Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A nucleic acid strand displacement system for the multiplexed detection of tuberculosis-specific mRNA using quantum dots†
H. D.
Gliddon‡
ab,
P. D.
Howes‡
a,
M.
Kaforou
b,
M.
Levin
b and
M. M.
Stevens
*a
aDepartment of Materials, Department of Bioengineering, and Institute of Biomedical Engineering, Imperial College London, UK. E-mail: m.stevens@imperial.ac.uk
bSection of Paediatrics and Wellcome Trust Centre for Global Health Research, Division of Infectious Diseases, Department of Medicine, Imperial College London, UK
First published on 18th April 2016
Abstract
The development of rapid, robust and high performance point-of-care diagnostics relies on the advancement and combination of various areas of research. We have developed an assay for the detection of multiple mRNA molecules that combines DNA nanotechnology with fluorescent nanomaterials. The core switching mechanism is toehold-mediated strand displacement. We have used fluorescent quantum dots (QDs) as signal transducers in this assay, as they bring many benefits including bright fluorescence and multiplexing abilities. The resulting assay is capable of multiplexed detection of long RNA targets against a high concentration of background non-target RNA, with high sensitivity and specificity and limits of detection in the nanomolar range using only a standard laboratory plate reader. We demonstrate the utility of our QD-based system for the detection of two genes selected from a microarray-derived tuberculosis-specific gene expression signature. Levels of up- and downregulated gene transcripts comprising this signature can be combined to give a disease risk score, making the signature more amenable for use as a diagnostic marker. Our QD-based approach to detect these transcripts could pave the way for novel diagnostic assays for tuberculosis.
The ‘-omics’ areas of biomedical research (transcriptomics, genomics, proteomics, metabolomics and lipidomics) are revealing ever more complex and subtle relationships between biomolecular signatures and health status. Such studies are identifying biomarkers with the potential to be developed into diagnostic tests. However, the detection of these biomarkers is often beyond the capabilities of current diagnostic technologies. The future of molecular diagnostics relies on technological developments in various research areas to create diagnostics that can make use of those biomarkers that are too hard to detect or otherwise beyond the reach of current diagnostic approaches in a clinical setting.
A vast amount of information concerning the health of an individual can be derived from studying their blood, and in particular its biomolecular make-up, which changes with health status. Studying the molecular content of blood (and other physiological samples) for signatures of biomolecular change is the principal behind molecular diagnostics, an approach that can yield vital information with minimal invasiveness. Of course molecular diagnostics is a firmly established art, and techniques such as enzyme-linked immunosorbent assays (ELISAs) for proteins and polymerase chain reaction (PCR)-based assays for nucleic acids offer viable ways of detecting and quantifying molecular biomarkers of disease in a clinical setting. There are however some impediments with these and related technologies. Critically, they are slow and cumbersome, and although medical practice can work with this in some situations, in others (such as time-dependent care and in resource-limited settings) they are a critical road block.
DNA nanotechnology, where nucleic acids are used to build structural and/or mechanistic systems, has emerged as an area that could prove transformative in the way we detect genomic biomarkers, both in vitro and in cell studies.1–4 Of specific interest here is a process called ‘toehold-mediated strand displacement’, which has become an important mechanism upon which to base dynamic DNA systems.5 Here, an incoming invading strand binds to an overhanging toehold region of a DNA duplex, then proceeds to displace the incumbent DNA strand by branch migration. The displacement is driven forward by the net gain in paired bases as the system naturally moves towards a lower energy configuration.6 This mechanism has been used in various systems related to DNA nanotechnology, including DNA logic circuits,7 mechanical structures,8 and nanomachines.9 Importantly, strand displacement mechanisms have been used to good effect in biosensing systems, and have been used to detect DNA,10 RNA,11 proteins and small molecules.12 Biosensing mechanisms must undergo a chemical or physical change upon interaction with their specific target, but must not respond to interaction with any non-target species. By nature of the Watson-Crick base pairing of the toehold and the target nucleic acid, toehold-mediated strand displacement is highly specific to its target. Furthermore, the inherent action of strand displacement leads to a structural change that can readily form the basis of a switching event. The switching event can either lead directly to a signal change itself,12 or it can initiate a downstream event that leads to a signal change.11
Another research area that has contributed to a push towards advanced molecular diagnostics is nanoparticle-based biological sensors.13,14 A key advantage of using nanoparticles as signal transducers is the ability to construct homogeneous (i.e. completely solution phase) assays, which typically offer greatly improved reaction kinetics and sensitivity compared to heterogeneous (i.e. surface-based) assays. Fluorescent inorganic semiconductor nanoparticles (so-called ‘quantum dots’, QDs)15 have been proven as effective signal transducers in highly sensitive biosensing systems.13,16–22 QDs offer extremely high fluorescence brightness, narrow emission and broad absorption spectra, large Stokes shift, high quantum yields and excellent resistance to photodegradation.23 As their emission spectra are extremely narrow, tunable by particle size (larger QDs are redder, smaller ones bluer), and absorbance spectra very wide, a mixed population of QDs can be excited with a single high-energy excitation source, and will emit in well-defined and spectrally separate emission bands. Linking these different colors of QDs to different target biomolecules in solution allows for multiplexed detection.24–31
Critically for the construction of QD biosensors, their emission characteristics (emission intensity, spectral profile or polarization) can be altered in response to interaction with target biomolecules in solution, creating the basis of a biosensing system. This is typically based on resonance energy transfer (RET) from the QD core to an external energy acceptor such as an organic molecular dye or a gold nanoparticle.32 A key task then is to develop mechanisms that can take advantage of their extraordinary properties. QDs can be readily conjugated with biomolecules using a large number of different methodologies,33,34 which allows us to build biomolecular-based sensing mechanisms directly onto compact QDs in solution.
A large variety of homogeneous QD assays for nucleic acid detection have been developed to date.35,36 These predominantly rely on sandwich-like detection mechanisms, where a dye-labeled signal probe is brought into close proximity of the QD surface by the formation of a surface-bound duplex.35 However, such mechanisms may suffer from steric hindrance and reduced reaction kinetics due to overloading nanoparticle surfaces, and the dye-QD separation distance is relatively large which is a significant drawback in such systems.
In this work we have combined the advantages of toehold-mediated strand displacement systems with those of QDs to produce a novel multiplexed assay for the detection of RNA. We demonstrate the efficacy of this system by detecting synthetic mRNA analogues as biomarkers of tuberculosis (TB) infection.
The control of TB is severely hindered by a lack of effective diagnostic tests. Current diagnosis depends on analysis of clinical and radiological signs, as well as other diagnostic tests, which are either insensitive, unspecific or time-consuming, leading to missed diagnoses and poor patient outcomes. In much of sub-Saharan Africa, HIV prevalence is also high, further complicating the diagnosis of TB. In patients co-infected with HIV and TB, the probability of developing extrapulmonary TB is increased,37 meaning sputum (mucus from the lower airways) samples and chest X-rays are of little use in diagnosing the disease. Unfortunately, most TB diagnostic assays in use today rely on sputum as a clinical sample. A more useful clinical sample would be whole blood, specifically purified RNA from whole blood.
Detection of mRNA is potentially a very powerful way of diagnosing a range of diseases. Studying gene expression patterns using microarray analysis has led to a deeper understanding of the pathogenesis of a number of diseases, as well as the identification of blood or tissue transcriptomic profiles.38,39 However, the use of these transcript signatures as diagnostic biomarkers has been limited by the large number of transcripts that comprise gene expression signatures and the technical and cost restraints required to measure gene expression. In an effort to reduce this complexity, a recent study successfully identified much reduced gene signatures for TB.40 A key advance in this work was the development of a ‘disease risk score’ (DRS) to simplify the analysis of the microarray intensities, where the summed intensities of downregulated genes were subtracted from the summed intensities of the upregulated genes. This allowed a single number to be calculated for each patient that indicated the probability that they had TB with high sensitivity and specificity, making the gene signatures much more amenable for use as biomarkers in diagnostic assays.
We selected two genes from the 44-gene signature for distinguishing TB from other diseases reported by this study to use as proof of principle biomarkers for our assay. These genes, GBP6 and TMCC1, are up- and downregulated respectively in patients with TB compared to patients with other diseases with similar clinical presentation.40
Our work addresses some key design challenges that must be overcome to realize effective multiplexed QD-based assays for detecting long nucleic acid targets as part of minimal gene panels. These are:
1. For large target molecules (e.g. long DNA/RNA strands), it is advantageous to have the target substrate decoupled from the nanoparticle surface, as steric hindrance of several large molecules interacting with a single nanoparticle may restrict reaction kinetics and curtail the advantages of colloidal systems.
2. For multiplexed measurements with QDs, each detection mechanism needs to be orthogonal such that a specific target interacts with a specific species of QD, and the mechanisms do not cross-react.
3. For Förster resonance energy transfer (FRET)-based sensing mechanisms, it is imperative to minimize the separation between the QD core and the FRET acceptor molecule. This is difficult to achieve with sandwich-type approaches.
4. For detection of nucleic acids in complex biological solutions (e.g. against a high concentration background of nucleic acids), where non-target interferent biomolecules are present in a vast abundance compared to any target molecules, it is challenging to create nanoparticle-based biosensing mechanisms that work effectively and avoid non-specific interactions.
5. Homogenous systems, where no washing or purification steps are required, are highly desirable to reduce the time and difficulty of running an assay. However this is challenging as detection and signaling must occur in complex matrices.
The QD-based assay described herein is capable of multiplexed detection of long synthetic RNA targets against a high concentration of background non-target RNA, with high sensitivity and specificity, and a low limit of detection (LOD), using a standard laboratory plate reader. Furthermore, the target RNA does not come into contact with the QDs, long RNA targets can be detected, orthogonal components allow multiplexing, QD–acceptor distance is minimized and no washing steps are required. We demonstrate the utility of our QD-based system for the detection of two genes selected from a TB-specific gene expression signature.
Materials and methods
Materials
Nuclease-free water (not DEPC treated), Qdot655 and Qdot525 ITK-streptavidin conjugate kits were purchased from Life Technologies. All other reagents were purchased from Sigma Aldrich, UK. Quencher probes (Qprobes) were purchased from IBA Life Sciences, Germany. All other oligonucleotides were purchased from Integrated DNA Technologies Inc., Belgium. All oligonucleotides were HPLC purified and freeze-dried by the supplier. Oligonucleotides were used as provided and dissolved in nuclease-free water to give stock solutions of 20 μM (probes) or 50 μM (targets). The sequences of the oligonucleotides used are detailed in Table 1. All assays were carried out in black polystyrene half volume Corning® 96 Well Plates (Sigma) and fluorescence emission was recorded using an EnSpire® Multimode Plate Reader (Perkin Elmer).| Name | Nucleic acid type | Sequence | Modifications | Length |
|---|---|---|---|---|
| GBP6 T | RNA | GCU UAU UAG AGG AUA UCA GUG CCU GGC CCA CAU GAG AGA ACA GAU CCA UAC ACA CUU UGA AAA ACU UUG U | — | 70 |
| GBP6 T* | DNA | TCA AAG TGT GTA TGG ATC TGT TCT CTC ATG TGG GCC AGG CAC TGA TAT CC | — | 50 |
| GBP6 BP | DNA | AAT CAA AGT ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) TG TAT GGA T TG TAT GGA T![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) T GTT CTC T T GTT CTC T |
Biotin at 5′ | 28 |
| GBP6 QProbe | DNA | AGA GAA CAG ATC CAT ACA CAC TTT GA | DYQ1 at 3′ | 26 |
| TMCC1 T | RNA | UGU AAU CUG ACA CAG GCA AAA UUA UGG UUC CCA CAC CCC AAC CCC AAA UGA AAC CUG GGA UUU UGA AUG U | — | 70 |
| TMCC1 T* | DNA | TCC CAG GTT TCA TTT GGG GTT TGG GTG TGG GAA CCA TAA TTT TGC CTG TG | — | 50 |
| TMCC1 BP | DNA | AAT CCC AG![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) TTT CAT TTG TTT CAT TTG ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GG TTT GGG T GG TTT GGG T |
Biotin at 5′ | 28 |
| TMCC1 QProbe | DNA | ACC CAA ACC CCA AAT GAA ACC TGG GA | DYQ2 at 3′ | 26 |
| GBP6 T* Comp | DNA | TGG TGG CCG GGT AGG ATG ATC CTG AGC ATC AGT GCC TGG CCC AGT ACA GCT TGA GAG ACC CCC TTC ACT C | — | 70 |
| GBP6 QProbe Comp | DNA | TGG CTC CCC GCC CCT CGG TCT CCA CGT GGT GTA TGG ATC TGT GGT CAT TGT CCC TCT GCA GAA TAA AGA T | — | 70 |
| GBP6 BP Comp | DNA | GGA CTA CGT CAA CAA GGA GCT GGC CCT GCA CAT CCA TAC AGA CAT CGA CAG CCA GGG TAT CTT CTT CAC A | — | 70 |
Kinetics of strand displacement
QProbe and T* (the detection complex), mixed at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, were first hybridized in Tris buffer (10 mM Tris-HCl, 150 mM NaCl, 5 mM MgCl2, pH 7.4) and were subjected to a temperature gradient (80 °C for 5 minutes, down to 25 °C, falling by 1 °C every 60 seconds). Then 8 pmoles of each hybridization reaction was added to a well, with SYBR Safe (Life Technologies) added to a final concentration of ×2.5 in the Tris buffer. One molar equivalent of target was added to make a final concentration of 80 nM target and the total volume was 100 μL. Fluorescence emission was read following excitation at 480 nm.
1 ratio, were first hybridized in Tris buffer (10 mM Tris-HCl, 150 mM NaCl, 5 mM MgCl2, pH 7.4) and were subjected to a temperature gradient (80 °C for 5 minutes, down to 25 °C, falling by 1 °C every 60 seconds). Then 8 pmoles of each hybridization reaction was added to a well, with SYBR Safe (Life Technologies) added to a final concentration of ×2.5 in the Tris buffer. One molar equivalent of target was added to make a final concentration of 80 nM target and the total volume was 100 μL. Fluorescence emission was read following excitation at 480 nm.
Analysing the kinetics of QProbe–CProbe hybridization
QDs were functionalized with a Capture Probe (Cprobe)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) QD ratio of 30
QD ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for TMCC1 and 20
1 for TMCC1 and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for GBP6, in borate buffer (50 mM borate, 150 mM NaCl, 0.01 % BSA (w/v), pH 8.3) for 60 minutes at room temperature in the dark. One molar equivalent of QProbe was added, bringing the final volume to 50 μL. Fluorescence was monitored for 70 minutes with excitation at 405 nm.
1 for GBP6, in borate buffer (50 mM borate, 150 mM NaCl, 0.01 % BSA (w/v), pH 8.3) for 60 minutes at room temperature in the dark. One molar equivalent of QProbe was added, bringing the final volume to 50 μL. Fluorescence was monitored for 70 minutes with excitation at 405 nm.
Assay protocol
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio was performed in Tris buffer. Solutions were heated to 80 °C for 5 minutes and then subjected to a temperature gradient to 25 °C, falling by 1 °C every 60 seconds to achieve maximum hybridization. QDs were functionalized with CProbe (at a CProbe
1 ratio was performed in Tris buffer. Solutions were heated to 80 °C for 5 minutes and then subjected to a temperature gradient to 25 °C, falling by 1 °C every 60 seconds to achieve maximum hybridization. QDs were functionalized with CProbe (at a CProbe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) QD ratio of 30
QD ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for TMCC1 and 20
1 for TMCC1 and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for GBP6) in borate buffer for 60 minutes at room temperature in the dark.
1 for GBP6) in borate buffer for 60 minutes at room temperature in the dark.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) QProbe ratio was 1
QProbe ratio was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to bring the total assay volume to 25 μL with a final QD concentration of 4 nM. Fluorescence emission was recorded using an excitation wavelength of 405 nm. Error bars in figures are calculated from technical triplicates. Dose–response curves were plotted in Origin 2015 (Origin Lab), and fitted using the dose–response function.
1) to bring the total assay volume to 25 μL with a final QD concentration of 4 nM. Fluorescence emission was recorded using an excitation wavelength of 405 nm. Error bars in figures are calculated from technical triplicates. Dose–response curves were plotted in Origin 2015 (Origin Lab), and fitted using the dose–response function.
Purification of RNA from whole blood
Whole blood taken from healthy volunteers was collected in PAXgeneTM tubes and stored at −80 °C. PAXgene™ Blood RNA Extraction Kits (Qiagen) were used to extract RNA before analyzing concentration and purity on a Nanodrop 1000 Spectrophotometer (Thermo Scientific).Ethics statement
Healthy Volunteers from the Department of Medicine, Imperial College London were recruited under an Imperial College Healthcare Tissue Bank (ICHTB) Human Tissue Act License (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 275) and a Research Ethics Committee Wales approval (12/WA/0196). Tissue Bank Application Number Project R13062; exploring genetic and cellular immune responses in children with life threatening infections. Written information was provided and all participants provided written consent.
275) and a Research Ethics Committee Wales approval (12/WA/0196). Tissue Bank Application Number Project R13062; exploring genetic and cellular immune responses in children with life threatening infections. Written information was provided and all participants provided written consent.
Results and discussion
Assay concept
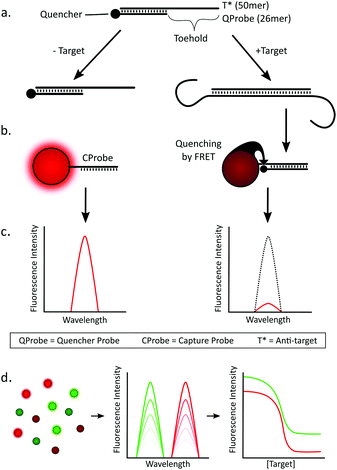
The core concept of our assay design is that the target nucleic acid does not come into direct contact with the QD (see Scheme 1). Instead, there is an intermediate DNA construct (termed the ‘detection complex’) that interacts directly with the target RNA, releasing a short quencher-labeled DNA probe (termed the ‘quencher probe’, QProbe) that subsequently binds to the QD. The detection complex consists of a 50mer DNA sequence (termed the anti-target or T*) that is complementary to a corresponding 50mer recognition sequence on the target RNA. Hybridized with this T* strand is the shorter 26mer QProbe, yielding a remaining 24mer ssDNA region that acts as the toehold for strand displacement upon binding to the RNA target. Additionally, these complexes are highly stable due to their relatively high Tm (ca. 65 °C for GBP6 and 73 °C for TMCC1, at 150 mM NaCl) therefore it should only dissociate in the presence of its specific target. The first step of the assay is to incubate the detection complex with the target, which induces strand displacement and yields a population of QProbes that are unbound in solution (Scheme 1a). In a second step, a population of QDs were functionalized with ssDNA surface probes (we term ‘capture probes’, CProbes) that are complementary to the QProbes (Scheme 1b). Specific hybridization of the QProbe and CProbe brings the dye into close proximity of the QD and allows FRET to occur from the QD (donor) to the dye (acceptor). The decrease in fluorescence of the QD is then the signal that the target has been detected (Scheme 1c). From a mixed population of QDs (Scheme 1d), simultaneous measurement of non-overlapping emission spectra can be made corresponding to different target mRNAs, which can then be converted into corresponding dose–response relationships.Melting temperatures for QProbe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CProbe hybridization were reduced by incorporating two mismatches into each CProbe, which reduced each Tm by approximately 10 °C (Fig. S1†). This resulted in the QProbe preferentially binding to the T* over the CProbe, reducing the likelihood of false positive results.
CProbe hybridization were reduced by incorporating two mismatches into each CProbe, which reduced each Tm by approximately 10 °C (Fig. S1†). This resulted in the QProbe preferentially binding to the T* over the CProbe, reducing the likelihood of false positive results.
With reference to the key design challenges that we identified above, our assay exhibits the following advantages: (1) the long RNA target does not come into direct contact with the QD, which negates steric hindrance issues that could occur with multiple long strand targets interacting at the surface of the nanoparticle. (2) The target induced signaling cascade is based purely on nucleic acid base pairing between complementary sequences, therefore many cascades can run in parallel without cross-reactivity. This makes the assay highly suitable for multiplexed detection using multiple colors of QD. (3) The donor–acceptor (QD–quencher) distance is minimized as the QProbe binds to the QD with the quencher at the proximal end of the CProbe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) QProbe duplex, allowing for maximal energy transfer and change in fluorescence signal per target present in the system. Such an orientation of probe is difficult to obtain using common ‘sandwich’ approaches. We note here that this distance could readily be decreased yet further using QDs with more compact capping layers, such as thiolated zwitterionic ligands.41 (4) As the signal cascade is based on highly specific nucleic acid hybridization there is little interference from an abundant background of non-target RNA or DNA. (5) This is a homogeneous two-step assay that requires no washing or purification steps, and functions well even in a background of non-target nucleic acids. Additionally – as there is no need to wash or purify – the assay time is limited only by the two incubation steps, which we show are both relatively fast.
QProbe duplex, allowing for maximal energy transfer and change in fluorescence signal per target present in the system. Such an orientation of probe is difficult to obtain using common ‘sandwich’ approaches. We note here that this distance could readily be decreased yet further using QDs with more compact capping layers, such as thiolated zwitterionic ligands.41 (4) As the signal cascade is based on highly specific nucleic acid hybridization there is little interference from an abundant background of non-target RNA or DNA. (5) This is a homogeneous two-step assay that requires no washing or purification steps, and functions well even in a background of non-target nucleic acids. Additionally – as there is no need to wash or purify – the assay time is limited only by the two incubation steps, which we show are both relatively fast.
Component testing
The assay relies on two main mechanisms. The first is the target-induced separation of the detection complex by toehold induced strand displacement. The second is the liberated QProbes binding to the QD. These two mechanisms were tested separately before constructing and testing the final assay.The QDs used in this work are coated with streptavidin (SA) to allow facile conjugation with biotinylated DNA. Each streptavidin tetramer has four biotin binding sites, but their availability on the QD surface is orientation-dependent. Additionally, different species of QD-SA conjugates are different sizes and therefore have varying capacities to bind biotin due to their variations in surface area. As there are multiple quenchers binding to a single QD, it is important to determine an upper limit for the ratio of quencher to QD in terms of signal delta for each additional quencher added i.e. we want to operate in a region where each additional quencher still has a significant quenching effect on the QD. By pre-hybridizing the QProbe and the CProbe, adding this into the QDs at increasing ratios and observing the quenching of the QDs, we determined this upper limit to be approximately 20 for QD525s and 30 for QD655s (Fig. S2†). Therefore in the assay, ratios of CProbe to QD were 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for the QD525s and 30
1 for the QD525s and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for the QD655s. The QD525s were paired with a DYQ1 quencher and the QD655s were paired with a DYQ2 quencher.
1 for the QD655s. The QD525s were paired with a DYQ1 quencher and the QD655s were paired with a DYQ2 quencher.
The theoretical limit of detection (LOD) of the assay is dependent on the QD fluorescence signal delta for binding a given number of QProbes. Additionally the concentration of QDs used is critical as there is direct linear proportionality between the target concentration and the QD fluorescence response – therefore the lower the concentration of QDs that can give a significant and repeatable signal intensity on a given fluorescence reader (in this case a commercial plate reader) the lower the ultimate LOD. We used a reaction volume of 25 μl and a QD concentration of 4 nM. The theoretical LOD for the quenching mechanism was determined by adding in an increasing ratio of QProbe to a QD solution, and subtracting three standard deviations from the mean of the control (no QProbe). Here we determined an LOD of 1.6 nM for QD525-DYQ1 (GBP6 QProbe detection) and 6.4 nM for QD655-DYQ2 (TMCC1 QProbe detection). Plots and calculations are shown in Fig. S3.†
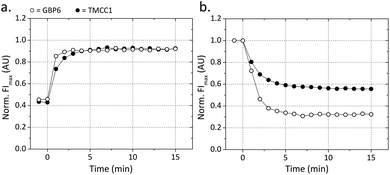
It is desirable for point-of-care applications to minimize assay run time. As incubation steps can take up a significant portion of assay time it is useful to have a thorough understanding of how long each reaction step needs in order to reach completion to avoid unnecessary time loss. In this assay there are two incubation steps: firstly the target addition to the detection complex, and secondly the addition of the QDs to the solution (to bind free QProbes). Two kinetic experiments were conducted to characterize these steps. For the target–detection complex step, the intercalating dye SYBR Safe was used to follow the kinetics of toehold-mediated strand displacement. When SYBR Safe is intercalated in dsDNA its fluorescence intensity (FI) increases by ca. four orders of magnitude, therefore it can be used to monitor the proportion of dsDNA versus ssDNA in a given system. Here, the detection complex – which contains 26 base pairs – was incubated with SYBR Safe. When the target was added (at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, target to detection complex), it displaced the QProbe and formed a 50 bp dsDNA duplex with the T* strand. This increase in total base pairs (24 per complex, ca. two-fold) led to a two-fold increase in overall FI (Fig. 1a). For the GBP6 target, the displacement reaction went to completion after only two minutes. For the TMCC1 target, displacement took slightly longer, but went to completion after only five minutes. To characterize the second incubation step, quencher probes were added to their respective QDs (at a ratio of 20
1, target to detection complex), it displaced the QProbe and formed a 50 bp dsDNA duplex with the T* strand. This increase in total base pairs (24 per complex, ca. two-fold) led to a two-fold increase in overall FI (Fig. 1a). For the GBP6 target, the displacement reaction went to completion after only two minutes. For the TMCC1 target, displacement took slightly longer, but went to completion after only five minutes. To characterize the second incubation step, quencher probes were added to their respective QDs (at a ratio of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for GBP6 QProbe
1 for GBP6 QProbe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) QD525 and 30
QD525 and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for TMCC1 QProbe
1 for TMCC1 QProbe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) QD655), and the fluorescence quenching of the QDs upon RET to the proximal acceptors was used to characterize the kinetics of this reaction. For both the QD525s and the QD655s, maximal quenching was reached after ca. 5 min (Fig. 1b). From these two experiments it is shown that incubation steps of little over 5 min are required during the assay.
QD655), and the fluorescence quenching of the QDs upon RET to the proximal acceptors was used to characterize the kinetics of this reaction. For both the QD525s and the QD655s, maximal quenching was reached after ca. 5 min (Fig. 1b). From these two experiments it is shown that incubation steps of little over 5 min are required during the assay.
Full assay
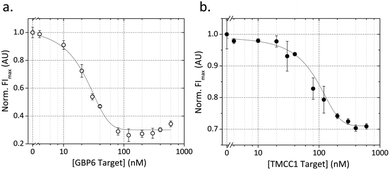
The full assay was tested in two stages. Firstly, the two targets were detected separately in monoplex assays and characterized as individual assays. Secondly, in order to demonstrate multiplexing with this system, we combined the two assays and showed that the two targets can be detected simultaneously in a single solution.The dose–response curves for the GBP6 and TMCC1 monoplex assays are shown in Fig. 2. In both cases, the QD fluorescence intensity (FI) is seen to decrease with increasing target concentration, due to target-induced displacement of QProbes and hybridization to the CProbe. Conducting experiments with higher resolution in the low target concentration regime, we determine the LODs to be below 10 nM for detection of both GBP6 and TMCC1 synthetic RNA targets (see Fig. S4†). As the response of the assays is sharp, the saturation point of each system is above 70 nM, resulting in a somewhat limited dynamic range. However, the assay could be readily tuned across a much larger dynamic range for specific targets by varying assay component concentrations and ratios.
The point of inflection of the dose–response curve of TMCC1 is shifted to a higher target concentration versus that of GBP6. We ascribe this to enhanced stability of the TMCC1 detection complex compared to the GBP6 detection complex. The melting temperature (Tm) of the TMCC1 detection complex is 73 °C compared to 65 °C for the GBP6 complex, making the strand displacement of the TMCC1 QProbe from T* less energetically favorable than it is for GBP6. The kinetics of strand displacement (Fig. 1a) show that strand displacement is slower for the TMCC1 complex, which supports our assertion that this complex is harder to disrupt than the GBP6 detection complex. Nevertheless, the detection mechanism is shown to work in each case with a low LOD.
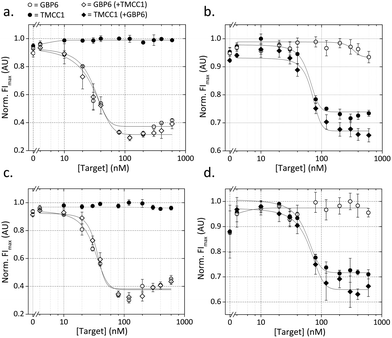
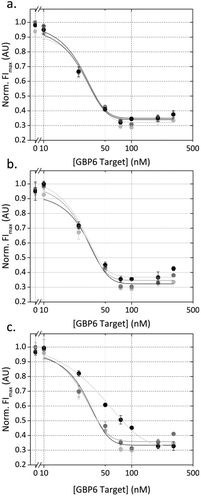
In order to demonstrate the potential for multiplexed analysis using this assay, the monoplex assays were combined and run simultaneously. Three conditions were explored for this experiment, with either target present or both targets present. Fig. 3a shows the response of the QD525 in the presence of either and both targets. When GBP6 is present, the QD fluorescence decreases in the same manner as the monoplex case. Fig. 3b shows the response of the QD655 for the same cases, and the QD fluorescence decreases as expected in the presence of TMCC1. Crucially, both QDs show no response to the presence of the ‘other’ target, with both showing flat response lines in Fig. 3a and b. This shows that there is no cross-reactivity in the system and that the two ‘cascades’ operate orthogonally to one another. Fig. 2a shows the appearance of an apparent ‘hook effect’ in the GBP6 assay, which could be caused by excess target RNA hybridizing to the CProbe, preventing it from hybridizing to the QProbe and thus reducing QD quenching. This could easily be overcome using multiple assay mixes, each optimized for the detection of a specific range of target concentrations.
Assay function against background RNA
Having shown that the assay mechanisms function well in pure buffer (i.e. target RNA present with no competing non-target RNA) we moved on to examine the behaviour of the assay in a more complex solution. For optimal assay function, it is imperative that non-target molecules in the solution do not interfere with the assay. For nanoparticle-based assays, such interference can cause false triggering or blocking of the sensing mechanism, or can directly affect the stability of the nanoparticles, resulting in either false positive or false negatives. In order to assess the resilience of our assay against interference from non-target nucleic acids, the assay was run in a solution containing 25 ng μL−1 concentration of RNA isolated from whole blood (this represents 25% reaction volume as standard yields are around 100 ng μL−1). This was designed to replicate the scenario of detecting target RNA directly from a total RNA isolation from whole blood. Fig. 3c and d shows the results of this experiment, and it can be seen that the form of the dose–response curves is well-matched to those conducted in pure buffer. This shows that the non-target RNAs are having little adverse affect on assay function, suggesting that the two detection cascades (for GBP6 and TMCC1) are functioning independently of one another and of other RNAs present in the solution.Assay function in presence of competing targets
The most likely clinical sample to be used in this assay is purified RNA from whole blood. This will contain a high concentration of other RNA molecules that could interfere with our detection system. We anticipated that false negatives could be caused by: (1) other nucleic acids blocking the toehold region of T*, which would prevent the target RNA from displacing the QProbe, or (2) other nucleic acids hybridizing to the CProbe or hybridizing to the QProbe (and thus preventing QProbe hybridizing to CProbe and causing quenching of the QD). This was investigated by designing GBP6 ‘competing targets’ for the toehold region of T*, the CProbe and the QProbe.These competing targets were designed by running a nucleotide BLAST (basic local alignment search tool) of each sequence against the Human genomic and transcript database. The result with the highest similarity that was complementary to the search sequence and that represented a coding RNA was used to design the competing target. The region of complementarity shown by the BLAST search (14–16 bp) was used, and equal sized flanking sequences were added either side, taken from the transcript sequence in question. Each resulting competing target was therefore a 70 nt RNA molecule. The GBP6 monoplex assay was run in the presence of 0, 10 or 100 nM of each of these competing targets. Both T* and CProbe competing targets had a negligible effect on the dose-response curve for GBP6 target detection (Fig. 4a and b). At 100 nM QProbe competing target, the dose-response curve was shifted slightly to the right, indicating that this RNA molecule was possibly hybridizing to the CProbe and partially inhibiting QProbe binding (Fig. 4c). However, 100 nM is well above the actual physiological concentration, so we consider this not to be problematic.
Sensitivity and specificity
The 50mer target recognition sequences used in these assays were selected on the basis of their lack of similarity to other transcripts within the human transcriptome, allowing specific detection of individual mRNA molecules. For this application, it is not necessary for the assay to discriminate between transcripts containing single nucleotide polymorphisms. Instead, we have detected the most biologically realistic interfering case of competing targets that could interfere with the assay (Fig. 4) at concentrations well above their physiological concentrations. The assay sensitivity required for the detection of individual transcripts is difficult to determine, especially because RNA purification yields can vary significantly between samples. Fortunately, the disease risk score method relies on the levels of upregulated transcripts compared to the levels of downregulated transcripts, so endogenous controls or ‘housekeeping genes’ should not be required when translating this into a point-of-care test.Conclusions
In this work, we developed a multiplexed assay capable of simultaneous detection of targets based on two transcripts selected from a gene expression signature, combining the advantages of toehold-mediated strand displacement with those of QDs to give an assay that is capable of sensitive and selective detection of synthetic RNA spiked in purified RNA from whole blood. With reference to the design challenges that we outlined at the end of the introduction, we explain how our system overcomes these: (1) the RNA targets do not come into direct contact with the QDs, which gets rid of steric hindrance issues and improves assay kinetics for long targets. (2) The system is completely reliant on highly specific nucleic acid hybridization and orthogonal cascades that allow for multiplexed detection of targets. (3) The detection mechanism on the surface is not a sandwich-type mechanism, instead the probe binds with the quencher at the proximal end of the surface bound duplex, thus maximizing energy transfer and minimizing the LOD of our system. This is a general characteristic that could be further improved using more compact nanoparticle capping layers. (4) The performance of the assay in pure buffer and against a high background of non-target RNA is shown to be highly similar, showing that the assay does not suffer from non-specific binding and interference. (5) The system is homogeneous as no washing or purification steps are required.Future high-performance diagnostic tools will likely combine several key enabling technologies. We envisage our QD-based toehold-mediated strand displacement assay could be combined with a simple enzyme-assisted isothermal amplification mechanism in order to achieve LODs low enough for direct detection of the target RNA transcripts from RNA isolations from whole blood. The assay that we have described could serve as a core concept upon which to build an advanced diagnostic test for TB diagnosis and monitoring.
Author contribution
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. M. L. is responsible for the ethics approvals for this project. Data requests can be made to m.stevens@imperial.ac.uk.Conflict of interest
The authors declare no competing financial interest.Acknowledgements
H. D. G. acknowledges support from the Medical Research Council (MRC, UK). P. D. H. acknowledges support from the Engineering and Physical Sciences Research Council (EPSRC Grant EP/K031953/1, UK). M. K. is supported by a European Union Framework Programme (TBVAC2020) grant (Ref no 643381). M. L. is a recipient of a NIHR Senior Fellowship Award, and supported by the Imperial College London Biomedical Research Centre (BRC), the Wellcome Trust Centre for Global Health Research, and a European Union Framework Programme (TBVAC2020) grant (Ref no 643381). M. M. S. acknowledges support from EPSRC through the Interdisciplinary Research Centre (IRC) “Early-Warning Sensing Systems for Infectious Diseases” (EP/K031953/1) and research grant “Bio-functionalised Nanomaterials for Ultrasensitive Biosensing” (EP/K020641/1). M. M. S. and M. L. were jointly awarded the Wellcome Trust Institutional Strategic Support Fund (ISSF): Networks of Excellence Award at Imperial College London under grant number 097816/Z/11/A. We thank Dr Lachlan Coin for useful scientific discussion and the feature selection software.Notes and references
- F. Zhang, J. Nangreave, Y. Liu and H. Yan, J. Am. Chem. Soc., 2014, 136, 11198–11211 CrossRef CAS PubMed.
- C. H. Lu, B. Willner and I. Willner, ACS Nano, 2013, 7, 8320–8332 CrossRef CAS PubMed.
- L. Qiu, C. Wu, M. You, D. Han, T. Chen, G. Zhu, J. Jiang, R. Yu and W. Tan, J. Am. Chem. Soc., 2013, 135, 12952–12955 CrossRef CAS PubMed.
- J. Hemphill and A. Deiters, J. Am. Chem. Soc., 2013, 135, 10512–10518 CrossRef CAS PubMed.
- J. Zhu, L. Zhang, S. Dong and E. Wang, ACS Nano, 2013, 7, 10211–10217 CrossRef CAS PubMed.
- G. Seelig, D. Soloveichik, D. Y. Zhang and E. Winfree, Science, 2006, 314, 1585–1588 CrossRef CAS PubMed.
- B. Yurke, A. J. Turberfield, A. P. Mills, F. C. Simmel and J. L. Neumann, Nature, 2000, 406, 605–608 CrossRef CAS PubMed.
- J. Bath and A. J. Turberfield, Nat. Nanotechnol., 2007, 2, 275–284 CrossRef CAS PubMed.
- D. A. Khodakov, A. S. Khodakova, A. Linacre and A. V. Ellis, J. Am. Chem. Soc., 2013, 135, 5612–5619 CrossRef CAS PubMed.
- S. Bi, J. Zhang, S. Hao, C. Ding and S. Zhang, Anal. Chem., 2011, 83, 3696–3702 CrossRef CAS PubMed.
- N. E. Larkey, C. K. Almlie, V. Tran, M. Egan and S. M. Burrows, Anal. Chem., 2014, 86, 1853–1863 CrossRef CAS PubMed.
- Z. Zhang, C. Hejesen, M. B. Kjelstrup, V. Birkedal and K. V. Gothelf, J. Am. Chem. Soc., 2014, 136, 11115–11120 CrossRef CAS PubMed.
- P. D. Howes, R. Chandrawati and M. M. Stevens, Science, 2014, 346, 1247390 CrossRef PubMed.
- P. D. Howes, S. Rana and M. M. Stevens, Chem. Soc. Rev., 2014, 43, 3835–3853 RSC.
- C. M. Tyrakowski and P. T. Snee, Phys. Chem. Chem. Phys., 2014, 16, 837–855 RSC.
- Z. W. Jin and N. Hildebrandt, Trends Biotechnol., 2012, 30, 394–403 CrossRef CAS PubMed.
- T. R. Pisanic Ii, Y. Zhang and T. H. Wang, Analyst, 2014, 139, 2968–2981 RSC.
- E. Petryayeva and W. R. Algar, Analyst, 2015, 140, 4037–4045 RSC.
- K. D. Wegner, Z. Jin, S. Linden, T. L. Jennings and N. Hildebrandt, ACS Nano, 2013, 7, 7411–7419 CrossRef CAS PubMed.
- W. Wang, A. Kapur, X. Ji, M. Safi, G. Palui, V. Palomo, P. E. Dawson and H. Mattoussi, J. Am. Chem. Soc., 2015, 137, 5438–5451 CrossRef CAS PubMed.
- J. C. Claussen, A. Malanoski, J. C. Breger, E. Oh, S. A. Walper, K. Susumu, R. Goswami, J. R. Deschamps and I. L. Medintz, J. Phys. Chem. C, 2015, 119, 2208–2221 CAS.
- L. Hu, X. Liu, A. Cecconello and I. Willner, Nano Lett., 2014, 14, 6030–6035 CrossRef CAS PubMed.
- E. Petryayeva, W. R. Algar and I. L. Medintz, Appl. Spectrosc., 2013, 67, 215–252 CrossRef CAS PubMed.
- E. R. Goldman, A. R. Clapp, G. P. Anderson, H. T. Uyeda, J. M. Mauro, I. L. Medintz and H. Mattoussi, Anal. Chem., 2003, 76, 684–688 CrossRef PubMed.
- E. Petryayeva and W. R. Algar, Anal. Chem., 2014, 86, 3195–3202 CrossRef CAS PubMed.
- S. B. Lowe, J. A. G. Dick, B. E. Cohen and M. M. Stevens, ACS Nano, 2012, 6, 851–857 CrossRef CAS PubMed.
- J. Park, Y. Park and S. Kim, ACS Nano, 2013, 7, 9416–9427 CrossRef CAS PubMed.
- O. Kovtun, X. Arzeta-Ferrer and S. J. Rosenthal, Nanoscale, 2013, 5, 12072–12081 RSC.
- D. Geißler, S. Stufler, H.-G. Löhmannsröben and N. Hildebrandt, J. Am. Chem. Soc., 2012, 135, 1102–1109 CrossRef PubMed.
- N. Hildebrandt, ACS Nano, 2011, 5, 5286–5290 CrossRef CAS PubMed.
- A. R. Clapp, I. L. Medintz, H. T. Uyeda, B. R. Fisher, E. R. Goldman, M. G. Bawendi and H. Mattoussi, J. Am. Chem. Soc., 2005, 127, 18212–18221 CrossRef CAS PubMed.
- W. R. Algar, H. Kim, I. L. Medintz and N. Hildebrandt, Coord. Chem. Rev., 2014, 263, 65–85 CrossRef.
- J. B. Blanco-Canosa, M. Wu, K. Susumu, E. Petryayeva, T. L. Jennings, P. E. Dawson, W. R. Algar and I. L. Medintz, Coord. Chem. Rev., 2014, 263, 101–137 CrossRef.
- K. E. Sapsford, W. R. Algar, L. Berti, K. B. Gemmill, B. J. Casey, E. Oh, M. H. Stewart and I. L. Medintz, Chem. Rev., 2013, 113, 1904–2074 CrossRef CAS PubMed.
- D. Zhou, Biochem. Soc. Trans., 2012, 40, 635–639 CrossRef CAS PubMed.
- S. Silvi and A. Credi, Chem. Soc. Rev., 2015, 44, 4275–4289 RSC.
- Z. Yang, Y. Kong, F. Wilson, B. Foxman, A. H. Fowler, C. F. Marrs, M. D. Cave and J. H. Bates, Clin. Infect. Dis., 2004, 38, 199–205 CrossRef PubMed.
- M. P. R. Berry, C. M. Graham, F. W. McNab, Z. Xu, S. A. A. Bloch, T. Oni, K. A. Wilkinson, R. Banchereau, J. Skinner, R. J. Wilkinson, C. Quinn, D. Blankenship, R. Dhawan, J. J. Cush, A. Mejias, O. Ramilo, O. M. Kon, V. Pascual, J. Banchereau, D. Chaussabel and A. O/'Garra, Nature, 2010, 466, 973–977 CrossRef CAS PubMed.
- L. J. van't Veer, H. Dai, M. J. van de Vijver, Y. D. He, A. A. M. Hart, M. Mao, H. L. Peterse, K. van der Kooy, M. J. Marton, A. T. Witteveen, G. J. Schreiber, R. M. Kerkhoven, C. Roberts, P. S. Linsley, R. Bernards and S. H. Friend, Nature, 2002, 415, 530–536 CrossRef PubMed.
- M. Kaforou, V. J. Wright, T. Oni, N. French, S. T. Anderson, N. Bangani, C. M. Banwell, A. J. Brent, A. C. Crampin, H. M. Dockrell, B. Eley, R. S. Heyderman, M. L. Hibberd, F. Kern, P. R. Langford, L. Ling, M. Mendelson, T. H. Ottenhoff, F. Zgambo, R. J. Wilkinson, L. J. Coin and M. Levin, PLoS Med., 2013, 10, e1001538 Search PubMed.
- K. Susumu, E. Oh, J. B. Delehanty, J. B. Blanco-Canosa, B. J. Johnson, V. Jain, W. J. Hervey, W. R. Algar, K. Boeneman, P. E. Dawson and I. L. Medintz, J. Am. Chem. Soc., 2011, 133, 9480–9496 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Base pair mismatch tuning of CProbes. Binding capacity of the QDs. Theoretical limit of detection (LOD) for the monoplex systems. Kinetics of strand displacement. Kinetics of QProbe–CProbe binding. LOD and saturation point calculations. See DOI: 10.1039/c6nr00484a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2016 |