Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Interplay between seven secondary metal uptake systems is required for full metal resistance of Cupriavidus metallidurans†
M.
Herzberg
,
L.
Bauer
,
A.
Kirsten
and
D. H.
Nies
*
Molecular Microbiology, Institute for Biology/Microbiology, Martin-Luther-University, Halle-Wittenberg, Germany. E-mail: d.nies@mikrobiologie.uni-halle.de
First published on 25th January 2016
Abstract
The beta-proteobacterium Cupriavidus metallidurans is able to grow in metal-contaminated environments due to having sophisticated metal efflux systems. Here, the contribution of all seven known secondary metal uptake systems (ZupT, PitA, CorA1, CorA2, CorA3, ZntB, HoxN) to metal resistance is characterized. In a strategic deletion approach, all ten double deletion mutants, a variety of triple and quadruple mutants, and from the Δ4 mutant (ΔzupT ΔcorA1 ΔcorA2 ΔcorA3) the mutants Δ5 (=Δ4 ΔpitA), Δ6 (=Δ4 ΔpitA ΔzntB), and finally Δ7 (ΔzupT ΔcorA1 ΔcorA2 ΔcorA3 ΔpitA ΔzntB ΔhoxN) were constructed. Metal resistance, metal content, and regulation of expression of these genes were characterized in these mutants. The ΔzupT single deletion strain exhibited an extended lag phase in Tris-buffered liquid mineral salts medium (TMM) compared to its parent strain AE104, indicating a decreased fitness level. Further deletions up to Δ6 did not influence growth in TMM without added metals but fitness of the Δ7 strain dropped to a lower level compared to Δ6, Δ5 and ΔzupT. The cells of the Δ7 multiple deletion strain still contained all essential metals, demonstrating that additional metal import systems must exist in C. metallidurans. PitA was an important contributor of metal:phosphate complexes to C. metallidurans. Up to Δ5 no evidence was found for increased expression of the transporter genes to recruit substitutes for the deleted importers. Only the hoxN-lacZ reporter gene fusion displayed a changed expression pattern in the Δ6 strain, indicating recruitment of HoxN. Metal resistance of the deletion strains decreased along the deletion series although all strains still contained metal efflux systems: up to the Δ6 mutant the overall fitness was kept at the ΔzupT mutant strain level at the cost of a diminished competence to handle μM concentrations of transition metals. Together, these data demonstrated an important contribution of the seven secondary metal import systems to metal homeostasis in this bacterium.
Introduction
The beta-proteobacterium Cupriavidus metallidurans owes its outstanding ability to survive in environments with high concentrations of transition metals to the possession of a variety of sophisticated heavy metal-resistance determinants.1–4 Central to these are metal efflux systems that have been characterized in detail in the last decades.5–7 Members of the PIB-type ATPase (TC#3.A.3), CDF (TC#2.A.4) or other transporter families {TC, transporter classification8–10} remove surplus transition metal cations from the cytoplasm to the periplasm. Transenvelope protein complexes containing RND proteins (TC#2.A.6), such as CzcCBA or CnrCBA as major component, are responsible in vivo for further export from the periplasm to the external medium.1,5,7,11 Substrates of these efflux systems are metal cations that have been imported by metal uptake systems, which are thus interaction partners of the exporters.In contrast to the efflux systems, contribution of the metal uptake systems to metal resistance in C. metallidurans has not been characterized in detail. The genome of C. metallidurans predicts at least seven secondary uptake systems for transition metals: (i) the ZIP protein ZupT (TC#2.A.5); (ii) the metal:phosphate importer PitA (TC#2.A.20); (iii) three members of the MIT family CorA1 through CorA3 (TC#1.A.35); (iv) the additional MIT protein ZntB that might be an uptake or efflux system; (v) and the NiCoT protein HoxN (TC#2.A.52) that might import additional Ni2+ cations into the cell for hydrogenase synthesis.1,11,12C. metallidurans13 does not possess orthologs of the ABC-type (TC#3.A.1) import systems ZnuABC and NikABC from E. coli for zinc or nickel import, respectively,14–16 so that the secondary transport systems should theoretically have an important role in the uptake of transition metal cations.
Using single gene deletion mutants and a ΔzupT ΔpitA double mutant, it could be demonstrated that neither of the five systems ZupT, PitA, CorA1, CorA2 or CorA3 alone was essential for the import of zinc or other transition metal ions.13 Moreover, none of these systems pairs up with an efflux system to form a regulated “shunt” of a metal through the cell. Such a shunt might considerably have maintained the cytoplasmic concentration of a metal in homeostasis, by either increased export or increased import of the metal if its concentration would be too high or too low, respectively. Instead, these five import systems possibly form a battery of redundant uptake systems with low and overlapping substrate specificities, which supplies metals to the cell as they become available, relying on the subsequent efflux system to cope with any surplus. In this way, the exact composition of the cytoplasmic metal content and mélange composition is maintained in a way that minimizes negative interference between the individual metals.13
Expression of zupT, corA1-corA3 and pitA is down-regulated by zinc excess. Only expression of zupT is up-regulated by zinc starvation via the Fur-type zinc uptake regulator Zur (previously FurC).17 Expression of corA1 is influenced by magnesium starvation.13 The pitA gene is up-regulated with increasing phosphate concentrations up to 5 mM phosphate, which complexes zinc ions and lowers its availability. At higher phosphate concentrations, pitA is down-regulated again but remains on a high expression level in a ΔzupT mutant,13 indicating a central function of zinc in control of expression of pitA; Zur is not involved in this process.17
The ZupT importer of Escherichia coli has a broad substrate specificity.18–20 Deletion of its ortholog in C. metallidurans does not impair net import of Zn(II) or any other metal13 at least not at the first glance. A more detailed analysis, however, reveals a pleiotropic effect of the ΔzupT deletion: (i) the mutant cell is no longer able to import Zn(II) at low zinc concentrations,13e.g. in the presence of the metal chelator ethylene-diamine-tetra-acetic acid (EDTA); (ii) the overall zinc content of the mutant cell cultivated in minerals salts medium amounts to about 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc per cell while the parent contains 70
000 zinc per cell while the parent contains 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc per cell;21 (iii) zinc cannot be efficiently allocated to the zinc-dependent beta-prime subunit RpoC of the RNA polymerase, leading to aggregation of RpoC into inclusion bodies;21 (iv) the central CzcA subunit of the RND-driven transenvelope complex CzcCBA is either not translated or is rapidly degraded;21 (v) surprisingly zinc is not efficiently allocated to the periplasmic Cu–Zn superoxide dismutase SodC;21,22 and (vi) a part of a genomic island that is silenced as a response to metal stress in the parent strain AE104 is un-silenced again in the ΔzupT mutant.23 So, the effect of the single ΔzupT gene deletion revealed interesting phenomena upon close inspection. We ask here what more can be learned if all seven currently known secondary metal uptake systems are removed from the C. metallidurans cell.
000 zinc per cell;21 (iii) zinc cannot be efficiently allocated to the zinc-dependent beta-prime subunit RpoC of the RNA polymerase, leading to aggregation of RpoC into inclusion bodies;21 (iv) the central CzcA subunit of the RND-driven transenvelope complex CzcCBA is either not translated or is rapidly degraded;21 (v) surprisingly zinc is not efficiently allocated to the periplasmic Cu–Zn superoxide dismutase SodC;21,22 and (vi) a part of a genomic island that is silenced as a response to metal stress in the parent strain AE104 is un-silenced again in the ΔzupT mutant.23 So, the effect of the single ΔzupT gene deletion revealed interesting phenomena upon close inspection. We ask here what more can be learned if all seven currently known secondary metal uptake systems are removed from the C. metallidurans cell.
Results
Deletion of seven genes for metal uptake systems decreases fitness of the resulting Δ7 mutant strain
All seven recognizable genes for secondary metal uptake systems (zupT, pitA, corA1, corA2, corA3, zntB, hoxN) were deleted from the chromosome of the plasmid-free C. metallidurans strain AE104. This plasmid-free derivative was used as parent instead of C. metallidurans CH34 wild type to avoid interference through the plasmid-encoded metal efflux systems, and because the czc-containing plasmid pMOL30 was unstable in a ΔzupT mutant.21 Starting with the five genes zupT, pitA, corA1, corA2 and corA3, all ten combinations of double mutants and four of the five possible quadruple mutants but only two triple mutants were constructed. During construction of the mutants, it was only once possible to obtain a “clean” (antibiotic marker-free deletion) ΔcorA1 deletion, in this case from the ΔzupT ΔcorA2 ΔcorA3 triple deletion strain leading to the ΔzupT ΔcorA1 ΔcorA2 ΔcorA3 quadruple mutant, designated Δ4. In all other cases, the corA1 gene had to be interrupted by insertion of a kanamycin resistance cassette. The “clean” quadruple deletion strain Δ4 was the parent of Δ5 (=Δ4 ΔpitA), Δ6 (=Δ4 ΔpitA ΔzntB) and finally Δ7 (=Δ4 ΔpitA ΔzntB ΔhoxN = ΔzupT ΔpitA ΔcorA1 ΔcorA2 ΔcorA3 ΔzntB ΔhoxN).The fitness of the mutant strains ΔzupT, Δ5, Δ6 and Δ7 was compared to that of the parent strain AE104 in Tris-buffered mineral salts medium (TMM) without added metals. As already reported,22 the ΔzupT strain showed an extended lag phase compared to strain AE104 (Fig. 1A and Fig. S1A, ESI†). The lag phase of the Δ7 (ΔzupT ΔpitA ΔcorA1 ΔcorA2 ΔcorA3 ΔzntB ΔhoxN) mutant strain was even more extended, while growth curves of the Δ5 (ΔzupT ΔpitA ΔcorA1 ΔcorA2 ΔcorA3) and Δ6 (ΔzupT ΔpitA ΔcorA1 ΔcorA2 ΔcorA3 ΔzntB) mutants were not different from that of the ΔzupT strain. While removal of the important zinc importer ZupT decreased fitness of C. metallidurans to a first level, the cells were able to compensate for deletion of an additional 5 genes leading to the Δ6 strain but not that of the 6th gene leading to the Δ7 strain. This decreased fitness to a second level, highlights the importance of the battery of seven metal importers for metal homeostasis of C. metallidurans.
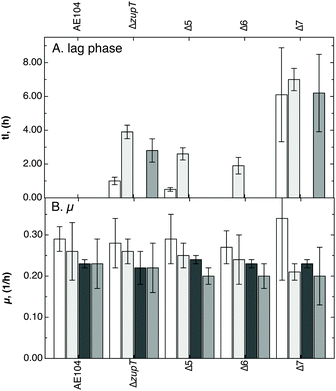 | ||
| Fig. 1 Growth impairment of the mutant strains. The strains AE104, ΔzupT, Δ5, Δ6 or Δ7 were cultivated in TMM at 30 °C without further additions (white bars) or in the presence of 10 μM Co(II) (light grey bars), 10 μM Zn(II) (dark grey bars), or 2.5 μM Cd(II) (medium grey bars) and growth was measured as turbidity at 600 nm. The duration of the lag phase (panel A) and the growth rates (panel B) were calculated. Growth curves in Fig. S1 (ESI†), data for the lag phase and growth rate in a table below Fig. S1 (ESI†). n ≥ 3, deviation bars indicated. | ||
Metal content of the deletion strains
The cells of the Δ7 multiple deletion strain still contained all essential metals, clearly indicating that additional metal import systems must exist in C. metallidurans (Fig. 2 and Table S1, ESI†). These are likely to be primary importers or members of currently unknown uptake systems.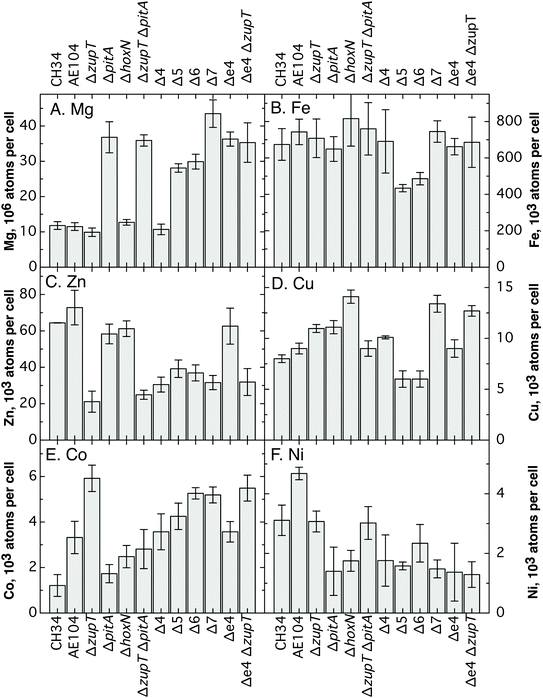 | ||
| Fig. 2 Metal content of C. metallidurans strains. As determined by ICP-MS. The data are also shown in Table S1 (ESI†). | ||
As published,21 deletion of zupT resulted in a decreased zinc content visible in all ΔzupT-containing multiple deletion strains up to Δ7 (Fig. 2). Except in these ΔzupT mutants, C. metallidurans cells were always able to compensate for the loss of a metal uptake system by adopting other systems, which altered the cellular metal content to some degree. First, also as published,13 deletion of pitA led to a 3-fold increase in magnesium (and phosphorus) content in the ΔzupT ΔpitA double deletion strain of AE104. An increased magnesium content was also a feature of all investigated ΔpitA-containing multiple deletion strains up to Δ7 (Fig. 2). Second, deletion of pitA in Δ4 leading to Δ5 additionally increased the zinc, but decreased the iron, copper and nickel content. This indicated some re-arrangement of metal import of Δ5 compared to Δ4 as a result of the pitA deletion. Importers of magnesium and zinc were increasingly used in the Δ5 strain while the decreased iron, copper and nickel content did not stimulate additional import of these metals, e.g. by up-regulation of iron import pathways via FurA or FurB.17 Third, while deletion of zntB from Δ5 (leading to Δ6) had no significant effect on the metal content (Fig. 2), deletion of hoxN in Δ6 (leading to Δ7) increased the magnesium content, restored the iron and copper content to the higher and the zinc content to the lower level of the Δ4 strain.
The cellular cobalt content decreased when pitA was deleted from the parent strain AE104. Together with the lowered iron, copper and nickel content of the Δ5 strain (=Δ4 ΔpitA) compared to Δ4, this indicated that PitA may be an important contributor of iron, copper, nickel and cobalt in the form of the metal:phosphate complexes to C. metallidurans.
Metal resistance of the deletion strains
The deletion strains up to Δ7 were still able to import all essential transition metals, probably due to use of other uptake pathways (Fig. 2 and Table S1, ESI†). All deletion mutants of the parent strain AE104 still contained the genes for the metal-exporting PIB2-type ATPases ZntA and CadA, for three copper-exporting PIB1-type ATPases such as CupA, and the two CDF proteins DmeF and FieF, which together should maintain a minimum zinc, cadmium, copper, cobalt and nickel resistance.7 If efficient physiological function of these efflux systems requires a balanced import of metals by the seven secondary importers, successive removal of importers should lead to increased imbalance in the import of transition metals, eventually to incompetence of the efflux systems, and ultimatively to loss of metal resistance. To test this hypothesis, metal resistance of the deletion strains was analyzed in growth curves, liquid culture and on solid agar medium.On solid TMM there was no significant effect in any of the studied multiple deletion strains on zinc, cadmium or copper resistance (Table S2, ESI†). Nickel resistance of all ΔcorA1-containing multiple mutant strains was enhanced. In addition, the ΔzupT ΔpitA ΔcorA2 ΔcorA3 mutant, which still possessed a functional corA1 gene, was also more resistant to nickel than the parent strain AE104, while its direct parent, the triple mutant ΔzupT ΔpitA ΔcorA3, was not (Table S2, ESI†). Together with the low nickel content of the Δ4 and Δ5 mutant strains cultivated in non-amended TMM (Fig. 2), this indicated that CorA1 was involved in nickel uptake but needed the CorA2 protein for full function. Due to the fact that CorA-like proteins are pentamers24 and TMM-grown C. metallidurans cells contain 117 ± 8 copies of CorA1 (Rmet_3052) per cell, 38 ± 19 copies of CorA2 (Rmet_0036) and 37 ± 5 copies of CorA3 (Rmet_3287), yielding a 3.2 ± 0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0 ± 0.5
1.0 ± 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ± 0.1 ratio of CorA1
1 ± 0.1 ratio of CorA1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CorA2
CorA2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CorA3,22 this could mean that the CorAs of C. metallidurans form heteromultimers, as has been described for other metal transporters.25
CorA3,22 this could mean that the CorAs of C. metallidurans form heteromultimers, as has been described for other metal transporters.25
The determination of the MIC values to cobalt in all ΔzupT ΔcorA and ΔpitA ΔcorA-containing double mutants were hampered by high deviations of the individual measurements. Cobalt resistance of two double mutants, ΔcorA1 ΔcorA2 and ΔcorA2 ΔcorA3 was very low, while that of the corA123 triple mutant was higher than that of the parent strain AE104. Resistance of the three single ΔcorA deletion strains was similar to the AE104 level (Table S2, ESI†). This could mean that a putative heteropentameric complex composed of two CorA species is able to function if PitA and ZupT are additionally present, whereas the presence of CorA3 or CorA1 alone results in a strongly decreased cobalt resistance. The three CorAs of C. metallidurans could be assigned to the import of nickel and cobalt, maybe in the form of a heteromultimeric complex CorA123, which interacts with ZupT and PitA as main metal supply routes in C. metallidurans.
Analysis of metal resistance of the mutants up to Δ5 on solid growth medium did not yield further insights and the study was continued using liquid TMM together with low concentrations of Zn(II), Co(II) or Cd(II) (Fig. 1 and Fig. S1, ESI†). The extended lag phase of the ΔzupT mutant compared to the parent AE104 increased when 10 μM Co(II) or 2.5 μM Cd(II) was added while addition of 10 μM Zn(II) ameliorated the effect of the ΔzupT deletion. Growth of the Δ5 and the Δ6 mutant strains was similar to that of the ΔzupT strain or was even better in the case of added cadmium. As in the case of non-amended medium (Fig. 1), deletion of pitA, corA123 and zntB in addition to ΔzupT did not impair fitness of the C. metallidurans strains further because the cells were able to compensate for the loss of these importers. However, in Δ7 resistance to cobalt and cadmium was significantly decreased (Fig. 1 and Fig. S1, ESI†), while zinc resistance was not so strongly affected. Addition of 10 μM Zn(II) partially restored fitness of the multiple deletion strains up to Δ7.
The mutant cells were cultivated in the presence of higher concentrations of transition metals in liquid TMM in dose–response experiments and the IC50 values were calculated (Fig. 3 and Table S3, ESI†). These experiments (up to 20 h) revealed more effectively the differences in metal resistance between the mutant strains than the long-term (up to 3 days) MIC determinations. Zinc, cobalt and cadmium resistance of the ΔzupT strain was decreased compared to the parent strain, and that of the Δ7 strain was decreased compared to ΔzupT (Fig. 3).
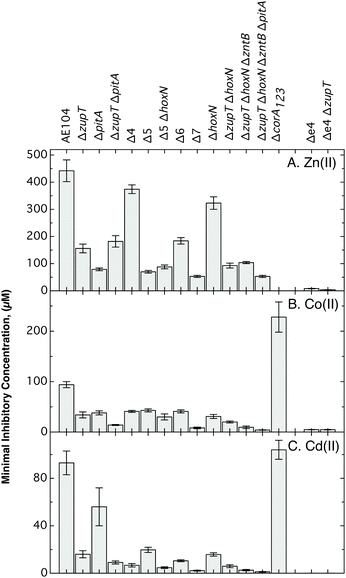 | ||
| Fig. 3 Metal resistance of deletion strains in liquid culture. Dose response experiments were performed (n > 3 per conditions) in the presence of Zn(II) (panel A), Co(II) (panel B) and Cd(II) (panel C), and the IC50 values calculated. The data are also shown in Table S3 (ESI†) for a better documentation of the differences in cobalt and cadmium resistance of the mutant strains. | ||
During the course of the deletion analysis it was observed that zinc resistance decreased in series from the parent strain AE104 to the ΔzupT and to the ΔpitA mutants (Fig. 3A). In the case of the ΔzupT ΔpitA mutant resistance remained at the ΔzupT level and increased to the level of the parent strain again in Δ4 (ΔzupT ΔcorA123). Decreased zinc resistance in the ΔpitA mutant strain was mediated partially by ZupT and decreased zinc resistance in the ΔzupT strain by CorA123. Zinc resistance of the Δ5 strain (ΔzupT ΔpitA ΔcorA123) was similar to that of the ΔpitA strain demonstrating that the interplay between these uptake systems, which substituted for each other, and the efflux systems was required for resistance to upper μM Zn(II) concentrations. Deletion of zntB in Δ5 generating Δ6 increased zinc resistance again. ZntB was partially responsible for the low zinc resistance level of the Δ5 strain, although this deletion did not change the zinc content of TMM-grown cells (Fig. 2C), Zinc resistance finally decreased to the lowest level in the Δ7 strain following deletion of hoxN, probably due to use of additional unknown zinc uptake systems that were not in balance with the efflux systems. This demonstrated that all seven systems were involved in zinc homeostasis, either as probable zinc importers (CorA123 in ΔzupT, ZntB in Δ5) or by interfering with activation of other uptake routes.
A second series of deletion mutants in the order ΔhoxN → ΔzupT → ΔzntB → ΔpitA also demonstrated this order of decreasing zinc resistance (Fig. 3A). Zinc resistance decreased in this order except when zntB was deleted in the ΔzupT ΔhoxN double mutant. The presence of ZntB was not required when PitA and CorA123 were still present, and it was recruited only in the Δ5 strain. These results also defined a ranking order of the zinc importers: first, PitA because ΔpitA had the strongest effect on zinc resistance and zinc resistance of Δ4 was close to that of the parent strain AE104; second, ZupT because of the required supply of zinc to zinc-dependent proteins; third, CorA123 which was partially responsible for zinc sensitivity of the ΔzupT strain; fourth, ZntB as metal importer in Δ5; and finally HoxN.
Additionally, cobalt and cadmium resistance decreased with each step along the second deletion series. With one exception, resistance to cadmium also decreased along the first deletion series (Fig. 3B, C and Fig. S3 (ESI†), upper part): it increased about three-fold when pitA was deleted from the Δ4 strain leading to Δ5 so that PitA was responsible for cadmium imbalance in the Δ4 strain, indicating that PitA also imported cadmium:phosphate.
Cobalt resistance was especially low in the ΔzupT ΔpitA double mutant and increased again when the corAs were deleted. Similar to what was observed with the experiments on solid TMM, the three CorAs seemed to be important cobalt uptake systems. In agreement with this, cobalt resistance of the ΔcorA123 triple mutant was twice as high in liquid culture as that of the parent strain AE104 (Fig. 3B).
Since the cobalt resistance level of most mutant strains was so surprisingly low, resistance of the uptake mutants was also compared to that of the quadruple efflux deletion mutant Δe4 (=ΔzntA ΔcadA ΔdmeF ΔfieF) and to that of the Δe4 ΔzupT mutant (Fig. 3). The IC50 of Δe4 for zinc was only 8 μM, much lower than that of the Δ7 mutant which had an IC50 of 53 μM. Deletion of zupT always decreased zinc resistance down to 1/2 or 1/3, in the AE104 background from 440 μM to 156 μM, in the ΔhoxN background from 323 μM to 93 μM, and in the Δe4 background from 8.3 to 3.9 μM. This occurred despite the fact that the cellular zinc content decreased (Fig. 2) and addition of zinc ameliorated the retarded growth of the ΔzupT mutant strain (Fig. 1). Because the CorAs were responsible for the decreased zinc resistance in the ΔzupT single mutant (Fig. 3), the cells were able to partially compensate for the loss of ZupT by use of the CorAs, which led to imbalanced zinc import that could not be compensated by the metal efflux systems.
Cobalt resistance of the Δe4, Δe4 ΔzupT and of the ΔzupT ΔhoxN ΔzntB ΔpitA quadruple mutant was similar with an IC50 of 4 to 5 μM Co(II). It doubled when the corAs were deleted in the quadruple mutant, leading to Δ7 (Fig. 3 and Table S3, ESI†). Again, balanced import was required to allow efficient export. Cadmium resistance of the Δe4 mutant was 60-times lower that that of the most sensitive ΔzupT ΔhoxN ΔzntB ΔpitA quadruple mutant showing the greater importance of the efflux systems compared to the uptake systems in resistance to this “toxic-only” metal cation. A balanced metal import and export was important for the “essential-but-also-toxic” metals but efflux was the main contributor to resistance to the “toxic-only” cadmium cation.
The Δ7 strain also displayed decreased resistance to copper, gold and nickel compared to strain AE104 (Fig. 4). While copper resistance of the ΔzupT, Δ5 and Δ6 strains was on the same level in-between Δ7 and AE104, nickel resistance increased from ΔzupT to Δ5 and Δ6, and decreased again from Δ6 to Δ7 as a result of the deletion of the nickel importer HoxN. Gold resistance of all mutant strains was similar and lower than that of the parent strain AE104 (Fig. 4). Resistance to the metal-complexing component EDTA decreased due to the ΔzupT deletion in the AE104 parent strain and in the Δe4 efflux mutant. Here, in the absence of efflux systems, EDTA resistance was 5-times higher than in AE104 (Fig. 4).
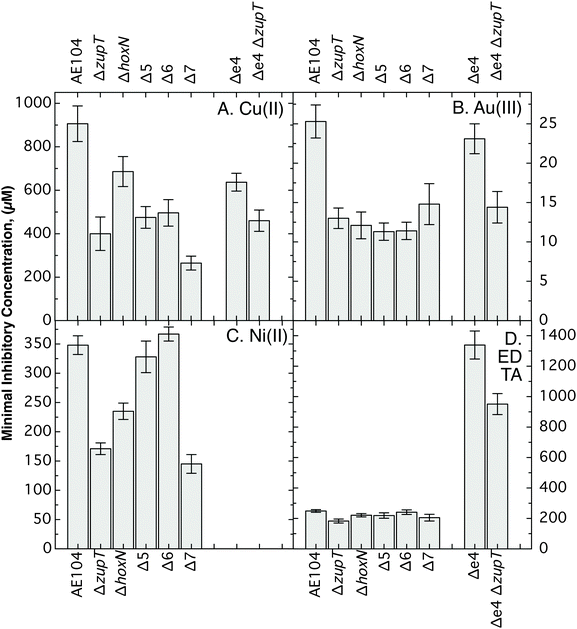 | ||
| Fig. 4 Metal resistance of deletion strains in liquid culture. Dose response experiments were performed (n > 3 per conditions) in the presence of Cu(II) (panel A), Au(III) (panel B), Ni(II) (panel C) and EDTA (panel D), and the IC50 values calculated. Bold-faced values indicate significant deviations from metal resistance of the direct parent strain. The data are also shown in Table S4 (ESI†) for a better visibility of the differences between the strains. | ||
When oxidative stress resistance of the mutant strains was examined as a marker of general stress tolerance (Fig. 5), Δ7 was as resistant to H2O2 and paraquat as the ΔzupT mutant. The ΔpitA and Δe4 strains demonstrated a higher paraquat, but not H2O2, resistance compared to the parent strain, which was decreased again by additional deletion of zupT.22 This demonstrates further the importance of ZupT for oxidative stress resistance.
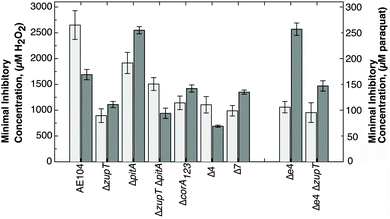 | ||
| Fig. 5 Oxidative stress resistance of deletion strains in liquid culture. Dose–response experiments were performed (n > 3 per conditions) with H2O2 (light grey bars) and paraquat (dark grey bars), and the IC50 values calculated. The data are also shown in Table S5 (ESI†). | ||
These data clearly demonstrate that the interaction between metal uptake and efflux systems, and not only the activity of the uptake systems, is responsible for a balanced supply of “essential-but-also-toxic” metals to C. metallidurans. Deletion of zupT resulted in decreased fitness of the mutant and use of other metal import systems, which subsequently resulted in imbalanced metal import and decreased overall metal resistance. PitA was involved in homeostasis of all metals tested, while the three CorAs were required for cobalt and nickel homeostasis but they were also responsible for the decreased zinc sensitivity of the ΔzupT mutant. Up to the Δ5 and Δ6 mutant, the fitness level of the mutant strains could be kept at that of the ΔzupT mutant but at the cost of increasing problems in handling higher (μM) concentrations of metals. This was first accomplished by PitA and the CorAs complementing loss of ZupT, subsequently the action of ZntB that was responsible for the low zinc resistance of the Δ5 strain, and finally of HoxN. When all seven systems were inactivated, overall fitness and metal resistance reached a low level, despite the presence of the metal efflux systems.
Regulation of expression of the genes for the secondary uptake systems
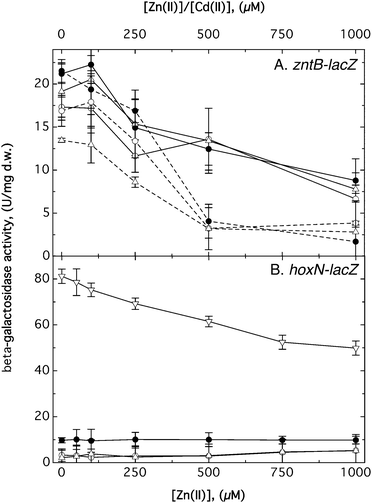
As published,13 expression of a zupT-lacZ reporter gene fusion was up-regulated under zinc starvation conditions, that of corA1-lacZ was down-regulated by increasing magnesium concentrations and that of pitA-lacZ by increasing phosphate concentrations. During the strategic construction of the deletion series, the lacZ gene was inserted downstream of all seven transporter genes mentioned in this study, always leaving the open reading frame of the transporter gene intact. These fusions were constructed in strain AE104 and several mutant strains, and regulation of expression of the reporter gene by divalent transition metal cations and EDTA was measured. With three exceptions, this did not yield any insights or differences in the regulatory patterns between the strains (data not shown). Exception 1 was the already known global down-regulation of most genes by increasing zinc concentrations, as published.13Exception 2 concerned expression of zntB-lacZ, which was introduced into AE104, ΔzupT, Δ5 (Fig. 6) and ΔpitA (data not shown). In all these mutants, addition of zinc chloride up to 1 mM Zn(II) decreased expression of the reporter construct by half. Addition of cadmium at 0.5 mM Cd(II) decreased zntB-lacZ expression down to 25% of the value without added metals. No difference was visible in zntB-lacZ expression between the AE104, ΔzupT and Δ5 derivatives (Fig. 6). EDTA, Co(II), or Mn(II) had no effect but Cu(II) also resulted in a down-regulation of zntB-lacZ expression (data not shown). Decreased expression at the high metal cation concentrations could also result from metal toxicity in addition to the metal-dependent changes. ZntB was regulated as would be expected for an uptake system, and was down-regulated when high concentrations of thiol-binding metals such as cadmium and copper were challenging the cells. Regulation was independent of the presence of ZupT, PitA or CorA123. Although ZntB was involved in the low zinc resistance level of the Δ5 strain, its gene was not up-regulated, nor in the double, triple or quadruple mutants tested. As the altered cellular metal content and metal resistance indicated that ZntB, the CorAs and PitA were subsequently used in the respective mutant strains, activation may have been on various levels, e.g. the translational level, by flux control or other post-transcriptional mechanisms in addition to the already published changes in zupT and pitA expression.13,17 This indicated a cross-talk between PitA, ZupT, CorA123 and ZntB, which organized metal import into C. metallidurans cells and was flexible enough to compensate for loss of some of these proteins, albeit not at higher metal concentrations.
The third exception was revealed by the hoxN-lacZ expression pattern (Fig. 6B): the gene was not regulated by EDTA, nickel or cobalt in all tested strains (data not shown), and was not regulated by zinc in strain AE104 (Fig. 6B), staying at a low specific activity of 9.4 ± 2.7 U mg−1 dry mass. In ΔzupT and Δ5, hoxN-lacZ was expressed on a low level but 2-fold up-regulated with increasing zinc concentrations up to 1 mM. Surprisingly, the operon fusion was strongly expressed in the Δ6 strain at 81 ± 5 U mg−1 and down-regulated 1.6-fold with increasing zinc concentrations (Fig. 6B). It is likely that HoxN was recruited for metal import but only in the Δ6 mutant when all other known secondary metal import systems were inactivated.
Although metal uptake systems were removed stepwise up to Δ5, the mutant cells remained at the fitness level of the ΔzupT mutant by use of substitute importers, members or non-members of the group of seven studied transport systems, but metal resistance decreased along the series of deletion strains despite the presence of metal efflux systems. In Δ6, increased expression of hoxN was required for the strain to remain at the ΔzupT fitness level. When hoxN was also gone, fitness and metal resistance decreased a second step to the level of the Δ7 mutant strain.
Intracellular zinc homeostasis
Deletion of zupT had two main effects: zinc could not be supplied efficiently to the zinc-dependent beta-prime subunit RpoC of the RNA polymerase, leading to accumulation of RpoC in inclusion bodies; and the central subunit CzcA of the CzcCBA transenvelope complex for cobalt, zinc, cadmium resistance could not be maintained in the cells although czcA was transcribed and constitutively expressed.21,22 Both effects were used here as reporters for the ability of the mutant strains to maintain their intracellular zinc homeostasis.Deletion of zupT always resulted in loss of the CzcA band, which was visible in the parent strain AE104 and the ΔpitA single deletion strain (Fig. 7). This corresponded to a zinc content below 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 atoms per cell (Fig. 2) in all these strains. The situation was more complicated concerning zinc allocation to RpoC. If this ability was disturbed, as in the ΔzupT single mutant, RpoC protein was transferred to inclusion bodies and the RpoC band was very intense in the Coomassie-stained polyacrylamide gels after separation of crude extracts (Fig. 7A, lane 2). Surprisingly, and accompanied by a decrease of the cellular zinc content from 73
000 atoms per cell (Fig. 2) in all these strains. The situation was more complicated concerning zinc allocation to RpoC. If this ability was disturbed, as in the ΔzupT single mutant, RpoC protein was transferred to inclusion bodies and the RpoC band was very intense in the Coomassie-stained polyacrylamide gels after separation of crude extracts (Fig. 7A, lane 2). Surprisingly, and accompanied by a decrease of the cellular zinc content from 73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 58
000 to 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 atoms per cell (Fig. 2), probably misfolded RpoC also accumulated in the ΔpitA strain (Fig. 7B, lane 3). In contrast, extracts of the ΔzupT ΔpitA double and the Δ4 quadruple mutant did not display a strong RpoC band, indicating efficient zinc allocation. Starting with the Δ5 mutant, crude extracts of all subsequent strains exhibited a strong RpoC band (Δ5 and Δ6 in lanes 6 and 7 of Fig. 7A, Δ7 not shown). Allocation of zinc to RpoC was efficient in the parent strain AE104, hampered when ZupT or when PitA were removed, efficient again when only CorA123 or only PitA was present, and no longer efficient in the subsequent higher order deletion strains. This could mean some negative interference between PitA and CorA123 on the one hand and of ZupT and CorA123 on the other, which prevented zinc allocation in the ΔzupT and ΔpitA mutant, respectively. On the other hand, PitA or CorA123 alone were able to substitute for ZupT when zinc allocation to RpoC was concerned, but their activity did not allow the stable presence of CzcA.
000 atoms per cell (Fig. 2), probably misfolded RpoC also accumulated in the ΔpitA strain (Fig. 7B, lane 3). In contrast, extracts of the ΔzupT ΔpitA double and the Δ4 quadruple mutant did not display a strong RpoC band, indicating efficient zinc allocation. Starting with the Δ5 mutant, crude extracts of all subsequent strains exhibited a strong RpoC band (Δ5 and Δ6 in lanes 6 and 7 of Fig. 7A, Δ7 not shown). Allocation of zinc to RpoC was efficient in the parent strain AE104, hampered when ZupT or when PitA were removed, efficient again when only CorA123 or only PitA was present, and no longer efficient in the subsequent higher order deletion strains. This could mean some negative interference between PitA and CorA123 on the one hand and of ZupT and CorA123 on the other, which prevented zinc allocation in the ΔzupT and ΔpitA mutant, respectively. On the other hand, PitA or CorA123 alone were able to substitute for ZupT when zinc allocation to RpoC was concerned, but their activity did not allow the stable presence of CzcA.
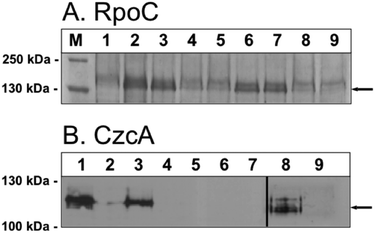 | ||
| Fig. 7 Presence of RpoC and CzcA proteins in mutant strains of C. metallidurans AE104. Accumulation of the RpoC subunit of the RNA polymerase (panel A, arrow) was demonstrated in crude extracts in Coomassie-stained SDS gel.21 Samples corresponding to 20 μg cellular dry mass was used per lane. Only the part of gel between the 130 kDa and the 250 kDa marker band is shown; the complete gels serving as loading control are shown in Fig. S2 (ESI†). The plasmid pDNA13026 that expressed the czcCBAD′ determinant constitutively in C. metallidurans cells had been transferred by conjugation into the mutant strains and their ability to produce the CzcA (panel B, arrow) central component of the CzcCBA efflux pump was determined in a Western blot using anti-CzcA antibodies, as published.21 Marker (M); AE104 negative control (1); ΔzupT positive control (2); ΔpitA (3); ΔzupT ΔpitA (4), Δ4 (5); Δ5 (6); Δ6 (7); Δe4 (8); Δe4 ΔzupT (9). As indicated by the bar, the experiment shown in panel B lanes 8 and 9 was on a different gel that that in B lanes 1 to 7. | ||
In agreement with a high cellular zinc content and resistance to EDTA, the Δe4 efflux mutant synthesized the CzcA protein and folded RpoC properly to prevent aggregation of this protein in inclusion bodies (Fig. 7, lanes 8). When zupT was deleted in Δe4 and the zinc content dropped to 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 atoms per cell, zinc allocation to RpoC was still efficient (Fig. 7A, lane 9), indicating that the negative interference of PitA and CorA123 did not occur in the Δe4 efflux mutant strain. The CzcA band was visible in Western blots of fresh cultures of the Δe4 ΔzupT mutant strains (data not shown) but similar to the situation in strain AE104 and its ΔzupT mutant, zinc resistance, as determined as IC50 values, decreased after more than 5 transfers on strain collection plates (Fig. S3, ESI†) and CzcA was no longer present in the cells (Fig. 7B, lane 9). While other import pathways could complement for a missing ZupT importer when it comes to cytoplasmic zinc allocation, the long-term presence of CzcA strictly depended on the presence of ZupT if the cells were not kept on media with increased zinc content.
000 atoms per cell, zinc allocation to RpoC was still efficient (Fig. 7A, lane 9), indicating that the negative interference of PitA and CorA123 did not occur in the Δe4 efflux mutant strain. The CzcA band was visible in Western blots of fresh cultures of the Δe4 ΔzupT mutant strains (data not shown) but similar to the situation in strain AE104 and its ΔzupT mutant, zinc resistance, as determined as IC50 values, decreased after more than 5 transfers on strain collection plates (Fig. S3, ESI†) and CzcA was no longer present in the cells (Fig. 7B, lane 9). While other import pathways could complement for a missing ZupT importer when it comes to cytoplasmic zinc allocation, the long-term presence of CzcA strictly depended on the presence of ZupT if the cells were not kept on media with increased zinc content.
Discussion
Heavy metal resistance can be defined as the ability to maintain metal homeostasis in the presence of high concentrations of metals or metal mixtures. In C. metallidurans, the plasmid-encoded RND-driven transenvelope systems such as CzcCBA and CnrCBA are responsible for the high resistance level to zinc and nickel, respectively.27–29 It is highly likely that efflux by these transenvelope protein complexes pre-adjusts the periplasmic metal content for subsequent uptake into the cytoplasm, or they transport metal ions to the outside that were previously exported from the cytoplasm to the periplasm by efflux systems of the inner membrane,5,6 such as P-type ATPases and CDF proteins. This demonstrates the importance of the metal transportome,30,31 the totality of all metal export and import systems, to metal resistance and homeostasis in C. metallidurans. The combined actions of metal uptake and efflux systems might result in a kinetic flow-equilibrium of the transition metal concentration in both cellular compartments,32 which adjusts individual concentrations and the composition of the overall mixture to prevent negative interference between “essential-but-also-toxic” metal cations such as Zn(II) and Ni(II).
E. coli does not possess CzcCBA- or CnrCBA-like efflux systems.33 In this gut bacterium, a controlled shunt of zinc import and export systems seems to be important for zinc homeostasis. Import is by a variety of systems with broad substrate specificity such as ZupT,18 complemented by the ABC transport system ZnuABC under zinc starvation conditions,14,15 or alternatively by ZntA-dependent export when the metal is in surplus.34–36 Expression of znuABC is under control of the Zur regulator of the Fur family of proteins,15,37 that of zntA for the zinc-transporting PIB2-type ATPases regulated by the MerR member ZntR.38 While zntA expression is 50% up-regulated at about 1 fM “free” zinc in the cytoplasm, that of znuABC is 50% up-regulated at about 0.2 fM “free” zinc, and both systems are below 25% expression level at about 0.5 fM.39 This equilibrium concentration of zinc in E. coli corresponds to a quota of 0.2 mM or 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 atoms per cell and a ratio of 2.5 × 10−12 between the zinc quota and “free” zinc. This can be explained by sequestration of Zn(II) in the cytoplasm by glutathione and zinc-binding proteins.22,40 This means that the distribution of Zn(II) between zinc-binding sites in ZntR, Zur and other proteins, e.g. those of the ribosome,41 may contribute to maintenance of the zinc homeostasis by activating alternatively zinc import by ZunABC or export by ZntA, so that this shunt controls the level of zinc in the cytoplasm of E. coli.
000 atoms per cell and a ratio of 2.5 × 10−12 between the zinc quota and “free” zinc. This can be explained by sequestration of Zn(II) in the cytoplasm by glutathione and zinc-binding proteins.22,40 This means that the distribution of Zn(II) between zinc-binding sites in ZntR, Zur and other proteins, e.g. those of the ribosome,41 may contribute to maintenance of the zinc homeostasis by activating alternatively zinc import by ZunABC or export by ZntA, so that this shunt controls the level of zinc in the cytoplasm of E. coli.
C. metallidurans cells contain a minimum of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc per cell, with approximately 70
000 zinc per cell, with approximately 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 when cultivated in TMM without added zinc. When zinc is added to the medium there are 120
000 when cultivated in TMM without added zinc. When zinc is added to the medium there are 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc per cell due to the action of the efflux systems, and the bacterium cannot grow when it harbors more than 250
000 zinc per cell due to the action of the efflux systems, and the bacterium cannot grow when it harbors more than 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc per cell.21 This corresponds to a quota of 58 μM, 204 μM, 350 μM and 728 μM, respectively (assumed cell volume of 0.57 fL42), so that the zinc quota of E. coli and C. metallidurans are similar. Unexpectedly, no evidence for a shunt was found in C. metallidurans.13 There is no ortholog in this bacterium for a ZnuABC uptake system but for a ZntA43 efflux pump. While expression of zupT in E. coli was constitutive,18zupT in C. metallidurans was controlled by its Zur ortholog.13,17 A main substrate and a role in basic resistance or high-level resistance could be assigned to all efflux systems in C. metallidurans;7 however this could not be done with the studied secondary metal import systems.13 This led to the hypothesis that a battery of redundant importers with low and overlapping substrate specificities was responsible for uptake of zinc and other transition metals in C. metallidurans.
000 zinc per cell.21 This corresponds to a quota of 58 μM, 204 μM, 350 μM and 728 μM, respectively (assumed cell volume of 0.57 fL42), so that the zinc quota of E. coli and C. metallidurans are similar. Unexpectedly, no evidence for a shunt was found in C. metallidurans.13 There is no ortholog in this bacterium for a ZnuABC uptake system but for a ZntA43 efflux pump. While expression of zupT in E. coli was constitutive,18zupT in C. metallidurans was controlled by its Zur ortholog.13,17 A main substrate and a role in basic resistance or high-level resistance could be assigned to all efflux systems in C. metallidurans;7 however this could not be done with the studied secondary metal import systems.13 This led to the hypothesis that a battery of redundant importers with low and overlapping substrate specificities was responsible for uptake of zinc and other transition metals in C. metallidurans.
Here, further evidence was found to support this hypothesis. It appears that C. metallidurans takes Zn(II), Ni(II) and Co(II) up as they become available. It relies on its efflux systems to remove surplus ions. Using the plasmid-encoded CzcCBA and CnrCBA systems it pre-adjusts the periplasmic metal composition, as well as concentration, prior to uptake across the inner membrane. The plasmid-free C. metallidurans derivative AE10427 is in this aspect an anomaly because it is not able to perform this periplasmic pre-adjustment but is forced to import zinc, cobalt, and nickel “as they come along”, while E. coli escapes this situation by using the ZnuABC/ZntA shunt for zinc, and maybe a similar shunt (NikABC/RcnA) for nickel.44,45 This also explains why strain AE104 suffers from transition metal stress in the cytoplasm, resulting in silencing of genomic islands even in TMM without added metals.23 However, it was not possible to construct similar deletion mutants in CH34 wild type because even deletion of zupT efficiently cures the czc-containing plasmid pMOL30,21 providing evidence that the interplay of metal uptake and efflux systems is important in C. metallidurans.
When the genes for the seven secondary uptake systems were deleted, the cells were able to compensate for some of the consequences of this, revealing flexibility of the metal uptake transportome. However if any other were removed no compensation was observe, pointing to unique and important contributions of individual systems. Removal of all seven importers did not impair the overall metal content of the cells (Fig. 2) so that additional metal uptake systems must exist. Up to the quintuple deletion mutant (Δ5) lacZ reporter gene fusions in various mutant strains did not show any up-regulation of the genes for the remaining uptake systems, nor was such a process evident on the transcriptome23 or proteome22 level of the ΔzupT deletion strain, despite the obvious zinc starvation conditions this strain was experiencing.23 Only in the sextuple deletion mutant, Δ6, was an altered expression pattern of the hoxN gene measured (Fig. 6B). This was due to recruitment of the predicted nickel importer HoxN for import and to compensate for the loss of the other systems. With the exception of HoxN, all the other known and unknown metal uptake systems were activated by means other than increased gene expression, such as flux control of, or protein–protein interaction with, already synthesized proteins. If this hypothesis is true, it would indicate the existence of a novel control mechanism of metal homeostasis in C. metallidurans.
Along the series of deletion mutants, the following observations were made: (i) deletion of zupT in the parent strain AE104 decreased fitness (Fig. 1); (ii) deletion of corA genes revealed an interplay of the CorA proteins (Table S2, ESI†); (iii) deletion of pitA increased the cellular magnesium content (Fig. 2) and influenced homeostasis of, and resistance to, most transition metals tested (Fig. 3). Nevertheless, deletion of these five genes did not decrease fitness (Fig. 1), only metal resistance (Fig. 3), indicating that use of other known (e.g. ZntB) or unknown importers compensated for the loss of the deleted genes. This also resulted in unbalanced metal import at μM metal concentrations that could no longer be compensated for by the efflux systems. Loss of zntB needed increased hoxN expression (Fig. 6B) to keep the fitness at the level of the ΔzupT strain (Fig. 1) but this was no longer possible in the Δ7 mutant (Fig. 1). Fitness and metal resistance dropped to a second, lower level (Fig. 1 and 3).
First, all these findings highlight the importance of ZupT among the other secondary metal uptake systems in C. metallidurans. ZupT was required in strain AE104 to allocate zinc to the client protein RpoC efficiently and this function could only partially be compensated for by an increased cellular zinc content in the ΔzupT mutant.21 Zinc imported by ZupT was required to allow a stable presence of the RND protein CzcA or the CzcCBA zinc efflux complex.21 While ZupT was absolutely essential for the long-term presence of CzcA, PitA and the CorAs were able to mediate efficient allocation of zinc to RpoC, but only when the respective other importer was not present in the cell (Fig. 7). Moreover, this negative interference between PitA and the CorAs did not occur in the ΔzupT mutant of the quadruple efflux mutant Δe4, which was able to fold RpoC efficiently but unable to harbor CzcA (Fig. 7). Replenishment of the zinc pool required for RpoC folding nevertheless was not sufficient to allow stable synthesis of CzcA.21 PitA and the CorAs must somehow interact, e.g. on the hypothetical post-translational control level of metal homeostasis in C. metallidurans.
Second, the three CorAs are able to provide Zn(II) to RpoC in the ΔzupT ΔpitA mutant (Fig. 7). Moreover, CorA1 and CorA2 are involved in nickel import. CorA1 and CorA3 have something to do with cobalt toxicity although CorA123 together functioned as cobalt importers. TMM-grown C. metallidurans cells contain 117 ± 8 copies of CorA1 (Rmet_3052) per cell, 38 ± 19 copies of CorA2 (Rmet_0036) and 37 ± 5 copies of CorA3 (Rmet_3287), yielding a 3.2 ± 0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0 ± 0.5
1.0 ± 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ± 0.1 ratio of CorA1
1 ± 0.1 ratio of CorA1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CorA2
CorA2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CorA3.22 Since CorA-like proteins are pentamers,24 this could mean that the CorA of C. metallidurans is actually a (CorA1)3(CorA2)1(CorA3)1 heteromultimer. CorA is the primary magnesium channel in Salmonella46 and essential for virulence.47 Metal uptake in the archaeum Methanococcus jannaschii is in the order Co(II) > Ni(II) > Mn(II) > Mg(II) > Ca(II).48 Co(II) and Ni(II) – but not Ca(II) and Fe(II/III) transport – was also demonstrated for a homolog from the plant Arabidopsis thaliana.49 In C. metallidurans, contribution of CorA123 to cobalt, zinc, nickel, and cadmium import explains the metal-resistance phenotypes of the deletion mutants.
CorA3.22 Since CorA-like proteins are pentamers,24 this could mean that the CorA of C. metallidurans is actually a (CorA1)3(CorA2)1(CorA3)1 heteromultimer. CorA is the primary magnesium channel in Salmonella46 and essential for virulence.47 Metal uptake in the archaeum Methanococcus jannaschii is in the order Co(II) > Ni(II) > Mn(II) > Mg(II) > Ca(II).48 Co(II) and Ni(II) – but not Ca(II) and Fe(II/III) transport – was also demonstrated for a homolog from the plant Arabidopsis thaliana.49 In C. metallidurans, contribution of CorA123 to cobalt, zinc, nickel, and cadmium import explains the metal-resistance phenotypes of the deletion mutants.
Import activity of CorA from T. maritima is under flux control. The pentameric complex is locked in a transport-incompetent conformation by allosteric, sequential binding of cytoplasmic Mg(II) cations to the five protomers.50 Because the amino acids required for transport and flux control by gating are all conserved within the MIT protein family, eukaryotic homologs included, this flux-controlled uptake of magnesium seems to be a general mechanism.51 This may indicate that the individual protomers of the possible heteromultimeric CorA123 importer in C. metallidurans could be under different flux control regimes, CorA1 by Mg(II), CorA2 by Ni(II) and CorA3 by Co(II). These CorA substrates were identified already a long time ago in E. coli and Aerobacter (=Enterobacter) aerogenes,52 nickel uptake by a magnesium uptake system in Cupriavidus necator (=Ralstonia eutropha, Alcaligenes eutrophus)53 and C. metallidurans.32,54 Moreover, expression of corA1 is up-regulated 3-fold under conditions of magnesium starvation while that of corA2 and of corA3 is slightly down-regulated,13 so that the complex may change from a homopentameric CorA1 under magnesium flux control at low (10 μM) magnesium concentrations to a heteropentameric CorA123 under combined magnesium, nickel and cobalt flux control at magnesium concentrations above 100 μM. Heteromultimers with different functions have also been described for other metal transporters25 so that this hypothesis should be tested.
Third, PitA was an important metal uptake system. In E. coli, PitA also seems to be involved in zinc uptake55 and some strains of this bacterium even contain a paralog of PitA, PitB.56 Consequently, mutations in pitA cause zinc resistance.57 PitA also imports phosphate complexes with magnesium and calcium, and Zn(II) competes with Mg(II) for import by PitA.57 This would be a very energy-efficient import route for essential micro- and macroelements. If, however, all 110 million phosphate molecules needed by C. metallidurans13 would be imported as metal:phosphate complexes by PitA, far too many metal cations would be imported into the cytoplasm, e.g. 76 million Mg(II) with 11 million actually needed. These surplus cations would have to be exported again.
As an alternative for energy-dependent efflux of surplus Mg(II) and Ca(II), metal:phosphate import by the low-affinity PitA transporter could be synchronized with phosphate import by the high-affinity ABC-type import system PstABC, which transports non-metallated H2PO4− and HPO42−.58,59C. metallidurans contains 1700 copies of the periplasmic-binding component PstS. Its genome possesses genes for two additional paralogs of this protein, one synthesized in 100 copies per cell.22 This argues for a strong participation of PstABC in phosphate import in TMM-grown cells, which may have the role to prevent any over-accumulation of metal cations. This scenario agrees with the fact that PitA is up-regulated with increasing phosphate concentrations13 starting at 100 μM phosphate and reaching maximum expression at 5 mM phosphate: at such a high phosphate concentration, use of the energy-efficient PitA system would be much more attractive than importing phosphate by the “expensive” PstACB pathway.
Last, there are also conflicting data concerning the 4th member of the MIT protein family in C. metallidurans, ZntB. In Salmonella, mutations in its zntB gene conferred decreased zinc and cadmium resistance and capacity for zinc efflux was demonstrated.60 The structure of ZntB is that of a pentameric funnel similar to CorA.61 In C. metallidurans, however, a zntB-lacZ fusion was down-regulated with increasing zinc and especially cadmium concentrations (Fig. 6). This regulatory pattern agrees more with that of an importer than with an exporter. The consequence of a zntB deletion, even from the genome of the Δ5 mutant with ZupT, PitA and CorA123 removed, was minor with the exception of a two-fold increase in zinc and cadmium resistance. This assigned to ZntB the role of a minor zinc and cadmium importer in C. metallidurans. Expression of a hoxN-lacZ fusion increased in the Δ6 strain compared to Δ5, indicating that additional deletion of zntB resulted in activation of the last known secondary import system with some potential in zinc import. Consequently, the Δ6 strain performed not much differently from the ΔzupT single deletion strain, while the growth problems of the Δ7 (=Δ6 ΔhoxB) were considerable.
Experimental
Materials and methods
| Name | Relevant markers, TCDB of the product70 | Ref. |
|---|---|---|
| Bacterial strains | ||
| Cupriavidus metallidurans | ||
| AE104 | Plasmid-free | 27 |
| DN515 | ΔzupT, (TC2.A.5) | 13 |
| DN527 | ΔpitA, (TC2.A.20) | 13 |
| DN675 | corA 1::kan, (TC1.A.35) | 13 |
| DN528 | ΔcorA2, (TC1.A.35) | 13 |
| DN529 | ΔcorA3. (TC1.A.35) | 13 |
| DN785 | ΔzntB, (TC1.A.35) | 13 |
| DN786 | ΔhoxN, (TC2.A.52) | 13 |
| DN578, Δe4 | ΔzntA Δcad ΔdmeF ΔfieF | 7 |
| DN579, Δe4z | Δe4 ΔzupT | 21 |
| DN532 | ΔzupT ΔpitA | 13 |
| DN674 | ΔzupT corA1::kan | This study |
| DN533 | ΔzupT ΔcorA2 | This study |
| DN534 | ΔzupT ΔcorA3 | This study |
| DN671 | ΔpitA corA1::kan | This study |
| DN530 | ΔpitA ΔcorA2 | This study |
| DN531 | ΔpitA ΔcorA3 | This study |
| DN666 | corA 1::kan ΔcorA2 | This study |
| DN667 | corA 1::kan ΔcorA3 | This study |
| DN787 | ΔcorA2 ΔcorA3 | This study |
| DN788 | ΔzupT ΔhoxN | This study |
| DN535 | ΔzupT ΔpitA ΔcorA3 | This study |
| DN789 | corA 1::kan ΔcorA2 ΔcorA3 | This study |
| DN790 | ΔzupT ΔhoxN ΔzntB | This study |
| DN580, Δ4 | ΔzupT ΔcorA1 ΔcorA2 ΔcorA3 | This study |
| DN791 | ΔzupT ΔpitA corA1::kan ΔcorA2 | This study |
| DN668 | ΔzupT ΔpitA corA1::kan ΔcorA3 | This study |
| DN792 | ΔzupT ΔpitA ΔcorA2 ΔcorA3 | This study |
| DN793 | ΔzupT ΔpitA ΔhoxN ΔzntB | This study |
| DN665, Δ5 | ΔzupT ΔpitA ΔcorA1 ΔcorA2 ΔcorA3 | This study |
| DN676, Δ6 | ΔzupT ΔpitA ΔcorA1 ΔcorA2 ΔcorA3 ΔzntB | This study |
| DN794 | ΔzupT ΔpitA ΔcorA1 ΔcorA2 ΔcorA3 ΔhoxN | This study |
| DN681, Δ7 | ΔzupT ΔpitA ΔcorA1 ΔcorA2 ΔcorA3ΔzntB ΔhoxN | This study |
| Conjugator strain | ||
| Escherichia coli | ||
| S17/1 | Plasmids | 63 |
| pECD794-1 | lacZ, derivate of pLO2 | 7 |
| pCM157 | Contains Cre recombinase | 65 |
| pECD1003 | Mutant lox sites, derivate of pECD889 | 7 |
| pVDZ2 | Broad host range expression vector | 71 |
| pDNA130 | pVDZ2::czcCBAD′ | 26 |
| pECD977 | pECD1003 construct for zupT deletion by cre-lox system | 13 |
| pECD981 | pECD1003 construct for pitA deletion by cre-lox system | 13 |
| pECD982 | pECD1003 construct for corA1 deletion by cre-lox system | 13 |
| pECD983 | pECD1003 construct for corA2 deletion by cre-lox system | 13 |
| pECD984 | pECD1003 construct for corA3 deletion by cre-lox system | 13 |
| pECD1260 | pECD1003 constr. for zntB deletion by cre-lox system | This study |
| pECD1144 | pECD1003 constr. for hoxN del. by cre-lox system | This study |
| pECD1259 | pECD794 constr. for corA1 disruption | 13 |
| pECD985 | pECD794 constr. for zupT LacZ reportergene | 13 |
| pECD986 | pECD794 constr. for pitA LacZ reportergene | 13 |
| pECD988 | pECD794 constr. for corA1 LacZ reportergene | 13 |
| pECD989 | pECD794 constr. for corA2 LacZ reportergene | 13 |
| pECD990 | pECD794 constr. for corA3 LacZ reportergene | 13 |
| pECD1486 | pECD794 constr. for zntB LacZ reportergene | This study |
| pECD1487 | pECD794 constr. for hoxN LacZ reportergene | This study |
| pECD1490 | pECD794 constr. for rpoB LacZ reportergene | This study |
| pECD1491 | pECD794 constr. for Enolase LacZ reportergene | This study |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and plated onto nutrient broth agar. After 2 d, the bacteria were suspended in TMM, diluted, and plated onto selective media as previously described.28 All primer pairs used were listed in Table S6 (ESI†). Plasmid pECD10037 was used to construct deletion mutants. It is a derivate of plasmid pECD88964 and therefore a derivative of plasmid pCM184.65 These plasmids harbor a kanamycin-resistance cassette flanked by loxP recognition sites. Plasmid pECD1003 additionally carries an exchange of 5 bp at each loxP-site. Using these mutant lox sequences, multiple gene deletions within the same genome are possible without secondary recombination events.66,67
1) and plated onto nutrient broth agar. After 2 d, the bacteria were suspended in TMM, diluted, and plated onto selective media as previously described.28 All primer pairs used were listed in Table S6 (ESI†). Plasmid pECD10037 was used to construct deletion mutants. It is a derivate of plasmid pECD88964 and therefore a derivative of plasmid pCM184.65 These plasmids harbor a kanamycin-resistance cassette flanked by loxP recognition sites. Plasmid pECD1003 additionally carries an exchange of 5 bp at each loxP-site. Using these mutant lox sequences, multiple gene deletions within the same genome are possible without secondary recombination events.66,67
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 in fresh medium and incubated for 24 h at 30 C and 250 rpm. This 24 h culture was diluted 1
20 in fresh medium and incubated for 24 h at 30 C and 250 rpm. This 24 h culture was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in fresh medium and used for streaking onto plates containing different concentration of the respective metal salts. The plates were incubated at 30 °C for 3 days and cell growth was monitored.
100 in fresh medium and used for streaking onto plates containing different concentration of the respective metal salts. The plates were incubated at 30 °C for 3 days and cell growth was monitored.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 in fresh medium and incubated for 24 h at 30 °C and 250 rpm. Overnight cultures were used to inoculate parallel cultures with increasing metal concentrations in 96-well plates (Greiner). Cells were cultivated for 20 h at 30 °C and 1300 rpm in a neoLab Shaker DTS-2 (neoLab, Heidelberg, Germany) and the optical density was determined at 500 nm or 600 nm as indicated in a TECAN infinite 200 PRO reader (TECAN, Männersdorf, Switzerland). To calculate the IC50 value (metal concentration that led to turbidity reduction by half) and the corresponding b-value (measure of the slope of the sigmoidal dose–response curve), the data were adapted to the formula OD(c) = OD0/{1 + exp((c − IC50)/b)}, which is a simplified version of a Hill-type equation as introduced by Pace and Scholtz (1997)68 as published.69 OD(c) is the turbidity at a given metal concentration; OD0, that at no added metal and c the metal concentration.
20 in fresh medium and incubated for 24 h at 30 °C and 250 rpm. Overnight cultures were used to inoculate parallel cultures with increasing metal concentrations in 96-well plates (Greiner). Cells were cultivated for 20 h at 30 °C and 1300 rpm in a neoLab Shaker DTS-2 (neoLab, Heidelberg, Germany) and the optical density was determined at 500 nm or 600 nm as indicated in a TECAN infinite 200 PRO reader (TECAN, Männersdorf, Switzerland). To calculate the IC50 value (metal concentration that led to turbidity reduction by half) and the corresponding b-value (measure of the slope of the sigmoidal dose–response curve), the data were adapted to the formula OD(c) = OD0/{1 + exp((c − IC50)/b)}, which is a simplified version of a Hill-type equation as introduced by Pace and Scholtz (1997)68 as published.69 OD(c) is the turbidity at a given metal concentration; OD0, that at no added metal and c the metal concentration.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with cracking buffer (final concentrations 125 mM Tris-HCl, pH 6.8, 20 g L−1 SDS; 0.5% (vol/vol) β-mercaptoethanol, 0.01 g L−1 bromophenol blue, 50% (vol/vol) glycerol), incubated for 30 min at 50 °C and loaded onto a SDS gel.73 The gel was stained with Coomassie Brilliant Blue or blotted (Trans-Blot, BioRad) onto a nitrocellulose membrane, dried by air and then blocked in PBS (4 mM KH2PO4, 16 mM Na2HPO4, 115 mM NaCl), 5 mL Tween20 and 50 g skimmed milk per l with shaking at 23 °C for 16 h. The membrane was washed 3 times for 5 min with PBS-Tween (PBS, 1 mL Tween20 L−1) at 23 °C. The membrane was incubated for 1 h at 23 °C with shaking with the first antibody {polyclonal CzcA-antibody 1
1 with cracking buffer (final concentrations 125 mM Tris-HCl, pH 6.8, 20 g L−1 SDS; 0.5% (vol/vol) β-mercaptoethanol, 0.01 g L−1 bromophenol blue, 50% (vol/vol) glycerol), incubated for 30 min at 50 °C and loaded onto a SDS gel.73 The gel was stained with Coomassie Brilliant Blue or blotted (Trans-Blot, BioRad) onto a nitrocellulose membrane, dried by air and then blocked in PBS (4 mM KH2PO4, 16 mM Na2HPO4, 115 mM NaCl), 5 mL Tween20 and 50 g skimmed milk per l with shaking at 23 °C for 16 h. The membrane was washed 3 times for 5 min with PBS-Tween (PBS, 1 mL Tween20 L−1) at 23 °C. The membrane was incubated for 1 h at 23 °C with shaking with the first antibody {polyclonal CzcA-antibody 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15
15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, diluted in PBS-Tween, antibody raised in rabbits after CzcA purification74}. The membrane was washed three times for 5 min with PBS-Tween and twice for 5 min with PBS, incubated with the second antibody {a monoclonal anti-rabbit-IgG conjugated with horseradish peroxidase (Sigma-Aldrich) diluted 1
000, diluted in PBS-Tween, antibody raised in rabbits after CzcA purification74}. The membrane was washed three times for 5 min with PBS-Tween and twice for 5 min with PBS, incubated with the second antibody {a monoclonal anti-rabbit-IgG conjugated with horseradish peroxidase (Sigma-Aldrich) diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in PBS-Tween} for 1 h at 23 °C with shaking. Unbound antibody was washed off the membrane 3 times for 5 min with PBS-Tween and once for 5 min with PBS. For detection, the membrane was incubated with 5 mL solution 1 {10 mM TrisHCl (pH 8.0), 150 mM NaCl, 0.4 mM p-coumaric acid, 0.25 mM luminol} and 5 mL solution 2 {10 mM TrisHCl (pH 8.0), 150 mM NaCl, 5.4 M H2O2} for 1 min. The excess liquid was wiped off and the membrane was exposed for 5 s to 5–15 min to chemo-luminescence detection film (Roche).
000 in PBS-Tween} for 1 h at 23 °C with shaking. Unbound antibody was washed off the membrane 3 times for 5 min with PBS-Tween and once for 5 min with PBS. For detection, the membrane was incubated with 5 mL solution 1 {10 mM TrisHCl (pH 8.0), 150 mM NaCl, 0.4 mM p-coumaric acid, 0.25 mM luminol} and 5 mL solution 2 {10 mM TrisHCl (pH 8.0), 150 mM NaCl, 5.4 M H2O2} for 1 min. The excess liquid was wiped off and the membrane was exposed for 5 s to 5–15 min to chemo-luminescence detection film (Roche).
Conclusions
The seven secondary metal uptake systems characterized in this study appear to communicate to form the core of the metal transportome30,31 of C. metallidurans AE104 by an interplay with the four metal efflux systems ZntA, CadA, DmeF and FieF. This is supported by a remarkable similarity of the metal content and cobalt resistance, a feature severely affected by deletion of metal import genes, of the Δ7 metal uptake and the Δe4 efflux mutant. Since Δ7 is to some degree still able to maintain its cellular metal composition, more metal uptake systems must exist in C. metallidurans beyond the seven secondary importers. Up to Δ6, the plasticity of the metal uptake transportome revealed here seems not to involve increased gene expression for substitute importers but rather post-transcriptional events take over. Taken together with the disappearance of CzcA in zupT mutants, an important post-transcriptional level of control of the metal transportome seems to exist in C. metallidurans.Acknowledgements
Funding for this work was provided by the Deutsche Forschungsgemeinschaft (Ni262/10). We thank Grit Schleuder for skilful technical assistance and Gary Sawers for helpful comments.Notes and references
- P. J. Janssen, R. Van Houdt, H. Moors, P. Monsieurs, N. Morin, A. Michaux, M. A. Benotmane, N. Leys, T. Vallaeys, A. Lapidus, S. Monchy, C. Medigue, S. Taghavi, S. McCorkle, J. Dunn, D. van der Lelie and M. Mergeay, PLoS One, 2010, 5, e10433 Search PubMed.
- L. Diels and M. Mergeay, Appl. Environ. Microbiol., 1990, 56, 1485–1491 CAS.
- F. Reith, S. L. Rogers, D. C. McPhail and D. Webb, Science, 2006, 313, 233–236 CrossRef CAS PubMed.
- C. Dressler, U. Kües, D. H. Nies and B. Friedrich, Appl. Environ. Microbiol., 1991, 57, 3079–3085 CAS.
- D. H. Nies, in Molecular Microbiology of Heavy Metals, ed. D. H. Nies and S. Silver, Springer-Verlag, Berlin, 2007, vol. 6, pp. 118–142 Search PubMed.
- D. H. Nies, in Microbial Efflux Pumps: Current Research, ed. E. W. Yu, Q. Zhang and M. H. Brown, Caister Academic Press, Norfolk, UK, 2013, pp. 79–122 Search PubMed.
- J. Scherer and D. H. Nies, Mol. Microbiol., 2009, 73, 601–621 CrossRef CAS PubMed.
- W. Busch and M. H. J. Saier, Crit. Rev. Biochem. Mol. Biol., 2002, 37, 287–337 CrossRef CAS PubMed.
- H. Nikaido and M. H. Saier, Jr., Science, 1992, 258, 936–942 CAS.
- M. H. J. Saier, C. V. Tran and R. D. Barabote, Nucleic Acids Res., 2006, 34, D181–D186 CrossRef CAS PubMed.
- T. von Rozycki and D. H. Nies, Antonie van Leeuwenhoek, 2009, 96, 115–139 CrossRef CAS PubMed.
- T. von Rozycki, D. H. Nies and M. H. J. Saier, Comp. Funct. Genomics, 2005, 6, 17–56 CrossRef CAS PubMed.
- A. Kirsten, M. Herzberg, A. Voigt, J. Seravalli, G. Grass, J. Scherer and D. H. Nies, J. Bacteriol., 2011, 193, 4652–4663 CrossRef CAS PubMed.
- S. I. Patzer and K. Hantke, Mol. Microbiol., 1998, 28, 1199–1210 CrossRef CAS PubMed.
- S. I. Patzer and K. Hantke, J. Biol. Chem., 2000, 275, 24321–24332 CrossRef CAS PubMed.
- C. Navarro, L. F. Wu and M. A. Mandrand-Berthelot, Mol. Microbiol., 1993, 9, 1181–1191 CrossRef CAS PubMed.
- C. Schmidt, C. Schwarzenberger, C. Grosse and D. H. Nies, J. Bacteriol., 2014, 196, 3461–3471 CrossRef PubMed.
- G. Grass, S. Franke, N. Taudte, D. H. Nies, L. M. Kucharski, M. E. Maguire and C. Rensing, J. Bacteriol., 2005, 187, 1604–1611 CrossRef CAS PubMed.
- G. Grass, M. D. Wong, B. P. Rosen, R. L. Smith and C. Rensing, J. Bacteriol., 2002, 184, 864–866 CrossRef CAS PubMed.
- N. Taudte and G. Grass, BioMetals, 2010, 23, 643–656 CrossRef CAS PubMed.
- M. Herzberg, L. Bauer and D. H. Nies, Metallomics, 2014, 6, 421–436 RSC.
- M. Herzberg, D. Dobritzsch, S. Helm, S. Baginski and D. H. Nies, Metallomics, 2014, 6, 2157–2165 RSC.
- M. Herzberg, M. Schüttau, M. Reimers, C. Grosse, H. G. Schlegel and D. H. Nies, Metallomics, 2015, 7, 632–649 RSC.
- V. Lunin, E. Dobrovetsky, G. Khutoreskaya, R. Zhang, A. Joachimiak, D. A. Doyle, A. Bochkarev, M. E. Maguire, A. M. Edwards and C. M. Koth, Nature, 2006, 440, 833–837 CrossRef CAS PubMed.
- Y. Golan, B. Berman and Y. G. Assaraf, J. Biol. Chem., 2015, 290, 9050–9063 CrossRef CAS PubMed.
- D. H. Nies, A. Nies, L. Chu and S. Silver, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 7351–7355 CrossRef CAS.
- M. Mergeay, D. Nies, H. G. Schlegel, J. Gerits, P. Charles and F. van Gijsegem, J. Bacteriol., 1985, 162, 328–334 CAS.
- D. Nies, M. Mergeay, B. Friedrich and H. G. Schlegel, J. Bacteriol., 1987, 169, 4865–4868 CAS.
- H. Liesegang, K. Lemke, R. A. Siddiqui and H.-G. Schlegel, J. Bacteriol., 1993, 175, 767–778 CAS.
- Y. Huang, P. Anderle, K. J. Bussey, C. Barbacioru, U. Shankavaram, Z. Y. Dai, W. C. Reinhold, A. Papp, J. N. Weinstein and W. Sadee, Cancer Res., 2004, 64, 4294–4301 CrossRef CAS PubMed.
- T. H. Mauchline, J. E. Fowler, A. K. East, A. L. Sartor, R. Zaheer, A. H. F. Hosie, P. S. Poole and T. M. Finan, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 17933–17938 CrossRef CAS PubMed.
- A. Legatzki, S. Franke, S. Lucke, T. Hoffmann, A. Anton, D. Neumann and D. H. Nies, Biodegradation, 2003, 14, 153–168 CrossRef CAS PubMed.
- D. H. Nies and G. Grass, in EcoSal – Escherichia coli and Salmonella: cellular and molecular biology, ed. A. Böck, R. Curtiss III, J. B. Kaper, F. C. Neidhardt, T. Nyström, K. E. Rudd and C. L. Squires, ASM Press, Washington, D.C., 2009, DOI: http://10.1128/ecosal.5.4.4.3 Search PubMed.
- C. Rensing, B. Mitra and B. P. Rosen, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 14326–14331 CrossRef CAS.
- R. Sharma, C. Rensing, B. P. Rosen and B. Mitra, J. Biol. Chem., 2000, 275, 3873–3878 CrossRef CAS PubMed.
- J. B. Liu, S. J. Dutta, A. J. Stemmler and B. Mitra, Biochemistry, 2006, 45, 763–772 CrossRef CAS PubMed.
- C. E. Outten, F. W. Outten and T. V. O'Halloran, J. Biol. Chem., 1999, 274, 37517–37524 CrossRef CAS PubMed.
- K. R. Brocklehurst, J. L. Hobman, B. Lawley, L. Blank, S. J. Marshall, N. L. Brown and A. P. Morby, Mol. Microbiol., 1999, 31, 893–902 CrossRef CAS PubMed.
- C. E. Outten and T. V. O'Halloran, Science, 2001, 292, 2488–2492 CrossRef CAS PubMed.
- K. Helbig, C. Bleuel, G. J. Krauss and D. H. Nies, J. Bacteriol., 2008, 190, 5431–5438 CrossRef CAS PubMed.
- M. P. Hensley, D. L. Tierney and M. W. Crowder, Biochemistry, 2011, 50, 9937–9939 CrossRef CAS PubMed.
- J. Goris, P. De Vos, T. Coenye, B. Hoste, D. Janssens, H. Brim, L. Diels, M. Mergeay, K. Kersters and P. Vandamme, Int. J. Syst. Evol. Microbiol., 2001, 51, 1773–1782 CrossRef CAS PubMed.
- A. Legatzki, A. Anton, G. Grass, C. Rensing and D. H. Nies, J. Bacteriol., 2003, 185, 4354–4361 CrossRef CAS PubMed.
- D. Koch, D. H. Nies and G. Grass, BioMetals, 2007, 20, 759–771 CrossRef CAS PubMed.
- A. Rodrigue, G. Effantin and M. A. Mandrand-Berthelot, J. Bacteriol., 2005, 187, 2912–2916 CrossRef CAS PubMed.
- J. Payandeh, C. Li, M. Ramjeesingh, E. Poduch, C. E. Bear and E. F. Pai, J. Biol. Chem., 2008, 283, 11721–11733 CrossRef CAS PubMed.
- K. M. Papp-Wallace, M. Nartea, D. G. Kehres, S. Porwollik, M. McClelland, S. J. Libby, F. C. Fang and M. E. Maguire, J. Bacteriol., 2008, 190, 6517–6523 CrossRef CAS PubMed.
- J. Kean, R. M. Cleverley, L. O'Ryan, R. C. Ford, S. M. Prince and J. P. Derrick, Mol. Membr. Biol., 2008, 25, 653–661 CrossRef CAS PubMed.
- S. Ishijima, Z. Shigemi, H. Adachi, N. Makinouchi and I. Sagami, Biochim. Biophys. Acta, Biomembr., 2012, 1818, 2202–2208 CrossRef CAS PubMed.
- R. Pfoh, A. Li, N. Chakrabarti, J. Payandeh, R. Pomes and E. F. Pai, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 18809–18814 CrossRef CAS PubMed.
- A. Guskov, N. Nordin, A. Reynaud, H. Engman, A. K. Lundback, A. J. O. Jong, T. Cornvik, T. Phua and S. Eshaghi, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 18459–18464 CrossRef CAS PubMed.
- M. Webb, Biochim. Biophys. Acta, 1970, 222, 428–439 CrossRef CAS.
- M. Lohmeyer and C. G. Friedrich, Arch. Microbiol., 1987, 149, 130–135 CrossRef CAS.
- D. H. Nies and S. Silver, J. Bacteriol., 1989, 171, 4073–4075 CAS.
- S. J. Beard, R. Hashim, G. H. Wu, M. R. B. Binet, M. N. Hughes and R. K. Poole, FEMS Microbiol. Lett., 2000, 184, 231–235 CrossRef CAS PubMed.
- R. M. Harris, D. C. Webb, S. M. Howitt and G. B. Cox, J. Bacteriol., 2001, 183, 5008–5014 CrossRef CAS PubMed.
- R. J. Jackson, M. R. B. Binet, L. J. Lee, R. Ma, A. I. Graham, C. W. McLeod and R. K. Poole, FEMS Microbiol. Lett., 2008, 289, 219–224 CrossRef CAS PubMed.
- Z. M. Wang, A. Choudhary, P. S. Ledvina and F. A. Quiocho, J. Biol. Chem., 1994, 269, 25091–25094 CAS.
- H. Rosenberg, R. G. Gerdes and K. Chegwidden, J. Bacteriol., 1977, 131, 505–511 CAS.
- A. J. Worlock and R. L. Smith, J. Bacteriol., 2002, 184, 4369–4373 CrossRef CAS PubMed.
- K. M. Tan, A. Sather, J. L. Robertson, S. Moy, B. Roux and A. Joachimiak, Protein Sci., 2009, 18, 2043–2052 CrossRef CAS PubMed.
- J. Sambrook, E. F. Fritsch and T. Maniatis, Molecular cloning, a laboratory manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y., 2nd edn, 1989 Search PubMed.
- R. Simon, U. Priefer and A. Pühler, Bio/Technology, 1983, 1, 784–791 CrossRef CAS.
- C. Große, S. Friedrich and D. H. Nies, J. Mol. Microbiol. Biotechnol., 2007, 12, 227–240 CrossRef PubMed.
- C. J. Marx and M. E. Lidstrom, Biotechniques, 2002, 33, 1062–1067 CAS.
- N. Suzuki, H. Nonaka, Y. Tsuge, M. Inui and H. Yukawa, Appl. Environ. Microbiol., 2005, 71, 8472–8480 CrossRef CAS PubMed.
- H. Albert, E. C. Dale, E. Lee and D. W. Ow, Plant J., 1995, 7, 649–659 CAS.
- C. N. Pace and M. J. Scholtz, in Protein Structure: A Practical Approach, ed. T. Creighton, IRL Press, Oxford, UK, 1997, pp. 299–322 Search PubMed.
- A. Anton, A. Weltrowski, J. H. Haney, S. Franke, G. Grass, C. Rensing and D. H. Nies, J. Bacteriol., 2004, 186, 7499–7507 CrossRef CAS PubMed.
- M. H. Saier, Jr., C. V. Tran and R. D. Barabote, Nucleic Acids Res., 2006, 34, D181–186 CrossRef PubMed.
- V. Deretic, S. Chandrasekharappa, J. F. Gill, D. K. Chatterjee and A. Chakrabarty, Gene, 1987, 57, 61–72 CrossRef CAS PubMed.
- D. H. Nies, J. Bacteriol., 1992, 174, 8102–8110 CAS.
- K. Weber, J. R. Pringle and M. Osborn, Methods Enzymol., 1972, 26, 3–27 CAS.
- M. Goldberg, T. Pribyl, S. Juhnke and D. H. Nies, J. Biol. Chem., 1999, 274, 26065–26070 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5mt00295h |
| This journal is © The Royal Society of Chemistry 2016 |