Open Access Article
Open Access ArticleMolecular interactions of graphene oxide with human blood plasma proteins
Kenry
abc,
Kian Ping
Loh
abd and
Chwee Teck
Lim
*abce
aNUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore 117456, Singapore. E-mail: ctlim@nus.edu.sg
bCentre for Advanced 2D Materials and Graphene Research Centre, National University of Singapore, Singapore 117543, Singapore
cDepartment of Biomedical Engineering, National University of Singapore, Singapore 117575, Singapore
dDepartment of Chemistry, National University of Singapore, Singapore 117543, Singapore
eMechanobiology Institute, National University of Singapore, Singapore 117411, Singapore
First published on 30th March 2016
Abstract
We investigate the molecular interactions between graphene oxide (GO) and human blood plasma proteins. To gain an insight into the bio-physico-chemical activity of GO in biological and biomedical applications, we performed a series of biophysical assays to quantify the molecular interactions between GO with different lateral size distributions and the three essential human blood plasma proteins. We elucidate the various aspects of the GO–protein interactions, particularly, the adsorption, binding kinetics and equilibrium, and conformational stability, through determination of quantitative parameters, such as GO–protein association constants, binding cooperativity, and the binding-driven protein structural changes. We demonstrate that the molecular interactions between GO and plasma proteins are significantly dependent on the lateral size distribution and mean lateral sizes of the GO nanosheets and their subtle variations may markedly influence the GO–protein interactions. Consequently, we propose the existence of size-dependent molecular interactions between GO nanosheets and plasma proteins, and importantly, the presence of specific critical mean lateral sizes of GO nanosheets in achieving very high association and fluorescence quenching efficiency of the plasma proteins. We anticipate that this work will provide a basis for the design of graphene-based and other related nanomaterials for a plethora of biological and biomedical applications.
Introduction
Graphene oxide (GO) is a two-dimensional hydrophilic nanomaterial decorated with epoxide and hydroxyl groups on its basal plane as well as carboxylic groups at its edges.1,2 With unique and exceptional bio-physico-chemical properties, such as excellent water dispersibility and superior in vitro and in vivo biocompatibility,3,4 the versatile GO has been increasingly explored for a wide array of biological and biomedical applications.5–9 Some of these include biological sensing,10–12 cellular imaging,13,14 drug delivery,15,16 anticancer therapy,17,18 and antithrombotic coating.19 A significant portion of these GO-based bioapplications is based on the molecular interactions between GO and proteins.As one of the fundamental elements of physiological systems, proteins play prominent roles in many biological functions. Physiological environments typically comprise a diverse range of proteins with different structures, compositions, functionalities, and concentrations. Upon entering a physiological environment, nanomaterials like GO will come into immediate contact and interact with proteins specific to that environment. Interactions with these proteins and their nature depend very much on the properties of nanomaterials, such as surface energy, surface charge, and hydrophobicity, and the eventual biological response towards nanomaterials will hinge on the outcome of these nanomaterial–protein interactions. One of the fundamental processes of the nanomaterial–protein interactions takes the form of association or binding of proteins onto the surface of nanomaterials. In general, protein binding to nanomaterials in biological fluids like serum or blood plasma will significantly affect the biological characteristics of nanomaterials.20,21 For example, surface-bound proteins have been demonstrated to enhance specific cellular uptake22,23 and activate intracellular signaling pathways.24 Consequently, a deeper understanding of the physiological effects of nanomaterials, in particular GO, requires the knowledge of the binding of proteins to GO in biological fluids. In fact, comprehensive studies on the GO–protein interactions are necessary for establishing the influence of GO on the structure and physiological activities of these proteins. For a majority of the GO-based in vivo biological applications, the utilization of GO will inevitably lead to it entering the blood circulation system and subsequently, interacting with blood plasma proteins.
Blood plasma consists of many distinct proteins carrying out unique and specific functionalities.25 Of all the proteins circulating in blood plasma, three major proteins stand out, i.e., albumin, globulin, and fibrinogen. Albumin is one of the smallest proteins and the most abundant plasma protein in the circulatory system. As a globular protein, albumin has a molecular weight of 66 kDa and constitutes approximately 55% of the blood plasma proteins. Known as a transporter molecule with multifunctionalities, an albumin molecule is capable of binding to a whole range of endogenous and exogenous compounds. The high conformational flexibility of the albumin molecule coupled with its labile binding characteristic endows it with diverse functionalities. As another important globular protein responsible for the molecular transport and body immune system, globulin forms the second most plentiful protein in the blood plasma. It constitutes about 38% of the plasma proteins. Globulin can be generally classified into four sub-categories, i.e., α1-globulin, α2-globulin, β-globulin, and γ-globulin, with α-globulin being the lightest globulin having a molecular weight of 92 kDa while the heaviest globulin molecule is the γ-globulin having a molecular weight of 120 kDa. Fibrinogen, on the other hand, is a large cylindrical plasma glycoprotein which possesses a molecular weight of 340 kDa and accounts for about 7% of the blood plasma. A crucial component in the blood coagulation process, fibrinogen is normally converted into fibrin by thrombin, leading to the formation of blood clots essential for wound healing.
Currently, there are still limited studies on the molecular interactions between GO and plasma proteins. Interestingly, most of these studies are based on GO with a particular lateral size distribution and are still focused on albumin.26–28 Currently, the effects of the lateral size distribution of GO on the interactions between GO and plasma proteins are still not well understood. In addition, the nature, specificity, and mechanisms driving the GO–plasma protein interactions remain to be elucidated. As such, we aim to investigate the molecular interactions between GO with different lateral size distributions and the three essential blood plasma proteins, i.e., albumin, fibrinogen, and globulin, and the biological effects GO induces on these proteins. By utilizing an array of biophysical techniques, such as atomic force microscopy (AFM), ultraviolet-visible (UV-Vis) absorption, fluorescence, and circular dichroism (CD) spectroscopy, we characterized the various aspects of the GO–protein interactions, notably protein adsorption on GO, GO–protein binding kinetics and equilibrium, and protein conformational stability. Based on these assays, fundamental properties, such as the morphology and size of the GO nanosheets, GO–protein association constants, GO–protein binding cooperativity, and the binding-induced protein structural changes, were determined. Furthermore, we proposed the existence of GO lateral size-dependent molecular interactions between GO nanosheets and the plasma proteins, and significantly, the presence of particular critical mean lateral sizes and size distributions of GO nanosheets in achieving the highest intrinsic fluorescence quenching efficiency of the plasma proteins. Significantly, through this work, the general perspective on the bio-physico-chemical effects of GO on the nature of plasma protein binding as well as the degree to which these GO-induced effects are protein-specific were established.
Results and discussion
Lateral size of GO nanosheets
To start with, we characterized the lateral size distribution of the as-prepared GO nanosheets (Fig. 1). As all the GO nanosheets possessed irregular shapes, here, we defined their lateral size as the longest distance between two points across a single nanosheet fragment, similar to that defined in ref. 29. The GO nanosheets in aqueous suspension were first synthesized based on Hummer's method, as reported in ref. 30. They were then subjected to ultrasonication treatment, similar to that used in ref. 31 for different durations of 10, 30, 60, 120, and 240 min (i.e., GO-10, GO-30, GO-60, GO-120, and GO-240, respectively) to yield GO nanosheets of different lateral size distributions. It is noteworthy that ultrasonication was used to produce GO nanosheets of different lateral sizes as this method would inflict negligible variations to the surface chemical properties of the GO nanosheets.31 An aqueous suspension of the different GO nanosheet samples was subsequently deposited on a freshly cleaved mica and the morphological features of the individual nanosheets were characterized using an atomic force microscope (AFM) operating under the tapping mode (Fig. 1a–f). For all GO samples from GO-0 to GO-240, we noted that the individual GO nanosheets possessed a thickness of around 1.3 nm, suggesting that all the as-prepared GO nanosheets were of single layer (Fig. 1g and h). | ||
| Fig. 1 Surface morphology and lateral size of the GO nanosheets. (a–f) Representative amplitude images showing the surface morphologies of the GO nanosheets subjected to ultrasonication for different durations, as obtained using AFM: (a) 0 min (GO-0), (b) 10 min (GO-10), (c) 30 min (GO-30), (d) 60 min (GO-60), (e) 120 min (GO-120), and (f) 240 min (GO-240). Scale bars represent 1 μm. (g) Representative height image of the GO-0 samples with (h) their corresponding sectional profiles (i.e., S1 and S2 in Fig. 1g). The thickness of the GO nanosheets based on the sectional height profiles was around 1.3 nm, indicating that the GO nanosheets were of single layer. (i) Lateral size distribution range of the six GO nanosheet samples. Inset illustrates their mean lateral sizes. (j–o) Histograms of the lateral size distribution of the GO nanosheets undergoing ultrasonication for (j) 0 (GO-0), (k) 10 (GO-10), (l) 30 (GO-30), (m) 60 (GO-60), (n) 120 (GO-120), and (o) 240 (GO-240) min. Insets show the enlarged version of the size distribution of the GO nanosheet samples. More than 400 GO sheets were evaluated for each sample to obtain the lateral size distribution. | ||
Next, to obtain the statistical data about the lateral size of the GO nanosheets with their size distribution, more than 400 GO nanosheets were evaluated. We observed that the lateral size distribution ranges of the GO nanosheets in GO-0, GO-10, GO-30, GO-60, GO-120, and GO-240 samples were 4.138, 2.199, 1.829, 1.175, 0.605, and 0.281 μm, respectively (Fig. 1i). Their mean lateral sizes were 0.917, 0.350, 0.304, 0.236, 0.126, and 0.099 μm, correspondingly (inset of Fig. 1i). More specifically, the lateral size distributions of the individual GO nanosheet samples are illustrated in Fig. 1j–o. Clearly, it is evident that the six samples possessed significantly different lateral size distributions and mean lateral sizes. Nevertheless, it is interesting to highlight that the lateral size distribution of GO-0 and GO-10 samples revealed the eminent presence of relatively large GO sheets (>1 μm). Furthermore, the size of the largest GO sheets of the GO-0 sample was larger by several orders of magnitude than that of the GO-240 sample. Importantly, we noted that with increasing ultrasonication time, the mean lateral size of the GO nanosheets decreased proportionately, suggesting that GO nanosheet samples with six distinct lateral mean sizes and size distributions were obtained.
Adsorption of plasma proteins
To start unraveling the molecular interactions between GO and plasma proteins, we sought to examine the adsorption of the plasma proteins on GO nanosheets with different lateral size distributions (Fig. 2). The loading capacity of the plasma proteins on GO was evaluated from the adsorption of the proteins pre- and post-incubation with GO nanosheets, similar to that reported previously.3 First, we obtained the free absorbance of albumin, fibrinogen, and globulin at different concentrations ranging from 0.2 to 20 mg mL−1 at the characteristic wavelength of 280 nm (Fig. 2a, c and e). The free absorbance of the plasma proteins at 280 nm against their concentration profiles was then plotted (insets of Fig. 2a, c and e), showing the linear relationship between these two entities of the three plasma proteins.Next, we measured the absorbance of the free plasma proteins after incubations with GO to evaluate the adsorption capacity of the plasma proteins on the GO nanosheet samples. Depending on the specific plasma proteins, we observed distinct GO lateral size-driven adsorption behaviors. For the albumin concentration ranging from 0.5 to 10 mg mL−1, we noted a highly consistent size-dependent adsorption behavior (Fig. 2b). More clearly, albumin adsorption increased with an increase in the lateral size distribution of the GO nanosheets, with GO-0 exhibiting the lowest free albumin absorbance while the highest absorbance was acquired from the GO-240 sample. This suggests that the GO-0 sample with the largest mean size and lateral size distribution facilitated the highest amount of albumin adsorption while the lowest amount of albumin adsorbed onto the GO-240 sample with the smallest mean lateral size and size distribution.
Interestingly, a reverse trend in the protein adsorption characteristic was noted from fibrinogen (Fig. 2d). At low fibrinogen concentrations ranging from 0.5 mg mL−1 to 2 mg mL−1, we observed that the free fibrinogen absorbance was not significantly different across all the GO nanosheet samples, suggesting that most fibrinogen adsorbed well on the GO nanosheets independent of their lateral size distribution. At a particular concentration of 5 mg mL−1, however, we noted that the free absorbance of fibrinogen on GO-0 increased while that on GO-10, GO-30, GO-60, GO-120, and GO-240 remained fairly unchanged. This indicates that all GO nanosheet samples, except GO-0, still exhibited maximum fibrinogen adsorption while that of GO-0 was significantly lower than the rest of the GO samples. The lower fibrinogen adsorption on GO-0 at this specific concentration could be due to the possibility that the loading capacity of fibrinogen on GO-0 had reached saturation. At a specific concentration of 10 mg mL−1, the GO lateral size-driven fibrinogen adsorption came into effect. Fibrinogen displayed an adsorption behavior which was strongly influenced by the lateral size distribution of the GO nanosheets. Particularly, fibrinogen adsorption increased with a decrease in the lateral size of the GO nanosheets and vice versa, with GO-0 exhibiting the highest free fibrinogen absorbance while the lowest free fibrinogen absorbance was observed from GO-240. This translates into the highest fibrinogen adsorption on GO-240 and lowest fibrinogen adsorption on GO-0.
While albumin and fibrinogen, to a certain degree, exhibited a strongly linear GO lateral size distribution-dependent adsorption characteristic, that of globulin was not really evident (Fig. 2f). At low globulin concentrations ranging from 0.2 mg mL−1 to 2 mg mL−1, GO nanosheets, irrespective of their lateral sizes and size distributions, displayed maximum globulin adsorption based on the minimum free globulin absorbance after incubations with the GO nanosheets. However, a certain amount of free globulin absorbance was detected on all GO nanosheet samples at higher globulin concentrations of 5 mg mL−1 and 10 mg mL−1. At these two globulin concentrations, GO-0 exhibited the lowest free globulin absorbance while the highest free absorbance was detected from GO-60. This highlights that the highest amount of globulin adsorbed on the GO-0 sample with the highest mean lateral size and size distribution while the lowest amount of globulin adsorbed on the GO-60 sample. Importantly, from all the plasma protein adsorption data, we deduced that all GO nanosheets readily interacted with the three plasma proteins and the loading capacities of the plasma proteins on the GO nanosheets were heavily dependent on the lateral sizes of the GO nanosheets and the specific types of the plasma proteins being adsorbed onto the GO surface.
Fluorescence quenching efficiency of plasma proteins
Proteins are polymeric complexes comprising unique and distinctive amino acids.32 The chromophore residues of proteins typically consist of tryptophan, tyrosine, and phenylalanine which serve as their intrinsic fluorescent probes. Intriguingly, these aromatic amino acid residues are extremely sensitive to the nature of their microenvironment. The emission characteristics of the amino acid residues are known to be correlated to changes in their microenvironment due to the binding and conformational changes of proteins upon their association with molecules and nanomaterials.33,34 Recent studies have revealed the fluorescence quenching capability of GO upon its interaction with emission-active biomolecules, such as amino acids, peptides, and proteins.26,35 Therefore, based on the variations in the fluorescence emission intensity, the highly sensitive fluorescence spectroscopy may be utilized to deduce the state and conformation of plasma proteins upon their binding with GO.Here, we characterized the intrinsic fluorescence quenching behavior of plasma proteins by GO nanosheets with different lateral size distributions and concentrations (Fig. 3). Subsequently, we derived the essential quantitative parameters correlated to the GO–protein interactions, particularly quenching efficiency, association and dissociation constants, and binding cooperativity. On the basis of the obtained experimental data, we observed that the maximum emission intensity Imax of all plasma proteins progressively decreased as concentrations of GO increased (Fig. 3a–c and g–i). The evident quenching effect in the presence of GO suggests the occurrence of direct interactions between GO and the chromophore residues of the three plasma proteins. In fact, decreases in the emission intensity of the plasma proteins could be attributed to their adsorption onto GO which resulted in the shortening of the relative distance between the quenching agent GO and the active fluorescence emitters of the plasma proteins. Since the steady-state fluorescence emission of the plasma proteins represents the total fluorescence signals from all amino acid residues present in the different parts of the proteins, changes in the fluorescence emission intensity suggests variations in the local molecular environment of the chromophore residues and possibly, a change in the tertiary structure of the plasma proteins. At the same time, the stronger fluorescence quenching could be driven by an increase in the amount of tryptophan and tyrosine residues of the bound proteins accessible to the GO nanosheet surface and possibly, the protein conformational change as a result of stronger association between GO nanosheets and these proteins.
We also noted gradual shifts in the emission maximum wavelength λmax of the plasma proteins with variations in the concentrations of the GO nanosheets (Fig. 3d–f and 3j–l). More specifically, we observed that albumin exhibited the largest emission red-shift compared to fibrinogen and globulin (Table 1) irrespective of the lateral size distribution of the GO nanosheets. Also, the emission red-shifts of both albumin and globulin were fairly constant regardless of the GO size distribution. In contrast, the emission red-shift characteristic of fibrinogen was uniquely different from those of the other two plasma proteins. In fact, fibrinogen displayed an increasing emission red-shift with a decrease in the lateral size distribution of the GO nanosheets (Fig. 3e, k and Table 1). In principle, a shift in the peak intensity wavelength λmax of the continuous fluorescence spectrum may reveal important information regarding the layer of proteins adsorbed on the GO nanosheets, variations in the microenvironment of the amino acid residues, and ultimately, changes in the protein tertiary structure.25 The red-shift of the maximum emission to higher wavelengths with the increasing GO concentration implies an increase in the polarity and hydrophilicity of the local molecular environment of the emission-active elements of the plasma proteins. Apparently, the microenvironment of the solution-dispersed protein was less polarized than the local dielectric microenvironment within the adsorbed protein layer. In addition, the red-shifted λmax shows that the amino acid residues were exposed to a more hydrophilic microenvironment possibly due to a change in the tertiary structure of the plasma proteins.
| GO nanosheets | ||||||
|---|---|---|---|---|---|---|
| GO-0 | GO-10 | GO-30 | GO-60 | GO-120 | GO-240 | |
| Albumin | ||||||
| K SV (mL μg−1) | 0.081 ± 0.031 | 0.140 ± 0.044 | 0.196 ± 0.049 | 0.115 ± 0.029 | 0.144 ± 0.007 | 0.127 ± 0.022 |
| Q max | 1.314 ± 0.212 | 1.088 ± 0.036 | 1.039 ± 0.016 | 1.042 ± 0.033 | 1.076 ± 0.021 | 1.075 ± 0.015 |
| n | 0.751 ± 0.328 | 0.865 ± 0.205 | 0.922 ± 0.042 | 1.226 ± 0.235 | 0.915 ± 0.093 | 0.994 ± 0.076 |
| K D (μg mL−1) | 27.887 ± 16.141 | 8.837 ± 3.573 | 5.836 ± 1.818 | 11.908 ± 2.259 | 8.694 ± 0.630 | 10.401 ± 2.096 |
| K A (mL μg−1) | 0.044 ± 0.021 | 0.128 ± 0.058 | 0.182 ± 0.051 | 0.086 ± 0.018 | 0.115 ± 0.008 | 0.099 ± 0.018 |
| Δλ (nm) | 34 ± 5.292 | 41.333 ± 5.033 | 41.333 ± 7.024 | 41.333 ± 3.055 | 38 ± 3.464 | 38.667 ± 3.055 |
| Fibrinogen | ||||||
| K SV (mL μg−1) | 0.086 ± 0.014 | 0.135 ± 0.007 | 0.134 ± 0.008 | 0.151 ± 0.048 | 0.171 ± 0.009 | 0.133 ± 0.031 |
| Q max | 0.942 ± 0.049 | 1.027 ± 0.016 | 1.026 ± 0.005 | 1.016 ± 0.004 | 1.023 ± 0.004 | 1.027 ± 0.010 |
| n | 1.342 ± 0.516 | 1.091 ± 0.148 | 1.158 ± 0.036 | 1.334 ± 0.083 | 1.187 ± 0.071 | 1.254 ± 0.053 |
| K D (μg mL−1) | 12.337 ± 2.547 | 8.534 ± 0.952 | 9.476 ± 0.677 | 9.639 ± 2.855 | 7.777 ± 0.465 | 9.854 ± 1.407 |
| K A (mL μg−1) | 0.084 ± 0.020 | 0.118 ± 0.013 | 0.106 ± 0.007 | 0.110 ± 0.030 | 0.129 ± 0.008 | 0.103 ± 0.014 |
| Δλ (nm) | 5.333 ± 1.155 | 11.333 ± 4.163 | 8.667 ± 3.055 | 12.667 ± 2.309 | 14.667 ± 6.110 | 20.667 ± 2.309 |
| Globulin | ||||||
| K SV (mL μg−1) | 0.101 ± 0.043 | 0.156 ± 0.019 | 0.141 ± 0.026 | 0.112 ± 0.034 | 0.123 ± 0.011 | 0.090 ± 0.016 |
| Q max | 1.043 ± 0.036 | 1.069 ± 0.032 | 1.051 ± 0.021 | 1.046 ± 0.020 | 1.051 ± 0.016 | 1.078 ± 0.010 |
| n | 1.248 ± 0.409 | 0.924 ± 0.111 | 1.049 ± 0.082 | 1.247 ± 0.301 | 1.126 ± 0.103 | 1.062 ± 0.037 |
| K D (μg mL−1) | 13.166 ± 5.051 | 7.796 ± 1.255 | 9.026 ± 3.023 | 12.114 ± 3.577 | 10.54 ± 0.694 | 13.175 ± 1.160 |
| K A (mL μg−1) | 0.087 ± 0.042 | 0.131 ± 0.023 | 0.122 ± 0.051 | 0.087 ± 0.023 | 0.095 ± 0.006 | 0.076 ± 0.007 |
| Δλ (nm) | 6.000 ± 2.000 | 15.333 ± 1.155 | 10.000 ± 3.464 | 14.000 ± 2.000 | 12.000 ± 2.000 | 10.667 ± 5.033 |
Next, we sought to determine the nature of the fluorescence quenching of the three plasma proteins by GO nanosheets with various lateral size distributions (Fig. 4). We observed that the fluorescence emission intensity I0/I ratios vs. GO concentration curves of the three proteins are distinctively different (Fig. 4a–f), in which I0 and I represent the maximum emission intensity of plasma proteins in the absence and presence of GO, respectively. In the presence of GO-0 nanosheets, the fluorescence intensity ratio of both albumin and globulin exhibited an exponential trend while that of fibrinogen was linear (Fig. 4a–c). This suggests that albumin and globulin experienced both static (i.e., ground-state complex formation) and dynamic (i.e., collisional process) fluorescence quenching whilst the fluorescence quenching of fibrinogen was primarily contributed by dynamic collisional quenching. Nevertheless, the fluorescence intensity ratio of fibrinogen adopted an exponential characteristic as the lateral size distribution of the GO nanosheets decreased, indicating the additional presence of static fluorescence quenching as the lateral size of the GO nanosheets decreased (Fig. 4e). As fluorescence quenching is primarily driven by diffusive transport at low nanomaterial concentrations,25 the relative kinetic efficiency of the fluorescence quenching of plasma proteins at these concentrations can be derived based on the non-equilibrium Stern–Volmer model, as explained in eqn (1),
 | (1) |
 | ||
| Fig. 4 Fluorescence quenching efficiency of human blood plasma proteins by GO nanosheets. (a–c) Stern–Volmer plots of the fluorescence quenching of (a) albumin, (b) fibrinogen, and (c) globulin in the presence of GO-0 nanosheets with various concentrations. (d–f) Stern–Volmer plots of the fluorescence quenching of (d) albumin, (e) fibrinogen, and (f) globulin in the presence of GO-240 nanosheets with various concentrations. Insets illustrate the linear regime of the Stern–Volmer plot at the low concentrations of the fluorescence quencher. Experimental data were fitted based on the Stern–Volmer mathematical model as represented by eqn (1). (g–i) Efficiency of the fluorescence quenching of the three plasma proteins of (g) albumin, (h) fibrinogen, and (i) globulin, as represented by KSV, in the presence of GO nanosheets with low concentrations and different lateral size distributions. The * and ** indicate statistically significant differences for p < 0.1 and 0.05, respectively. All statistical analyses were made based on the two-tailed Student's t-test. | ||
Based on the mathematical fitting of the experimental data to eqn (1) (insets of Fig. 4a–f), we derived the Stern–Volmer quenching constants. Interestingly, we noted that the quenching efficiency of the GO nanosheets towards the three plasma proteins, as represented by KSV, was size-dependent and distinctively unique for specific plasma proteins (Fig. 4g–i). For all proteins, GO-0 with the largest mean lateral size exhibited the lowest fluorescence quenching efficiency while the highest quenching efficiency was manifested by different GO nanosheet samples for different plasma proteins. For albumin, GO-30 exhibited the highest quenching efficiency. GO-120, on the other hand, quenched the intrinsic fluorescence of fibrinogen more effectively than the rest of the GO samples. For globulin, GO-10 and GO-30 displayed higher quenching efficiencies compared to other GO nanosheets.
It is generally known that the larger the lateral size of the quenching agent, the stronger the fluorescence quenching observed. A larger lateral size of the fluorescence quenchers implies a higher relative surface area of the quenchers. As such, an increase in the surface area of the quenching agents allows the accommodation of a larger number of proteins around the nanosheets, resulting in a more efficient fluorescence quenching. On the contrary, the smaller surface area of the smaller quenching agents reduced the amount of plasma proteins being adsorbed onto their surface, leading to less efficient fluorescence quenching. However, in our study, the linear relationship between the lateral size of the quenching agent and the fluorescence quenching efficiency was not observed. In fact, a recent study has reported that the unique nanometer size-dependent fluorescence quenching efficiency of GO in which GO with three different average lateral sizes of 500, 200, and 40 nm were prepared and their DNA fluorescence quenching efficiencies were assessed.36 It was revealed that GO with an average lateral size of about 200 nm, instead of that of 500 nm, possessed the highest efficiency of fluorescence quenching while that of 40 nm demonstrated a lower fluorescence quenching efficiency. As such, our observations on the different intrinsic fluorescence quenching efficiencies exhibited by the various GO nanosheet samples further suggested the possible existence of critical lateral sizes of GO nanosheets in inducing the most effective fluorescence quenching of specific plasma proteins.
Binding of plasma proteins
Subsequent to evaluating the fluorescence quenching efficiency of plasma proteins by GO nanosheets at low concentrations, we sought to examine the effect of the lateral size distribution of the GO nanosheets on the relative strength and cooperativity of the GO–protein interactions (Fig. 5). This could be achieved by quantifying several key parameters describing the association between plasma proteins and GO nanomaterials, i.e., binding dissociation constant KD, binding association constant KA, and the Hill coefficient n, assuming that the binding of the plasma proteins to GO nanosheets occurred under equilibrium conditions. Using the non-linear curve fitting of the fluorescence quenching of albumin, fibrinogen, and globulin by the GO nanosheets (Fig. 5a–f), we derived the necessary parameters based on the Hill equation, as represented in eqn (2) and (3), | (2) |
 | (3) |
 | ||
| Fig. 5 Fluorescence quenching properties of human blood plasma proteins by GO nanosheets and the associated GO–protein complexes binding parameters. (a–c) Hill plots of the fluorescence quenching of (a) albumin, (b) fibrinogen, and (c) globulin, in the presence of GO-0 nanosheets with increasing concentrations. (d–f) Hill plots of the fluorescence quenching of (d) albumin, (e) fibrinogen, and (f) globulin, in the presence of GO-240 nanosheets with increasing concentrations. Experimental data were fitted based on the Hill mathematical model as represented by eqn (2) and (3). (g–i) Dissociation constant, KD, of (g) GO–albumin, (h) GO–fibrinogen, and (i) GO–globulin complexes. (j–l) Association constant, KA, of (j) GO–albumin, (k) GO–fibrinogen, and (l) GO–globulin complexes. (m–o) Hill coefficient, n, of (m) GO–albumin, (n) GO–fibrinogen, and (o) GO–globulin complexes. The * and ** indicate statistically significant differences for p < 0.1 and 0.05, respectively. All statistical analyses were made based on the two-tailed Student's t-test. | ||
To elucidate the binding parameters between the plasma proteins and GO nanosheets as a function of the lateral size distribution of GO nanosheets, we first estimated the equilibrium dissociation constant, KD, of the various GO–plasma protein complexes (Fig. 5g–i). For albumin, the GO-30–albumin complex exhibited the lowest KD while GO-0–albumin displayed the highest (Fig. 5g). For fibrinogen, the dissociation constant displayed by the GO-0–fibrinogen complex was comparatively higher than the rest of the GO–fibrinogen samples while that of GO-120–fibrinogen was the lowest (Fig. 5h). For globulin, GO-10–globulin displayed the lowest KD whereas those of GO-0–globulin and GO-240–globulin complexes were comparatively higher than the other GO–globulin samples (Fig. 5i). Following the derivation of KD, we quantitatively computed the equilibrium binding association constant, KA, between the three plasma proteins and GO nanosheets (Fig. 5j–l). As KA is defined as the reciprocal of KD, a reverse trend was anticipated. Indeed, for albumin, the KA displayed by the GO-0–albumin complex was the lowest while that of the GO-30–albumin complex was the highest (Fig. 5j), indicating that albumin had the weakest association with GO-0 while GO-30 possessed the strongest association with albumin. For fibrinogen, the KA of the GO-120–fibrinogen complex was apparently higher but similarly, that of GO-0–fibrinogen was the lowest (Fig. 5k). This translates into the possibility that fibrinogen possessed the highest and the lowest association with GO-120 and GO-0, respectively. In the case of globulin, GO-10–globulin and GO-30–globulin complexes displayed higher KA compared to the rest of the GO–globulin complexes while the KA of GO-0–globulin and GO-240–globulin complexes were lower (Fig. 5l).
Here, we noted that the obtained experimental results on the GO–protein binding interactions as a function of the mean lateral size and size distribution of GO coincided with that on the fluorescence quenching efficiency of the plasma proteins by GO nanosheets. In fact, the GO–protein binding interaction data provided an insight into the observed trend of stronger fluorescence quenching efficiency of GO nanosheets with certain average lateral sizes and size distributions. For example, albumin associated most strongly with the GO-30 sample. At the same time, the highest efficiency in quenching the intrinsic fluorescence of albumin could be achieved with GO-30. On the other hand, the weakest association and subsequently, the weakest fluorescence quenching of albumin were noticed with the addition of GO-0. Similar correlation between GO–protein association and fluorescence quenching could be noticed from both fibrinogen and globulin. All these demonstrated that a higher rate of fluorescence quenching (as represented by a higher KSV) could be ascribed to the closer proximity of the chromophore residues of the bound protein to the adsorbing surface of a particular GO nanosheet (as represented by a higher KA).
To gain a better understanding on the scale of the GO–plasma protein binding interactions, we examined the multifunctional and cooperative nature of the binding between the GO nanosheets and plasma proteins. As it is highly likely that the plasma proteins possess multiple binding sites37,38 on the GO nanosheets, we anticipated the binding between the GO nanosheets and plasma proteins to exhibit a certain level of cooperativity. We elucidated this by examining the Hill constants, n, of the different GO–protein complex formations, which is a critical measure describing the binding cooperativity or the degree of independence of molecular interactions between multiple binding sites (Fig. 5m–o). The Hill constants of the GO–albumin, GO–fibrinogen, and GO–globulin complexes were extracted from the fluorescence quenching data based on eqn (2) and (3). For albumin, we observed that, except for that of GO-60, all the other GO–albumin complexes displayed anti-cooperative binding or negative cooperativity (n < 1). On the contrary, for both fibrinogen and globulin, except for the GO-10–globulin complex, all GO–fibrinogen and GO–globulin complexes exhibited cooperative binding with positive cooperativity (n > 1).
Generally, a Hill coefficient of n < 1 denotes a negatively cooperative reaction. This means that as an increasing number of proteins adsorb onto the nanomaterial surface, the binding strength between the proteins and the surface of that nanomaterial weakens progressively. Contrastingly, for a positively cooperative reaction, as represented by a Hill coefficient of n > 1, once a single protein molecule adsorbs onto the nanomaterial surface, its affinity and association energy per protein molecule for the nanomaterials strengthen progressively with further protein adsorption. The anti-cooperativity exhibited by the GO–albumin complexes could possibly point to changes in the physicochemical properties of the GO nanosheets with progressive adsorption of albumin. Specifically, the ionic or electrostatic binding energy between GO nanosheets and albumin could possibly be reduced with the adsorption of albumin onto the GO surface, resulting in a corresponding decrease in the entropy and enthalpy of the albumin binding.39 Therefore, the adsorption of albumin onto GO nanosheets exhibited an anti-cooperative binding characteristic. On the other hand, the positive cooperativity of the GO–fibrinogen and GO–globulin interactions could be attributed to the multiple binding sites of both proteins with GO. Fibrinogen and globulin might possess multiple binding sites with the GO nanosheets and some of these binding sites could be buried within the protein core structures. As soon as a fibrinogen or globulin molecule binds to GO, the corresponding interaction might induce protein conformational changes. This led to the exposures of the inner binding sites of the proteins and eventually, resulted in the progressively increasing binding affinity between the two plasma proteins and GO nanosheets and the eventual cooperative binding interactions.
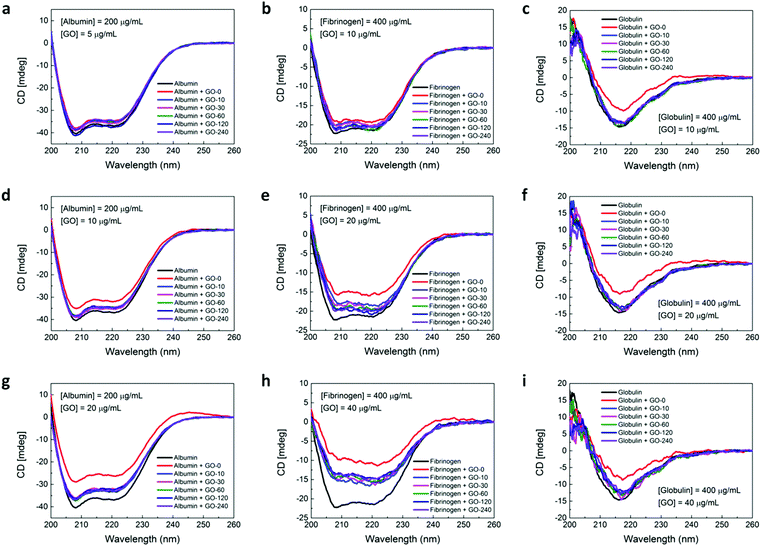
Conformational stability of plasma proteins
One of the primary processes of the nanomaterial–protein interactions involves the adsorption of proteins onto the nanomaterial surface. While some proteins retain their native-like structure upon surface binding, other proteins may experience conformational changes or denaturation of their tertiary and/or secondary structures. In fact, most of the nanomaterial–protein interactions will induce a certain degree of alterations or conformational changes to the protein structures. Furthermore, these proteins may bind to the nanomaterial surface in a specific orientation and the extent of the alterations will be heavily influenced by the surface chemistry of the nanomaterials. These conformational changes, in turn, are capable of influencing the functionality of proteins and induce their aggregation, leading to the loss of regular biological activity and unregulated interaction with other cellular components. On this basis, it is vital to address in greater detail the nature of the conformational changes experienced by the plasma proteins upon their interaction with the GO nanosheets. Therefore, we utilized CD spectroscopy to assess more specifically the degree of denaturation of the secondary structures of the three plasma proteins upon association with GO nanosheets with different lateral size distributions.CD spectroscopy is a robust analytical tool for probing the conformation and secondary structure of proteins in solutions or when they adsorbed onto other biomolecules or nanomaterials. In fact, the CD spectra in the far UV region from 190 to 260 nm may be utilized to quantify variations in the secondary structure of the proteins as a result of the molecular interactions between proteins and other objects. Here, albumin, fibrinogen, and globulin were prepared at concentrations of 200, 400, and 400 μg mL−1, respectively, and the concentration ratios of the plasma proteins to GO were fixed at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.025, 1
0.025, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.05, and 1
0.05, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1. The far UV CD spectra of the plasma proteins in the absence and presence of GO nanosheets with different lateral sizes were then recorded (Fig. 6). The secondary structures of albumin and fibrinogen were predominantly α-helical and their CD spectra exhibited two distinct negative bands in the UV region at the specific wavelengths of 208 and 222 nm. In contrast, the secondary structure of globulin was mainly β-sheet and an evident negative band was noticed from its CD spectrum at approximately 216 nm.
0.1. The far UV CD spectra of the plasma proteins in the absence and presence of GO nanosheets with different lateral sizes were then recorded (Fig. 6). The secondary structures of albumin and fibrinogen were predominantly α-helical and their CD spectra exhibited two distinct negative bands in the UV region at the specific wavelengths of 208 and 222 nm. In contrast, the secondary structure of globulin was mainly β-sheet and an evident negative band was noticed from its CD spectrum at approximately 216 nm.
The stability of the secondary structures of the plasma proteins in the absence and presence of GO nanosheets can be assessed by evaluating the ellipticity of the CD spectra of the plasma proteins at the characteristic wavelengths. From the obtained results, we observed that in the presence of increasing concentrations of GO nanosheets, the ellipticity of the CD spectra of the three plasma proteins decreased progressively. This indicates the strong molecular interactions between the GO nanosheets and plasma proteins.
For albumin, in the presence of 5 μg mL−1 GO nanosheets with different lateral sizes, all recorded CD spectra coincided closely (Fig. 6a). However, as the GO concentrations were increased to 10 and 20 μg mL−1 (Fig. 6d and g), a decrease in the ellipticity of the CD spectra in inverse proportion to the lateral size distribution of the GO nanosheets became apparent. More clearly, the CD spectrum of albumin exhibited the highest decrease in its ellipticity in the presence of GO-0 with the highest lateral size and size distribution while the lower decreases in the ellipticity of the CD spectra were noted in the presence of GO nanosheets with smaller lateral size distributions. However, although the intensity of the two characteristic bands at 208 and 222 nm reduced upon the conjugation of albumin with GO nanosheets, importantly, these negative bands were still evident with their α-helical structures. In other words, albumin preserved its conformational stability by retaining its overall secondary structure upon the interactions with GO nanosheets.
For fibrinogen, in contrast, in the presence of GO nanosheets with a concentration of 10 μg mL−1, the two apparent α-helical bands experienced a slight decreased ellipticity in inverse proportion to the lateral size distribution of the GO nanosheets (Fig. 6b). As the concentration of the GO nanosheets was increased to 20 μg mL−1, a further decrease in the ellipticity of the CD spectra of the fibrinogen was evident (Fig. 6e). Additionally, we observed that the negative peak at 208 nm began attenuating, suggesting a probable disturbance to the secondary structure of the fibrinogen upon its binding with GO nanosheets at this particular concentration. This might be ascribed to the unraveling of the α-helix domains of fibrinogen. The attenuation of the 208 nm α-helical band of fibrinogen was significantly enhanced as the GO concentration was increased to 40 μg mL−1 (Fig. 6h). This indicated a major disruption to the secondary structure of fibrinogen leading to possible denaturation of fibrinogen.
Interestingly, for globulin, we noted that the ellipticity of its CD spectra decreased at a similar magnitude in the presence of GO-0 irrespective of its concentration (Fig. 6c, f and i). On the other hand, the CD spectra of globulin in the presence of GO nanosheets with other lateral size distributions (i.e., GO-10 to GO-240) followed that of pure globulin closely. In fact, a slight decrease in the spectra ellipticity could only be observed at the specific GO concentration of 40 μg mL−1. It is noteworthy that despite a reduced ellipticity of the 216 nm negative bands, the β-sheet and overall secondary structure of the globulin were still highly maintained in the presence of GO nanosheets with various lateral sizes and concentrations. Eventually, from all the CD spectra of the three plasma proteins, we clearly observed the lateral size and concentration-dependent effects induced by the GO nanosheets in influencing the secondary structures and overall conformational stability of the plasma proteins upon their interactions with GO nanosheets.
Altogether, based on the obtained molecular binding parameters, we proposed the existence of the molecular interactions between GO nanosheets and plasma proteins which are strongly dependent on the lateral sizes of the GO nanosheets (Scheme 1). GO-0, with its highest mean lateral size and size distribution, displayed the lowest fluorescence quenching efficiency (Fig. 4g–i) and association constant (Fig. 5j–l) for the three plasma proteins albeit the highest loading capacity of both albumin and globulin on GO-0 (Fig. 2b and f). In contrast, for fibrinogen, GO-0 displayed the lowest adsorption capacity (Fig. 2d). Along with the lowest fluorescence quenching efficiency and with increasing concentrations, GO-0 induced the lowest red-shift of the maximum fluorescence emission. Contrastingly, the highest fluorescence quenching efficiency, maximum emission red-shift, and association constant were exhibited by different GO nanosheet samples for different plasma proteins. Specifically, for albumin, the GO-30 sample with a mean size of 304 nm displayed the highest quenching efficiency (Fig. 4g) and association constant (Fig. 5j). For fibrinogen, the highest quenching efficiency (Fig. 4h) and association constant (Fig. 5k) were achieved with GO-120 with an average lateral size of 126 nm. On the other hand, the GO-10 sample with a mean lateral size of 350 nm quenched the intrinsic fluorescence of globulin (Fig. 4i) and associated with it (Fig. 5l) most effectively.
 | ||
| Scheme 1 Proposed distinct molecular interactions between GO nanosheets and the three plasma proteins based on the binding parameters of the three GO–protein complexes. | ||
It is worth noting that the highest loading capacity of albumin and globulin on GO-0 did not translate directly into the highest association and intrinsic fluorescence quenching efficiency. In fact, the protein association and quenching efficiency were at their lowest with GO-0. Conversely, GO-30 and GO-10, with a lower loading capacity for albumin and globulin, respectively, exhibited higher association and fluorescence quenching efficiency for the two plasma proteins. It is important to highlight that the intrinsic fluorescence of the plasma proteins is normally contributed by their aromatic amino acid residues of tryptophan, tyrosine, and phenylalanine.32 Adsorption of proteins onto GO generally shortens the relative distance between GO and the active intrinsic fluorescence emitters and this will lead to the quenching of the protein fluorescence. Fluorescence quenching will be enhanced if a particular protein possesses multiple binding sites with GO nanosheets. Moreover, some of these chromophore residues may be buried inside the hydrophobic protein cores and when exposed to their hydrophilic microenvironment post protein adsorption, further fluorescence quenching will occur. Variations in the fluorescence emission intensity, therefore, serve as one of the indicators of the changes in the local molecular microenvironment of the chromophore residues and probably, a conformational change of the plasma proteins. As such, the lower intrinsic fluorescence quenching exhibited by GO-0 could be attributed to the adsorption of more albumin and globulin with fewer binding sites coupled with minimal exposure of the aromatic residues of the plasma proteins to their local hydrophilic microenvironment.
On the contrary, GO-30 and GO-10, despite their lower albumin and globulin adsorption, respectively, quenched the intrinsic fluorescence of these proteins more efficiently, suggesting that the chromophore residues of both proteins were more exposed to the local microenvironment, and possibly a certain degree of conformational changes was experienced by albumin and globulin on GO-30 and GO-10, correspondingly. This was actually supported by our observation on the CD spectra (Fig. 6) as well as the red-shift of the maximum fluorescence emission of the plasma proteins induced by GO-0, GO-10, and GO-30 (Table 1). The maximum fluorescence emission wavelength serves as a crucial indicator of the changes of the local microenvironment of the protein chromophore residues.25 Specifically, the smallest red-shift of the maximum emission of the three plasma proteins was driven by GO-0 whilst the largest red-shift was induced by GO-30 and GO-10 on albumin and globulin, respectively. Intriguingly, the conventional direct correlation between a high adsorption and a high association and fluorescence quenching efficiency could only be noticed from fibrinogen when it bound to the GO-120 nanosheet sample. On the whole, based on the obtained GO–protein binding data, we rationalized the presence of critical mean lateral sizes of the GO nanosheets in achieving the highest association with the plasma proteins as well as the highest efficiency in quenching their intrinsic fluorescence, specific to albumin, fibrinogen, and globulin.
Plasma protein-mediated aggregation of GO nanosheets
Following the establishment of the biological effect induced by the GO nanosheets to the plasma proteins, we sought to determine the effect induced by plasma proteins on the physical state of the GO nanosheets. This effect was evaluated through the measurement of the absorbance of GO nanosheets in the absence and presence of plasma proteins (Fig. 7).From the experimental data, we noted that the absorption spectra of all GO nanosheet samples broadened in the presence of plasma proteins (Fig. 7a–f). This broadened spectra effect increased progressively with a corresponding decrease in the lateral size distribution of the GO nanosheets. More clearly, the absorption spectra of the GO–protein complexes of the GO-240 samples were much broader than those of other GO–protein complexes of other GO nanosheet samples. Nevertheless, we noticed that the broadening of the absorption spectra was not dependent on the types of the plasma proteins and their concentrations. In fact, a recent study has reported that the broadening of the absorption spectra of solid-state nanomaterials is directly correlated to the formation of nanomaterial aggregates.40 As such, here, we proposed that the broadening of the absorption spectra of the GO nanosheets with the addition of plasma proteins might be due to the formation of aggregates comprising either solely the GO nanosheets or the GO–protein complexes. Moreover, the broader spectra of the GO nanosheets with smaller lateral size distributions suggest that the formation of aggregates was significantly dependent on the lateral size of the GO nanosheets and that aggregation of GO nanosheets in the presence of plasma proteins might occur more easily for the small-sized GO nanosheets.
Interestingly, the plasma protein-induced broadening effect of the absorption spectra of all GO nanosheet samples was much more pronounced on Day 3 (Fig. 7g–l). It is evident that for a specific GO nanosheet sample, the absorption spectra of the GO–protein complexes were broader on Day 3 than on Day 0. Additionally, a similar trend of increased broadening of absorption spectra with decreased lateral size distributions of GO samples was also observed on Day 3. This indicates that the formation of the GO nanosheet or GO–protein complex aggregates in the presence of plasma proteins was probably time-dependent and could increase over time.
In short, from all the characterization data on the molecular interactions between GO and plasma proteins, we derived that as GO nanosheets induced particular biological effects on the plasma proteins, plasma proteins concurrently exerted certain influences on the physical state of GO nanosheets, notably on the formation of GO nanosheets or GO–protein aggregates.
Conclusions
In summary, we investigated the molecular interactions between GO with different lateral size distributions and the three essential human blood plasma proteins. On the basis of the different qualitative and quantitative data, we have illuminated the various facets of the GO–plasma protein interactions. Importantly, we have demonstrated that the lateral size distribution of the GO nanosheets plays a significant role in controlling the biological outcomes of the GO–plasma protein interactions. We foresee that this work will provide a better understanding of the dynamics of the GO–plasma protein interactions for improved design and utilization of graphene-based nanomaterials for different in vivo bioapplications. For instance, with the higher adsorption of albumin and lower adsorption of fibrinogen on GO nanosheets with large mean lateral sizes, one can possibly utilize these GO nanosheets with high molecular hemocompatibility for antithrombotic coating applications for decreasing the formation and occurrence of undesirable blood clots. Nonetheless, further works on the molecular interactions between plasma proteins and GO nanosheets with narrower (<100 nm) and non-overlapping size distributions are necessary and warranted. Despite challenges in developing facile synthesis of GO nanosheets with uniform and narrow size distributions which do not overlap with each other, we anticipate these to be overcome in the near future and further elucidations of the size-dependent effects and the specific underlying mechanisms of the GO–plasma protein interactions are expected.Methods
Materials
Albumin (A9511), fibrinogen (F4883), and globulin (G4386) in powder form were used directly without further purification (Sigma-Aldrich, St Louis, MO) and were prepared in phosphate-buffered saline (PBS 1×). The GO nanosheet sample, GO-0, was prepared according to Hummer's method. GO nanosheets with different lateral size distributions were then prepared by ultrasonication for 10, 30, 60, 120, and 240 min using an ultrasonic processor (SONICS, VCX-130).GO lateral size and size distribution characterization
GO suspensions were first dropped onto a freshly cleaved mica, followed by air drying. The surface topography of the as-deposited GO nanosheets was characterized using the tapping mode AFM (Bruker, Billerica, MA). The lateral size distribution of the GO nanosheets was subsequently evaluated by analyzing the obtained AFM images using the ImageJ software (NIH, US). More than 400 GO nanosheets were examined and measured to obtain the lateral size distributions and mean lateral sizes.Absorbance characterization
Absorption spectra of albumin, fibrinogen, globulin, and GO nanosheets were recorded using a UV-Vis spectrophotometer (NanoDrop 2000, Thermo Scientific). For free protein absorbance measurements, the plasma proteins were prepared at concentrations of 0.2, 0.4, 1, 2, 3, 4, 10, and 20 mg mL−1. For the measurement of the protein adsorption on GO, GO nanosheets of different lateral sizes were fixed at 200 μg mL−1. Plasma proteins were fixed at different concentrations of 0.2, 0.5, 1, 2, 5, and 10 mg mL−1. The plasma proteins (30 μL) were then mixed with GO nanosheets (30 μL) and vortexed for equilibration. Incubation of plasma proteins on GO nanosheet samples was performed at room temperature for 10 min. The mixture was subsequently centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for a duration of 7 min. The supernatant was subsequently obtained for spectrophotometric measurements at 280 nm to acquire the adsorption isotherm of all plasma proteins. The amount of plasma proteins adsorbed was determined from variations in the adsorption of proteins pre- and post-addition of the GO nanosheet samples. For the GO–protein aggregation absorbance measurement, the plasma protein concentrations were fixed at 5 μg mL−1 and 50 μg mL−1. Each spectrum was obtained as an average of at least three independent readings at room temperature.
000g for a duration of 7 min. The supernatant was subsequently obtained for spectrophotometric measurements at 280 nm to acquire the adsorption isotherm of all plasma proteins. The amount of plasma proteins adsorbed was determined from variations in the adsorption of proteins pre- and post-addition of the GO nanosheet samples. For the GO–protein aggregation absorbance measurement, the plasma protein concentrations were fixed at 5 μg mL−1 and 50 μg mL−1. Each spectrum was obtained as an average of at least three independent readings at room temperature.
Fluorescence quenching characterization
Intrinsic tryptophan fluorescence of proteins in the presence and absence of GO with different concentrations was recorded using a fluorescence microplate reader (Infinite M200, Tecan) in a black 96-well microplate (Nunc MicroWell, Thermo Scientific). Fluorescence excitation was set at 280 nm while emission was recorded from 306 to 450 nm. The plasma protein concentration was fixed at 500 μg mL−1 while that of GO was varied from 5, 10, 20, 25, 50, 100, 150, 200, to 250 μg mL−1. Three independent readings were performed at room temperature to obtain all quantitative parameters.Circular dichroism characterization
The structural stability of the plasma proteins was assessed utilizing circular dichroism (CD) spectroscopy. Using a quartz cell with a 1 mm optical path length, the CD spectrum was obtained using a Jasco J-810 spectropolarimeter. The range of wavelength was fixed from 190 to 260 nm while the scanning speed was fixed at 50 nm min−1. All CD spectra were collected as an accumulation of three successive scans at room temperature. The concentrations of albumin, fibrinogen, and globulin were set at 200, 400, and 400 μg mL−1, respectively. In contrast, the GO concentrations were varied progressively such that the concentration ratios of the plasma proteins to GO were fixed at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.025, 1
0.025, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.05, and 1
0.05, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1.
0.1.
Statistical analysis
Experimental data were presented as mean with standard deviation (mean ± standard deviation). Statistical differences between the data were evaluated based on Student's t-test and the statistically significant difference was represented by * for p < 0.1 and ** for p < 0.05 (i.e., 90% and 95% confidence intervals, respectively).Acknowledgements
This research was supported by the National Research Foundation, Prime Minister's Office, Singapore, under its medium-sized centre programme, Centre for Advanced 2D Materials, and its Research Centre of Excellence, Mechanobiology Institute, of the National University of Singapore. The National Research Foundation Competitive Research Program Funding is also gratefully acknowledged.References
- D. R. Dreyer, A. D. Todd and C. W. Bielawski, Chem. Soc. Rev., 2014, 43, 5288–5301 RSC.
- S. Eigler and A. Hirsch, Angew. Chem., Int. Ed., 2014, 53, 7720–7738 CrossRef CAS PubMed.
- W. C. Lee, C. H. Lim, Kenry, C. Su, K. P. Loh and C. T. Lim, Small, 2015, 11, 963–969 CrossRef CAS PubMed.
- S. A. Sydlik, S. Jhunjhunwala, M. J. Webber, D. G. Anderson and R. Langer, ACS Nano, 2015, 9, 3866–3874 CrossRef CAS PubMed.
- Y. Zhang, T. R. Nayak, H. Hong and W. Cai, Nanoscale, 2012, 4, 3833–3842 RSC.
- C. Chung, Y.-K. Kim, D. Shin, S.-R. Ryoo, B. H. Hong and D.-H. Min, Acc. Chem. Res., 2013, 46, 2211–2224 CrossRef CAS PubMed.
- D. Bitounis, H. Ali-Boucetta, B. H. Hong, D.-H. Min and K. Kostarelos, Adv. Mater., 2013, 25, 2258–2268 CrossRef CAS PubMed.
- Y. Yang, A. M. Asiri, Z. Tang, D. Du and Y. Lin, Mater. Today, 2013, 16, 365–373 CrossRef CAS.
- K. Kostarelos and K. S. Novoselov, Science, 2014, 344, 261–263 CrossRef CAS PubMed.
- C.-H. Lu, H.-H. Yang, C.-L. Zhu, X. Chen and G.-N. Chen, Angew. Chem., Int. Ed., 2009, 48, 4785–4787 CrossRef CAS PubMed.
- Y. Liu, D. Yu, C. Zeng, Z. Miao and L. Dai, Langmuir, 2010, 26, 6158–6160 CrossRef CAS PubMed.
- A. M. H. Ng, Kenry, C. Teck Lim, H. Y. Low and K. P. Loh, Biosens. Bioelectron., 2015, 65, 265–273 CrossRef CAS PubMed.
- X. Sun, Z. Liu, K. Welsher, J. Robinson, A. Goodwin, S. Zaric and H. Dai, Nano Res., 2008, 1, 203–212 CrossRef CAS PubMed.
- Q. Liu, L. Wei, J. Wang, F. Peng, D. Luo, R. Cui, Y. Niu, X. Qin, Y. Liu, H. Sun, J. Yang and Y. Li, Nanoscale, 2012, 4, 7084–7089 RSC.
- L. Zhang, J. Xia, Q. Zhao, L. Liu and Z. Zhang, Small, 2010, 6, 537–544 CrossRef CAS PubMed.
- C. L. Weaver, J. M. LaRosa, X. Luo and X. T. Cui, ACS Nano, 2014, 8, 1834–1843 CrossRef CAS PubMed.
- J. T. Robinson, S. M. Tabakman, Y. Liang, H. Wang, H. Sanchez Casalongue, D. Vinh and H. Dai, J. Am. Chem. Soc., 2011, 133, 6825–6831 CrossRef CAS PubMed.
- M. Fiorillo, A. F. Verre, M. Iliut, M. Peiris-Pagés, B. Ozsvari, R. Gandara, A. R. Cappello, F. Sotgia, A. Vijayaraghavan and M. P. Lisanti, Graphene oxide selectively targets cancer stem cells, across multiple tumor types: Implications for non-toxic cancer treatment, via “differentiation-based nano-therapy”, 2015 Search PubMed.
- Kenry, K. P. Loh and C. T. Lim, Small, 2015, 11, 5105–5117 CrossRef CAS PubMed.
- I. Lynch and K. A. Dawson, Nano Today, 2008, 3, 40–47 CrossRef CAS.
- A. E. Nel, L. Madler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- B. D. Chithrani and W. C. W. Chan, Nano Lett., 2007, 7, 1542–1550 CrossRef CAS PubMed.
- A. Lesniak, A. Campbell, M. P. Monopoli, I. Lynch, A. Salvati and K. A. Dawson, Biomaterials, 2010, 31, 9511–9518 CrossRef CAS PubMed.
- Z. J. Deng, M. Liang, M. Monteiro, I. Toth and R. F. Minchin, Nat. Nanotechnol., 2011, 6, 39–44 CrossRef CAS PubMed.
- S. H. D. P. Lacerda, J. J. Park, C. Meuse, D. Pristinski, M. L. Becker, A. Karim and J. F. Douglas, ACS Nano, 2010, 4, 365–379 CrossRef PubMed.
- S. Li, A. N. Aphale, I. G. Macwan, P. K. Patra, W. G. Gonzalez, J. Miksovska and R. M. Leblanc, ACS Appl. Mater. Interfaces, 2012, 4, 7069–7075 CAS.
- Z. Ding, H. Ma and Y. Chen, RSC Adv., 2014, 4, 55290–55295 RSC.
- Z. Ding, Z. Zhang, H. Ma and Y. Chen, ACS Appl. Mater. Interfaces, 2014, 6, 19797–19807 CAS.
- C. G. Salzmann, V. Nicolosi and M. L. H. Green, J. Mater. Chem., 2010, 20, 314–319 RSC.
- L. A. L. Tang, W. C. Lee, H. Shi, E. Y. L. Wong, A. Sadovoy, S. Gorelik, J. Hobley, C. T. Lim and K. P. Loh, Small, 2012, 8, 423–431 CrossRef CAS PubMed.
- S. Liu, M. Hu, T. H. Zeng, R. Wu, R. Jiang, J. Wei, L. Wang, J. Kong and Y. Chen, Langmuir, 2012, 28, 12364–12372 CrossRef CAS PubMed.
- C. A. Royer, Chem. Rev., 2006, 106, 1769–1784 CrossRef CAS PubMed.
- P. Roach, D. Farrar and C. C. Perry, J. Am. Chem. Soc., 2005, 127, 8168–8173 CrossRef CAS PubMed.
- Q. Xiao, S. Huang, Z.-D. Qi, B. Zhou, Z.-K. He and Y. Liu, Biochim. Biophys. Acta, 2008, 1784, 1020–1027 CrossRef CAS PubMed.
- X. Ling, L. Xie, Y. Fang, H. Xu, H. Zhang, J. Kong, M. S. Dresselhaus, J. Zhang and Z. Liu, Nano Lett., 2010, 10, 553–561 CrossRef CAS PubMed.
- H. Zhang, S. Jia, M. Lv, J. Shi, X. Zuo, S. Su, L. Wang, W. Huang, C. Fan and Q. Huang, Anal. Chem., 2014, 86, 4047–4051 CrossRef CAS PubMed.
- Z. J. Deng, M. Liang, I. Toth, M. J. Monteiro and R. F. Minchin, ACS Nano, 2012, 6, 8962–8969 CrossRef CAS PubMed.
- J. Deng, M. Sun, J. Zhu and C. Gao, Nanoscale, 2013, 5, 8130–8137 RSC.
- I. Lynch, T. Cedervall, M. Lundqvist, C. Cabaleiro-Lago, S. Linse and K. A. Dawson, Adv. Colloid Interface Sci., 2007, 134–135, 167–174 CrossRef CAS PubMed.
- L. Yang, Y. Yu, J. Zhang, F. Ge, J. Zhang, L. Jiang, F. Gao and Y. Dan, Chem. – Asian J., 2015, 10, 1215–1224 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2016 |