 Open Access Article
Open Access ArticleSupramolecular encapsulation of benzocaine and its metabolite para-aminobenzoic acid by cucurbit[7]uril
Shengke
Li
a,
Hang
Yin
a,
Gudrun
Martinz
a,
Ian W.
Wyman
b,
David
Bardelang
c,
Donal H.
Macartney
b and
Ruibing
Wang
*a
aState Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau, Taipa, Macau, China. E-mail: rwang@umac.mo
bDepartment of Chemistry, Queen's University, Kingston, ON K7L 3N6, Canada
cAix-Marseille Université, CNRS, Institut de Chimie Radicalaire, UMR 7273, 13013 Marseille, France
First published on 11th February 2016
Abstract
An ester-type local anesthetic agent, benzocaine (BZC), and its metabolite, para-aminobenzoic acid (PABA), both form 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host–guest complexes with cucurbit[7]uril (CB[7]) in aqueous solution and has been observed by 1H NMR, UV-visible spectroscopic titrations (including Job's plot), electrospray ionization (ESI) mass spectrometry, and density functional theory (DFT) molecular modeling. The host–guest binding affinities are (2.2 ± 0.2) × 104 M−1 and (1.5 ± 0.2) × 104 M−1 for the protonated BZC and PABA, respectively, in acidic solutions. The binding constants decrease by ∼100-fold to approximately 300 and 200 M−1 for BZC and PABA, respectively, upon deprotonation of these guest molecules in PBS buffered solution (pH = 7.4). However, the encapsulation of these guest molecules by CB[7] only resulted in very moderate pKa shifts. This supramolecular encapsulation of BZC and PABA could potentially find applications in drug formulation for the purpose of enhancing bio-absorption as well as reducing methemoglobinemia and allergic reactions caused by the derivation of PABA during the metabolism of BZC.
1 host–guest complexes with cucurbit[7]uril (CB[7]) in aqueous solution and has been observed by 1H NMR, UV-visible spectroscopic titrations (including Job's plot), electrospray ionization (ESI) mass spectrometry, and density functional theory (DFT) molecular modeling. The host–guest binding affinities are (2.2 ± 0.2) × 104 M−1 and (1.5 ± 0.2) × 104 M−1 for the protonated BZC and PABA, respectively, in acidic solutions. The binding constants decrease by ∼100-fold to approximately 300 and 200 M−1 for BZC and PABA, respectively, upon deprotonation of these guest molecules in PBS buffered solution (pH = 7.4). However, the encapsulation of these guest molecules by CB[7] only resulted in very moderate pKa shifts. This supramolecular encapsulation of BZC and PABA could potentially find applications in drug formulation for the purpose of enhancing bio-absorption as well as reducing methemoglobinemia and allergic reactions caused by the derivation of PABA during the metabolism of BZC.
1. Introduction
It is generally believed that synthetic macrocyclic molecules may mimic the non-covalent encapsulation behaviour of enzymes.1–3 Particularly, a novel family of macrocyclic hosts known as cucurbit[n]urils (CB[n]s, n = 5–8, 10 and 14) has shown remarkable potential in molecular encapsulation and enzymes biomimetics.4,5 CB[n] compounds consist of n glycoluril units connected by 2n methylene groups, and they possess not only a hydrophobic internal cavity that can encapsulate guest molecules, but also two carbonyl-fringed portals that are good cation receptors. The chemical activity of a guest molecule is often altered upon encapsulation by CB[n].1,6 Among all of the members of the CB[n] family, CB[7] (Fig. 1), in particular, has been widely adopted as a promising molecular capsule of drug molecules because of both its good water-solubility and its capacity to encapsulate organic molecules of a wide range of sizes with binding affinities of up to 1017 M−1.7–10 Following the seminal work on the use of CB[7] for the encapsulation of drug molecules,11,12 numerous reports regarding the encapsulation of bioactive molecules by CB[7] have been published and well-reviewed in recent years.1,6,7,13 CB[7] encapsulation often leads to improved solubility, chemical stability, and therapeutic efficacy as well as reduced side effects. Our research groups have investigated the biocompatibility14 of CB[7] and its complexation with a variety of drug molecules and medically important molecules during the past decade, which have included imidazolium- and thiazolium-based model drugs,15,16 ranitidine,17 coumarin,18 vitamin B12 and its coenzyme,19 and MPTP/MPP+.20 Very recently, we reported the inclusion of a general anesthetic tricaine for fish by CB[7] and evaluated its biological effects in vivo.21 As a synthetic receptor, CB[7] accelerated the recovery of anesthetized zebrafish by encapsulating tricaine in vivo.21 We recently extended our efforts to study the CB[7] encapsulation of another well-known structurally relevant anesthetic agent, benzocaine (BZC) (Fig. 1), and its metabolite para-aminobenzoic acid (PABA) (Fig. 1).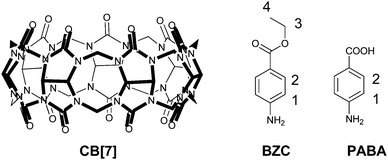 | ||
| Fig. 1 Molecular structures of CB[7] and the two guest molecules, BZC and PABA. The protons of BZC and PABA molecules are numerically labelled. | ||
As an ester-type local anesthetic, BZC is frequently utilized to relieve topical pain and reduce local sensitivity.22 Due to the low pKa value of its protonated form, BZC is always present as a neutral species under physiological conditions23,24 and therefore its corresponding poor solubility in aqueous solution has limited its application in superficial treatments.25 Additionally, the common problem encountered with local anesthetics is their short duration,26 and the increasing dosage of BZC in particular often leads to systemic side-effects, as a high concentration of BZC in plasma would be metabolized by esterases generating toxic PABA and its derivatives thus causing methemoglobinemia as well as allergic reactions.22,27 In order to improve its solubility and bio-availability as well as to decrease the side-effects, various approaches including nanocapsule and liposome delivery formulations have been developed.25,27–29 In addition to the aforementioned methods, supramolecular encapsulation may also provide a promising pathway toward achieving such a goal, and it has been reported that BZC complexation with cyclodextrin and calixarene derivatives might have this potential.22,30,31 However, the supramolecular encapsulation of BZC by cucurbit[n]urils is yet to be investigated. Our aim is (1) to obtain a clear understanding of the complexation behaviour between CB[7] and BZC as well as its metabolite PABA and (2) to determine the pKa shifts of BZC and PABA upon binding to the synthetic receptor CB[7]. It is anticipated that such a supramolecular encapsulation may find potential applications in the formulation of this local anesthetic agent for improved delivery with reduced side-effects.
2. Experimental
2.1. Materials
The host molecule CB[7] was prepared according to a literature method.32 BZC, PABA and PBS pellets were used as received from Sigma-Aldrich. The buffer solutions for the pKa investigation were prepared by using the buffer pairs of NaOAc/HOAc (0.2 M).2.2. Instrumentation
The 1D 1H NMR spectra were recorded using a BRUKER ULTRA SHIELD 400 PLUS NMR spectrometer. Meanwhile, the ESI-MS spectra were acquired using THERMO LTQ ORBITRAP XL equipped with an ESI/APcI multiprobe. The UV-visible spectra were all acquired on a HACH DR6000 UV-visible spectrometer using quartz cells with a 1.0 cm path length. The modelled structures of the host–guest complexes involved in this investigation were calculated by energy-minimizations using DFT (B3LYP/6-31G(d)) level of theory using the Gaussian09 Rev. D01 package program, performed at the computing facility of Aix-Marseille Université.2.3. Preparation of solutions of the complexes
In order to prepare solutions for 1H NMR characterization, a 1 mM solution of BZC and PABA in a D2O/DCl solution (with a pD of ∼2.0) was simply mixed with various amounts of CB[7] without changing the BZC and PABA concentrations. The solutions were sonicated for 3 min before they were characterized using 1H NMR spectroscopy.In order to perform the UV-visible spectroscopic titrations to determine the binding constant, aqueous solutions of BZC (or PABA) were prepared in PBS buffer solution (pH = 7.4) and acidic solution of HCl (pH = 2). These BZC (or PABA) solutions were subsequently titrated with various volumes of solutions containing the same concentration BZC (or PABA) and an excess of CB[7] to achieve different ratios of BZC (or PABA)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CB[7], while allowing the BZC (or PABA) concentration to remain constant during the entire titration.
CB[7], while allowing the BZC (or PABA) concentration to remain constant during the entire titration.
In order to prepare solutions for continuous variation titrations for Job's plot, solutions with the appropriate total concentrations of BZC (or PABA) and CB[7] were prepared. Among these solutions, the ratio of CB[7]/([Guest] + CB[7]) was varied from 0 to 1.0 in steps of 0.1.
In order to conduct pH-dependent UV-visible titration for the determination of pKa values, 0.2 M solutions of NaOAc and HOAc were prepared, and a total of 10 mL of buffer solution with different volumes of the two solutions was utilized to dilute a certain amount of BZC (or PABA) in the absence and in the presence of a large excess of CB[7], with pH values range from 2.5 to 5.8. The solutions with pH values lower than 2.5 were prepared by the direct dilution of a concentrated HCl solution.
3. Results and discussion
By following the experimental methods described above, the host–guest binding behaviours of CB[7] with BZC and PABA were examined, respectively, in detail, and the pKa shifts of both these guest molecules upon encapsulation by CB[7] were also determined via UV-vis spectroscopic titrations under various pH conditions.3.1. Host–guest binding behaviour study
Considering the BZC metabolite PABA (Fig. 2b) in a D2O solution at pD = 2, the upfield shift of the resonances of the two aromatic protons (H(1) and H(2) protons) on the benzene ring indicates its location inside the cavity of CB[7]. However, the H(1) protons and H(2) protons exhibit different CIS values of 0.55 ppm and 0.85 ppm, respectively, implying that H(2) is situated deeper within the cavity than H(1), which differs slightly from the case of BZC encapsulation. Similar to BZC, the broadened (even disappearing when a limiting amount of CB[7] was added) proton signals that do not exhibit splitting into free and bound species also demonstrate an intermediate exchange rate of complexation–decomplexation processes between the free and bound PABA species on the 1H NMR timescale. Continuous titration of BZC and PABA guests with increasing amounts of CB[7] (up to 3.0 equivalents) did not yield additional CIS values, providing supportive evidence of the exclusive 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry with relatively strong binding affinity.
1 binding stoichiometry with relatively strong binding affinity.
In addition to 1H NMR titration, the gas-phase structures of the two host–guest complexes, as calculated by virtue of Density Functional Theory (DFT B3LYP/6-31 basis**), were also employed to clearly describe the binding geometry of CB[7] on BZC and PABA molecules, respectively. The molecular modelling provided further evidence of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host–guest complex formation, as well as a further indication of the guest binding geometries within the host cavity. As demonstrated in Fig. 3 (left), the full encapsulation of PABA within the cavity of CB[7], presumably through the hydrophobic effect, seems to be consistent with the 1H NMR results (Fig. 2). As expected, the cationic ammonium group is situated at one of the two carbonyl-lined portals via ion–dipole interactions, further stabilizing the host–guest complexation. In the case of BZC (Fig. 3 right), similar to PABA, the entire aromatic ring of the guest is preferentially encapsulated by the CB[7] cavity, with the cationic ammonium group sitting at the portal, leaving the ethyl group outside of the cavity. This seems to contradict the 1H NMR results, where the ethyl group seems to be shielded by the hydrophobic cavity of CB[7]. This is not surprising, as the 1H NMR results give an average binding geometry in aqueous solution whereas the DFT modelling proposes one single geometry at a minimized energy moment. Very likely, the host is shuttling between the ethyl and aromatic groups due to the hydrophobic effect, with the majority of the time being spent with the benzene ring due to the close proximity with the ammonium group. The CB[7] host molecule acts not only as a steric barrier (including the whole aromatic ring) for the guest molecules, but also as a hydrogen bond acceptor for the nitrogen-based proton and hydroxyl protons, by positioning the guests within its cavity to facilitate the optimal hydrogen bonding and cation–dipole interactions.
1 host–guest complex formation, as well as a further indication of the guest binding geometries within the host cavity. As demonstrated in Fig. 3 (left), the full encapsulation of PABA within the cavity of CB[7], presumably through the hydrophobic effect, seems to be consistent with the 1H NMR results (Fig. 2). As expected, the cationic ammonium group is situated at one of the two carbonyl-lined portals via ion–dipole interactions, further stabilizing the host–guest complexation. In the case of BZC (Fig. 3 right), similar to PABA, the entire aromatic ring of the guest is preferentially encapsulated by the CB[7] cavity, with the cationic ammonium group sitting at the portal, leaving the ethyl group outside of the cavity. This seems to contradict the 1H NMR results, where the ethyl group seems to be shielded by the hydrophobic cavity of CB[7]. This is not surprising, as the 1H NMR results give an average binding geometry in aqueous solution whereas the DFT modelling proposes one single geometry at a minimized energy moment. Very likely, the host is shuttling between the ethyl and aromatic groups due to the hydrophobic effect, with the majority of the time being spent with the benzene ring due to the close proximity with the ammonium group. The CB[7] host molecule acts not only as a steric barrier (including the whole aromatic ring) for the guest molecules, but also as a hydrogen bond acceptor for the nitrogen-based proton and hydroxyl protons, by positioning the guests within its cavity to facilitate the optimal hydrogen bonding and cation–dipole interactions.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry has also been confirmed by the continuous variation titration method. For the UV-visible version of this method, the mole fractions of the guest and the host were varied in aqueous solution, while the total concentrations of both the guest and the host were kept constant. If the Job plot exhibits a maximum at 0.5 (the concentration ratio of CB[7] to the sum of the concentrations of CB[7] and the guest), it is indicative of a 1
1 binding stoichiometry has also been confirmed by the continuous variation titration method. For the UV-visible version of this method, the mole fractions of the guest and the host were varied in aqueous solution, while the total concentrations of both the guest and the host were kept constant. If the Job plot exhibits a maximum at 0.5 (the concentration ratio of CB[7] to the sum of the concentrations of CB[7] and the guest), it is indicative of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry between the host and the guest.34,35 The Job plot for the BZC@CB[7] system (with [CB[7]] + [BZC] fixed to 0.05 mM), as monitored using UV-visible spectroscopy (Fig. 4a) at 226 nm, reached a maximum at a ratio of 0.5 for [[CB[7]]/[CB[7]] + [BZC]]. This behaviour thus indicated that the 1
1 binding stoichiometry between the host and the guest.34,35 The Job plot for the BZC@CB[7] system (with [CB[7]] + [BZC] fixed to 0.05 mM), as monitored using UV-visible spectroscopy (Fig. 4a) at 226 nm, reached a maximum at a ratio of 0.5 for [[CB[7]]/[CB[7]] + [BZC]]. This behaviour thus indicated that the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes between CB[7] and BZC represented the dominant species in this concentration range. For PABA, in a similar manner, the peak exhibited a maximum at a ratio of 0.5 at 232 nm for [[CB[7]]/[CB[7]] + [PABA]] (with [CB[7]] + [PABA] fixed to 0.2 mM), demonstrating the formation of 1
1 complexes between CB[7] and BZC represented the dominant species in this concentration range. For PABA, in a similar manner, the peak exhibited a maximum at a ratio of 0.5 at 232 nm for [[CB[7]]/[CB[7]] + [PABA]] (with [CB[7]] + [PABA] fixed to 0.2 mM), demonstrating the formation of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes between CB[7] and PABA (Fig. 4b).
1 complexes between CB[7] and PABA (Fig. 4b).
Additionally, ESI-MS was performed to further verify the formation of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host–guest complexes as well, as this method is often considered to provide direct evidence of the binding stoichiometry. As was expected, the ionized host–guest complex between CB[7] and BZC was detected, and the singly charged m/z peak (experimental m/z = 1328.4359) corresponds to a 1
1 host–guest complexes as well, as this method is often considered to provide direct evidence of the binding stoichiometry. As was expected, the ionized host–guest complex between CB[7] and BZC was detected, and the singly charged m/z peak (experimental m/z = 1328.4359) corresponds to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding ratio, conforming to the calculated m/z value of 1328.4303. Similarly, the ESI-MS spectrum of the PABA@CB[7] complex also showed a singly charged peak with a m/z value of 1300.3981, indicative of a 1
1 binding ratio, conforming to the calculated m/z value of 1328.4303. Similarly, the ESI-MS spectrum of the PABA@CB[7] complex also showed a singly charged peak with a m/z value of 1300.3981, indicative of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding ratio and consistent with the calculated m/z value of 1300.3990.
1 binding ratio and consistent with the calculated m/z value of 1300.3990.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry model and provides a binding constant Ka of ∼300 M−1 (Fig. 5a inset). Under acidic conditions (pH = 2), the absorbance of BZC experienced a very moderate hypsochromic shift and decreased dramatically to nearly 5-fold (ε ∼ 3.3 × 103 M−1 cm−1) with respect to that observed at neutral pH (Fig. 5b).
1 binding stoichiometry model and provides a binding constant Ka of ∼300 M−1 (Fig. 5a inset). Under acidic conditions (pH = 2), the absorbance of BZC experienced a very moderate hypsochromic shift and decreased dramatically to nearly 5-fold (ε ∼ 3.3 × 103 M−1 cm−1) with respect to that observed at neutral pH (Fig. 5b).
After gradual addition of CB[7] to the BZC solution, the absorbance peak of BZC at 278 nm decreased further, along with a modest blue shift in accordance with the encapsulation of the protonated BZC inside of the cavity of CB[7] facing a macrocyclic microenvironment that is different from that of the bulk surroundings. The plot of the absorbance of BZC at 278 nm as a function of the CB[7] concentration in acidic solution (pH = 2) exhibited a non-linear least-squares fit, which is consistent with the previously discussed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry model (Fig. 5b inset), with a binding constant of Ka = 2.2 ± 0.2 × 104 M−1. Such a dramatic difference in the binding affinities between BZC@CB[7] and BZCH@CB[7] is likely attributed to the ion–dipole interaction between the protonated nitrogen and the carbonyl portal of CB[7]. Additionally, the presence of sodium cations in the PBS solution may have had some negative effects on the binding strength between the neutral BZC and CB[7].39
1 binding stoichiometry model (Fig. 5b inset), with a binding constant of Ka = 2.2 ± 0.2 × 104 M−1. Such a dramatic difference in the binding affinities between BZC@CB[7] and BZCH@CB[7] is likely attributed to the ion–dipole interaction between the protonated nitrogen and the carbonyl portal of CB[7]. Additionally, the presence of sodium cations in the PBS solution may have had some negative effects on the binding strength between the neutral BZC and CB[7].39
In both PBS buffered solution and acidic solution (pH = 2), PABA@CB[7], the binding behaviours were found to be similar to those of BZC-CB[7] (Fig. 5c and d). The binding constants Ka of PABA and protonated PABA with CB[7] were ∼200 and 1.5 ± 0.2 × 104 M−1, respectively.
3.2. CB[7]-induced pKa shifts
The two carbonyl-fringed portals of CB[7] are considered as cation receptors that may preferentially bind to cationic species such as protonated amines, thus the pKa values of the guest molecules often shift upwards upon encapsulation by CB[7].1 With a thorough understanding of the binding behaviours between CB[7] and the two guest molecules of BZC and PABA, the effect of the inclusion of CB[7] on the pKa values of the two guest molecules was investigated by a pH titration method via UV spectroscopic measurements.17 The absorbance changes of the guest or the guest–host complexes were monitored with varying pH in aqueous solutions. In particular, the pKa values of both BZC and PABA were examined in the absence and presence of a large excess equivalents of CB[7]. As illustrated in Fig. 6a, from the plot of absorbance of free BZC at 278 nm against a range of pH values, the pKa of BZC in the absence of CB[7] was determined to be 2.71 ± 0.03, which is consistent with the literature reported value.36 From the plot of the absorbance of BZC@CB[7] complexes at 280 nm against a range of pH values (Fig. 6b), the pKa value of BZC in the presence of CB[7] was determined to be 2.80 ± 0.04, which is shifted negligibly (only ∼0.1 pKa unit) from that of the free BZC compound. This extremely negligible pKa shift may be ascribed to the shuttling of CB[7] between the benzene and ethyl groups of BZC, as discussed previously, thus resulting in less influential interactions on the cationic amino group of BZC.With regards to PABA, the pH dependent absorbance in the presence and absence of CB[7] showed similar phenomena to that of BZC. As shown in Fig. 6c and d, the pKa of PABA in the absence of CB[7] is 2.63 ± 0.03, which is consistent with the reported result,37 while the pKa in the presence of a large excess of CB[7] shifted to 3.12 ± 0.07 (approximately 0.5 pKa units). Such a pKa shift of PABA is more pronounced than that of BZC, which may be attributed to the full encapsulation of PABA within the cavity of CB[7], thus optimizing the electrostatic interactions and hydrogen bonding interactions between the host carbonyl portal and the guest cationic amino group.
Although it has been recognized that a cation-receptor driven pKa shift is difficult to predict, theoretically the large differences (∼100-fold) of binding affinities (Ka), between the protonated and deprotonated host–guest species, might result in approximately a 2 pKa unit shift.1 However, the binding strength differences observed in this study might be a result of the different mediums, as it has been known that competitive salts (e.g. sodium salt in buffer) may intervene CB[n] based on the host–guest complexation.39 On the other hand, the medium was consistently acetate buffered during pH titrations, rendering reliable determinations of pKa values and the associated pKa shifts.
4. Conclusions
In summary, we have investigated the host–guest binding behaviours of benzocaine and its metabolite para-aminobenzoic acid with cucurbit[7]uril in aqueous solution at different pH values. The protonated guests formed relatively tight host–guest complexes with binding affinities in the order of 104 M−1. Conversely, the binding affinities decrease by ∼100-fold when the guest molecules are in their neutral forms, in comparison with those of the corresponding protonated forms in notably different media. Upon encapsulation by cucurbit[7]uril, benzocaine showed a negligible pKa shift, while para-aminobenzoic acid exhibited a moderate pKa shift of 0.5 units. The benzocaine and para-aminobenzoic acid supramolecular complexation with cucurbit[7]uril may have potential applications in the development of benzocaine-based drug formulations, drug delivery, as well as the minimization of benzocaine's side effects induced by para-aminobenzoic acid through supramolecular encapsulation in plasma. We are currently working along this line of research.Acknowledgements
The University of Macau Research Fund (SRG2014-00025), Macau Science and Technology Development Fund (FDCT/020/2015/A1) and the Natural Sciences and Engineering Research Council Grant (NSERC, Canada) are gratefully acknowledged for providing financial support.Notes and references
- I. Ghosh and W. M. Nau, Adv. Drug Delivery Rev., 2012, 64, 764–783 CrossRef CAS PubMed.
- B. Honig and A. Nicholls, Science, 1995, 268, 1144–1149 CAS.
- C. S. Mahon and D. A. Fulton, Nat. Chem., 2014, 6, 665–672 CrossRef CAS PubMed.
- L. Zheng, S. Sonzini, M. Ambarwati, E. Rosta, O. A. Scherman and A. Herrmann, Angew. Chem., Int. Ed., 2015, 54, 13007–13011 CrossRef CAS PubMed.
- Y. Jang, R. Natarajan, Y. H. Ko and K. Kim, Angew. Chem., Int. Ed. Engl., 2014, 53, 1003–1007 CrossRef CAS PubMed.
- D. H. Macartney, Isr. J. Chem., 2011, 51, 600–615 CrossRef CAS.
- D. Shetty, J. K. Khedkar, K. M. Park and K. Kim, Chem. Soc. Rev., 2015, 44, 8747–8761 RSC.
- E. Masson, X. Ling, R. Joseph, L. Kyeremeh-Mensah and X. Lu, RSC Adv., 2012, 2, 1213–1247 RSC.
- K. I. Assaf and W. M. Nau, Chem. Soc. Rev., 2015, 44, 394–418 RSC.
- L. Cao, M. Sekutor, P. Y. Zavalij, K. Mlinaric-Majerski, R. Glaser and L. Isaacs, Angew. Chem., Int. Ed. Engl., 2014, 53, 988–993 CrossRef CAS PubMed.
- Y. J. Jeon, S. Y. Kim, Y. H. Ko, S. Sakamoto, K. Yamaguchi and K. Kim, Org. Biomol. Chem., 2005, 3, 2122–2125 CAS.
- N. J. Wheate, A. I. Day, R. J. Blanch, A. P. Arnold, C. Cullinane and J. G. Collins, Chem. Commun., 2004, 1424–1425 RSC.
- S. Walker, R. Oun, F. J. McInnes and N. J. Wheate, Isr. J. Chem., 2011, 51, 616–624 CrossRef CAS.
- H. Chen, J. Y. W. Chan, X. Yang, I. W. Wyman, D. Bardelang, D. H. Macartney, S. M. Y. Lee and R. Wang, RSC Adv., 2015, 5, 30067–30074 RSC.
- R. Wang, L. Yuan and D. H. Macartney, Chem. Commun., 2006, 2908–2910 RSC.
- S. Li, X. Miao, I. W. Wyman, Y. Li, Y. Zheng, Y. Wang, D. H. Macartney and R. Wang, RSC Adv., 2015, 5, 56110–56115 RSC.
- R. Wang and D. H. Macartney, Org. Biomol. Chem., 2008, 6, 1955–1960 CAS.
- X. Miao, Y. Li, I. Wyman, S. M. Y. Lee, D. H. Macartney, Y. Zheng and R. Wang, Med. Chem. Commun., 2015, 6, 1370–1374 RSC.
- R. Wang, B. C. Macgillivray and D. H. Macartney, Dalton Trans., 2009, 3584–3589 RSC.
- S. Li, H. Chen, X. Yang, D. Bardelang, I. W. Wyman, J. Wan, S. M. Y. Lee and R. Wang, ACS Med. Chem. Lett., 2015, 6, 1174–1178 CrossRef CAS PubMed.
- H. Chen, J. Y. W. Chan, S. Li, J. J. Liu, I. W. Wyman, S. M. Y. Lee, D. H. Macartney and R. Wang, RSC Adv., 2015, 5, 63745–63752 RSC.
- L. M. Pinto, L. F. Fraceto, M. H. Santana, T. A. Pertinhez, S. O. Junior and E. de Paula, J. Pharm. Biomed. Anal., 2005, 39, 956–963 CrossRef CAS PubMed.
- S. Li and D. Li, Spectrochim. Acta, Part A, 2011, 82, 396–405 CrossRef CAS PubMed.
- R. D. Porasso, W. F. Bennett, S. D. Oliveira-Costa and J. J. Lopez Cascales, J. Phys. Chem. B, 2009, 113, 9988–9994 CrossRef CAS PubMed.
- N. F. De Melo, D. R. De Araujo, R. Grillo, C. M. Moraes, A. P. De Matos, E. de Paula, A. H. Rosa and L. F. Fraceto, J. Pharm. Sci., 2012, 101, 1157–1165 CrossRef PubMed.
- C. M. Moraes, A. P. de Matos, E. de Paula, A. H. Rosa and L. F. Fraceto, Mater. Sci. Eng., B, 2009, 165, 243–246 CrossRef CAS.
- N. F. de Melo, R. Grillo, V. A. Guilherme, D. R. de Araujo, E. de Paula, A. H. Rosa and L. F. Fraceto, Pharm. Res., 2011, 28, 1984–1994 CrossRef PubMed.
- P. Mura, F. Maestrelli, M. L. Gonzalez-Rodriguez, I. Michelacci, C. Ghelardini and A. M. Rabasco, Eur. J. Pharm. Biopharm., 2007, 67, 86–95 CrossRef CAS PubMed.
- C. Puglia, M. G. Sarpietro, F. Bonina, F. Castelli, M. Zammataro and S. Chiechio, J. Pharm. Sci., 2011, 100, 1892–1899 CrossRef CAS PubMed.
- L. M. Arantes, E. V. Varejao, K. J. Pelizzaro-Rocha, C. M. Cereda, E. de Paula, M. P. Lourenco, H. A. Duarte and S. A. Fernandes, Chem. Biol. Drug Des., 2014, 83, 550–559 CAS.
- E. Iglesias, Photochem. Photobiol. Sci., 2011, 10, 531–542 CAS.
- A. Day, A. P. Arnold, R. J. Blanch and B. Snushall, J. Org. Chem., 2001, 66, 8094–8100 CrossRef CAS PubMed.
- R. Wang, D. Bardelang, M. Waite, K. A. Udachin, D. M. Leek, K. Yu, C. I. Ratcliffe and J. A. Ripmeester, Org. Biomol. Chem., 2009, 7, 2435–2439 CAS.
- K. Hirose, J. Inclusion Phenom. Macrocyclic Chem., 2001, 39, 193–209 CrossRef CAS.
- J. S. Renny, L. L. Tomasevich, E. H. Tallmadge and D. B. Collum, Angew. Chem., Int. Ed., 2013, 52, 11998–12013 CrossRef CAS PubMed.
- J. Bezencon, M. B. Wittwer, B. Cutting, M. Smiesko, B. Wagner, M. Kansy and B. Ernst, J. Pharm. Biomed. Anal., 2014, 93, 147–155 CrossRef CAS PubMed.
- S. Shaomin, Y. Yu and P. Jinghao, Anal. Chim. Acta, 2002, 458, 305–310 CrossRef.
- P. Thordarson, Chem. Soc. Rev., 2011, 40, 1305–1323 RSC.
- W. Ong and A. E. Kaifer, J. Org. Chem., 2004, 69, 1383–1385 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2016 |





