur (BD, USA). A total of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were analyzed for each sample. The samples that were prepared without the addition of antibodies were used to control the cell autofluorescence. All samples were performed in duplicate. The data were analyzed using WinMDI 2.8 software (The Scripps Research Institute, USA).
000 cells were analyzed for each sample. The samples that were prepared without the addition of antibodies were used to control the cell autofluorescence. All samples were performed in duplicate. The data were analyzed using WinMDI 2.8 software (The Scripps Research Institute, USA).
Fluorescence spectroscopy
The cell suspensions were seeded into flat-bottomed 96-well plates at a concentration of 2.5 × 104 cells per ml and cultured to confluence. Then the cells were washed with PBS, fixed for 2 h in 1% paraformaldehyde, washed twice with PBS and stained with 50 μg ml−1 Trastuzumab-FITC in PBS with 1% BSA for 40 min at room temperature. Then the cells were washed twice with PBS with 1% BSA and analyzed by using an Infinite M1000 PRO microplate reader (Tecan, Austria) in bottom fluorescence mode at excitation and emission wavelengths of 490 nm and 515 nm, respectively, at 400 Hz in flash frequency mode with a gain of 217.Conflict of interest
Petr I. Nikitin is a named inventor on a patent on MPQ.Acknowledgements
This research was supported by the Russian Science Foundation grants no. 14-24-00106 (cell culture, flow cytometry, fluorescence spectroscopy) and no. 16-19-00131 (nanoparticles synthesis and characterization, immune-enzyme analysis, optical microscopy).References
- A. Lesniak, A. Salvati, M. J. Santos-Martinez, M. W. Radomski, K. A. Dawson and C. Åberg, J. Am. Chem. Soc., 2013, 135, 1438–1444 CrossRef CAS PubMed.
- M. P. Nikitin, V. O. Shipunova, S. M. Deyev and P. I. Nikitin, Nat. Nanotechnol., 2014, 9, 716–722 CrossRef CAS PubMed.
- A. Verma and F. Stellacci, Small, 2010, 6, 12–21 CrossRef CAS PubMed.
- H. D. Summers, M. R. Brown, M. D. Holton, J. A. Tonkin, N. Hondow, A. P. Brown, R. Brydson and P. Rees, ACS Nano, 2013, 7, 6129–6137 CrossRef CAS PubMed.
- A. J. Giustini, I. Perreard, A. M. Rauwerdink, P. J. Hoopes and J. B. Weaver, Integr. Biol., 2012, 4, 1283–1288 RSC.
- H. Lee, E. Sun, D. Ham and R. Weissleder, Nat. Med., 2008, 14, 869–874 CrossRef CAS PubMed.
- L. Treuel, S. Brandholt, P. Maffre, S. Wiegele, L. Shang and G. U. Nienhaus, ACS Nano, 2014, 8, 503–513 CrossRef CAS PubMed.
- C. Schweiger, R. Hartmann, F. Zhang, W. J. Parak, T. H. Kissel and G. P. Rivera, J. Nanobiotechnol., 2012, 10, 1–11 CrossRef PubMed.
- E. C. Costa, V. M. Gaspar, J. G. Marques, P. Coutinho and I. J. Correia, PLoS One, 2013, 8, e70072 CAS.
- M. P. Nikitin, T. A. Zdobnova, S. V. Lukash, O. A. Stremovskiy and S. M. Deyev, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 5827–5832 CrossRef CAS PubMed.
- S. A. James, B. N. Feltis, M. D. de Jonge, M. Sridhar, J. A. Kimpton, M. Altissimo, S. Mayo, C. Zheng, A. Hastings, D. L. Howard, D. J. Paterson, P. F. Wright, G. F. Moorhead, T. W. Turney and J. Fu, ACS Nano, 2013, 7, 10621–10635 CrossRef CAS PubMed.
- W. J. Wang, X. Yu, S. V. Boriskina and B. M. Reinhard, Nano Lett., 2012, 12, 3231–3237 CrossRef PubMed.
- J. G. Rouse, J. Yang, A. R. Barron and N. A. Monteiro-Riviere, Toxicol. In Vitro, 2006, 20, 1313–1320 CrossRef CAS PubMed.
- C. Rosman, S. Pierrat, A. Henkel, M. Tarantola, D. Schneider, E. Sunnick, A. Janshoff and C. Sönnichsen, Small, 2012, 8, 3683–3690 CrossRef CAS PubMed.
- I. Lubitz and A. Kotlyar, Bioconjugate Chem., 2011, 22, 482–487 CrossRef CAS PubMed.
- K. Maximova, A. Aristov, M. Sentis and A. V. Kabashin, Nanotechnology, 2015, 26, 065601 CrossRef CAS PubMed.
- U. F. Aghayeva, M. P. Nikitin, S. V. Lukash and S. M. Deyev, ACS Nano, 2013, 7, 950–961 CrossRef CAS PubMed.
- D. Fayol, N. Luciani, L. Lartigue, F. Gazeau and C. Wilhelm, Adv. Healthcare Mater., 2013, 2, 313–325 CrossRef CAS PubMed.
- C. Wilhelm, F. Gazeau and J. C. Bacri, Eur. Biophys. J., 2002, 31, 118–125 CrossRef CAS PubMed.
- R. M. Zucker, K. M. Daniel, E. J. Massaro, S. J. Karafas, L. L. Degn and W. K. Boyes, Cytometry, Part A, 2013, 83, 962–972 CAS.
- M. Gaumet, R. Gurny and F. Delie, Eur. J. Pharm. Sci., 2009, 36, 465–473 CrossRef CAS PubMed.
- B. D. Chithrani, A. A. Ghazani and W. C. Chan, Nano Lett., 2006, 6, 662–668 CrossRef CAS PubMed.
- A. M. Rad, B. Janic, A. S. Iskander, H. Soltanian-Zadeh and A. S. Arbab, BioTechniques, 2007, 43, 627–628 CrossRef CAS PubMed.
- R. A. Smith and T. D. Giorgio, Cytometry, Part A, 2009, 75, 465–474 CrossRef CAS PubMed.
- M. Helou, M. Reisbeck, S. F. Tedde, L. Richter, L. Bär, J. J. Bosch, R. H. Stauber, E. Quandt and O. Hayden, Lab Chip, 2013, 13, 1035–1038 RSC.
- O. M. Girard, R. Ramirez, S. McCarty and R. F. Mattrey, Contrast Media Mol. Imaging, 2012, 7, 411–417 CrossRef CAS PubMed.
- C. Freese, C. Uboldi, M. I. Gibson, R. E. Unger, B. B. Weksler, I. A. Romero, P. O. Couraud and C. J. Kirkpatrick, Part. Fibre Toxicol., 2012, 9, 23 CrossRef CAS PubMed.
- V. Ström, K. Hultenby, C. Grüttner, J. Teller, B. Xu and J. Holgersson, Nanotechnology, 2004, 15, 457–466 CrossRef.
- F. Réty, O. Clément, N. Siauve, C. A. Cuénod, F. Carnot, M. Sich, A. Buisine and G. Frija, J. Magn. Reson. Imaging, 2000, 12, 734–739 CrossRef.
- P. Oswald, O. Clement, C. Chambon, E. Schouman-Claeys and G. Frija, Magn. Reson. Imaging, 1997, 15, 1025–1031 CrossRef CAS PubMed.
- P. I. Nikitin and P. M. Vetoshko, Russian Patent, RU2177611, 2000 Search PubMed.
- P. I. Nikitin, P. M. Vetoshko and T. I. Ksenevich, J. Magn. Magn. Mater., 2007, 311, 445–449 CrossRef CAS.
- A. V. Orlov, J. A. Khodakova, M. P. Nikitin, A. O. Shepelyakovskaya, F. A. Brovko, A. G. Laman, E. V. Grishin and P. I. Nikitin, Anal. Chem., 2013, 85, 1154–1163 CrossRef CAS PubMed.
- M. P. Nikitin, M. Torno, H. Chen, A. Rosengart and P. I. Nikitin, J. Appl. Phys., 2008, 103, 07A304 CrossRef.
- A. V. Orlov, V. A. Bragina, M. P. Nikitin and P. I. Nikitin, Biosens. Bioelectron., 2016, 79, 423–429 CrossRef CAS PubMed.
- M. A. Lemmon, J. Schlessinger and K. M. Ferguson, Cold Spring Harbor Perspect. Biol., 2014, 6, a020768 CrossRef PubMed.
- Y. Yarden and M. Sela, Clin. Cancer Res., 2015, 21, 4030–4032 CrossRef CAS PubMed.
- I. Beyer, Z. Li, J. Persson, Y. Liu, R. van Rensburg, R. Yumul, X. B. Zhang, M. C. Hung and A. Lieber, Mol. Ther., 2011, 19, 479–489 CrossRef CAS PubMed.
- B. Dreier, G. Mikheeva, N. Belousova, P. Parizek, E. Boczek, I. Jelesarov, P. Forrer, A. Plückthun and V. Krasnykh, J. Mol. Biol., 2011, 405, 410–426 CrossRef CAS PubMed.
- S. J. Osterfeld, H. Yu, R. S. Gaster, S. Caramuta, L. Xu, S. J. Han, D. A. Hall, R. J. Wilson, S. Sun, R. L. White, R. W. Davis, N. Pourmand and S. X. Wang, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 20637–20640 CrossRef CAS PubMed.
- H. L. Grossman, W. R. Myers, V. J. Vreeland, R. Bruehl, M. D. Alper, C. R. Bertozzi and J. Clarke, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 129–134 CrossRef CAS PubMed.
- J. M. Perez, F. J. Simeone, Y. Saeki, L. Josephson and R. Weissleder, J. Am. Chem. Soc., 2003, 125, 10192–10193 CrossRef CAS PubMed.
- L. Lenglet, J. Magn. Magn. Mater., 2009, 321, 1639–1643 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6nr03507h |
| This journal is © The Royal Society of Chemistry 2016 |