FecB, a periplasmic ferric-citrate transporter from E. coli, can bind different forms of ferric-citrate as well as a wide variety of metal-free and metal-loaded tricarboxylic acids†
Sambuddha
Banerjee
,
Subrata
Paul
,
Leonard T.
Nguyen
,
Byron C. H.
Chu
and
Hans J.
Vogel
*
Biochemistry Research Group, Department of Biological Sciences, University of Calgary, 2500 University Dr. NW, Calgary, Alberta T2N 1N4, Canada. E-mail: vogel@ucalgary.ca; Fax: +1-(403)-289-9311; Tel: +1-(403)-220-6006
First published on 13th November 2015
Abstract
The Escherichia coli Fec system, consisting of an outer membrane receptor (FecA), a periplasmic substrate binding protein (FecB) and an inner membrane permease-ATPase type transporter (FecC/D), plays an important role in the uptake and transport of Fe3+-citrate. Although several FecB sequences from various organisms have been reported, there are no biophysical or structural data available for this protein to date. In this work, using isothermal titration calorimetry (ITC), we report for the first time the ability of FecB to bind different species of Fe3+-citrate as well as other citrate complexes with trivalent (Ga3+, Al3+, Sc3+ and In3+) and a representative divalent metal ion (Mg2+) with low μM affinity. Interestingly, ITC experiments with various iron-free di- and tricarboxylic acids show that FecB can bind tricarboxylates with μM affinity but not biologically relevant dicarboxylates. The ability of FecB to bind with metal-free citrate is also observed in 1H,15N HSQC-NMR titration experiments reported here at two different pH values. Further, differential scanning calorimetry (DSC) experiments indicate that the ligand-bound form of FecB has greater thermal stability than ligand-free FecB under all pH and ligand conditions tested, which is consistent with the idea of domain closure subsequent to ligand binding for this type of periplasmic binding proteins.
Introduction
Iron is the second most abundant metal in the earth's crust and it plays a crucial role in biology owing to its versatile redox chemistry which in turn is controlled by the ligands present in its first and second coordination shells.1 In spite of its extreme usefulness in biology, the paradox of iron in biology is its high insolubility and toxic free radical forming ability, especially under neutral and oxic conditions.1 Hence biological systems have evolved mechanisms to sequester iron tightly by expressing proteins that will bind it effectively to slow down redox reactions and precipitation. Like higher organisms most bacterial species, with the exception of Lactobacillus, require iron for their growth and survival.2,3 Sequestration of iron in living systems leads to a very low concentration of available free iron that can be used by invading bacterial species.4 For many bacterial species the acquisition of iron from the host system determines pathogenesis and hence this has been reported as the iron-bio-fulcrum by previous workers.5Non-heme iron uptake systems in Gram negative bacteria are comprised mainly of two distinct pathways. Many bacteria produce and secrete low molecular weight, yet high affinity Fe3+ binding molecules called siderophores which are sent out into the host environment and compete for and sequester Fe3+ bound to host proteins. Several bacterial species have also evolved mechanisms to utilize siderophores produced by other bacterial species (Xenosiderophores) living in the same biological niche. The ferric-siderophores are then recognized and transported across the outer membrane (OM) using specific TonB-dependent receptor proteins (ESI,† Fig. S1A,4–8). Alternatively, some pathogenic bacteria can express cognate transferrin (Tf) or lactoferrin (Lf) binding proteins on the OM which bind to holo-Tf/Lf and steal iron from these host proteins. Iron removed in this manner from the host proteins is then transported via a cognate TonB-dependent receptor (ESI,† Fig. S1B,9–13). Once the ferric-siderophore complex/inorganic iron reaches the periplasm, the cargo is then picked up by a soluble periplasmic binding protein (PBP) and is transported to the inner membrane ATPase–permease complex and ultimately shuttled into the cytosol.14–16
The soluble PBPs ascribed to the transport of different types of cargos in Gram negative systems are associated with diverse functions in addition to transmembrane ligand transport, including signal transduction or chemoreception.17 Although the PBPs have very low sequence identity in general they essentially consist of two lobes (N and C) connected by a hinge region (either an α-helix or a β-sheet) and undergo a hinge rotation motion concomitant with ligand binding.18,19 In the literature, the PBPs have been classified based on different characteristics, such as sequence alignment or cargo specificity of the proteins17 and the number or types of hinges connecting the two lobes.20–22 The PBPs that have a small β-sheet connecting between the N and C lobes are classified as Class I and Class II proteins whereas PBPs with a single α-helical hinge connecting these two lobes are classified as Class III protein.20–22 The Class III proteins (e.g. E. coli FhuD and FepB) show almost negligible “hinge motions” upon ligand binding due to the rigidity of the α-helical linker14,21,23 compared to the other two classes of PBPs.
E. coli, a Gram negative pathogenic bacterium requires iron as an essential nutrient and acquires this element by turning on various iron regulated genes like fep, fhu and fec.24,25 Of these three systems, the first two are involved in the uptake of the catecholate (enterobactin) and hydroxymate (ferrichrome) siderophores, respectively, whereas the third one is involved with Fe3+-citrate uptake. Citrate is a primary metabolite that is produced in the tricarboxylic acid cycle by all aerobic species and is often used as a carbon and energy source.26 Citrate can act as a versatile metal ion chelator and bind Fe3+ using its carboxylate and hydroxyl groups.27 This ubiquitous anion under aerobic conditions can be converted in vivo to iso-citrate (via an intermediate cis-aconitate) both of which can act as metal ion chelators (see ESI,† Fig. S2). In addition to citrate and its isomer iso-citrate, the tricarboxylic acid cycle also produces two other related dicarboxylate metabolites, succinate and malate which have some structural similarity to citrate. The versatility of the metal binding ability is perhaps the reason why the citrate backbone is preserved at the core of many other siderophores produced by bacteria.28–31
The Fec system found in E. coli consists of a TonB-dependent outer membrane (OM) receptor (FecA), a PBP to carry the Fe3+-citrate across the periplasm (FecB) and an inner membrane (IM) ATPase–permease complex for uptake into the cytosol (FecC/D) (Fig. S1C, ESI,†32,33). So far, FecA has been crystallized in both the apo- and holo-form34,35 but at this time very little structural or biophysical data is available for the FecB or FecC/D systems.36 Recently, the crystal structure of HtsA, a related protein to FecB found in the Gram positive bacterium Staphylococcus aureus, transporting the siderophore staphyloferrin A (SA) has been reported.37 HtsA shows ∼35% identity with E. coli FecB, and has been shown to be specific to SA, having citrate at the metal binding core.37 Multiple sequence alignments between HtsA and FecB sequences from different bacterial species show that several arginine residues involved in SA binding in HtsA are also conserved in FecB proteins, which might indicate the presence of a similar ligand binding pocket in FecB.37
As was mentioned before, only limited biophysical data for the ability of FecB to interact with Fe3+-citrate have been reported in the literature so far. The complex speciation chemistry of Fe3+-citrate which is heavily dependent on the solution pH and the concentrations of individual constituents, make ligand binding experiments for FecB complicated.38,39 In addition, the importance of Fe3+-citrate as an iron source has also been debated in the literature for a long time. However, a recent review reported that the main species of non-transferrin bound iron (NTBI) in bio-fluids could be Fe3+-citrate,40 although the possibility of the existence of other acetate/albumin or mixed species of Fe3+ cannot be underestimated. Moreover, the exact speciation of NTBI in bio-fluids may be variable.40 Based on this information it is logical to hypothesize that different physiological conditions determined by pH, “free” Fe3+ and citrate concentrations, may give rise to different Fe3+-citrate species in bio-fluids. Thus it is reasonable to assume that the invading bacterial species, that uses Fe3+-citrate as its iron source, has the potential to encounter several iron-citrate species in the host. Interestingly, structural data for FecA, the OM receptor for Fe3+-citrate present in E. coli, have indicated that it can bind to both iron-free citrate and diferric dicitrate.35 Additionally, the authors also suggest that due to the availability of ferric dicitrate in vivo, in addition to diferric dicitrate, FecA is likely to bind and transport both these Fe3+-citrate species in E. coli, however they were not able to crystallize ferric dicitrate with FecA.35
In this work we have investigated the ability of FecB to interact and bind with various species of Fe3+-citrate as well as iron-free citrate under different conditions of pH using isothermal titration calorimetry (ITC). Moreover, in this contribution we also investigated whether FecB could interact with several representative iron-free di-, tri- and poly-carboxylates (in addition to iron-free citrate) using ITC, to determine if FecB can discriminate between biologically relevant tri-carboxylates and di-/poly-carboxylates. We also explored if FecB was able to bind with various Fe3+-mimics (Al3+, Ga3+, Sc3+, In3+) in complex with the citrate anion as well as the potential for this protein to interact with Mg2+-citrate using ITC. The ability of FecB to interact with metal-free and metal-loaded citrate was also investigated using 1H,15N HSQC fingerprint NMR experiments. Finally, we also report the folding stability of apo-FecB and FecB in complex with various metal-free and metal-bound anions using differential scanning calorimetry (DSC).
Materials and methods
All chemicals and materials used in this study are of the highest purity grade and most chemicals were procured from Sigma-Aldrich.Cloning and protein purification
The mature FecB protein (residues 1–279), without the periplasmic signal sequence, was amplified from the E. coli K12 genome through polymerase chain reaction (PCR) with oligonucleotides containing 5′ NdeI and 3′ XhoI restriction sites and cloned into the pET30b vector (New England Biolabs). For overexpression of FecB, E. coli BL21 (DE3) cells with the vector of interest were grown in 1 L of Luria-Bertani (LB) broth in the presence of 30 μg mL−1 of kanamycin at 37 °C until the OD600 reached ∼0.8. For the 1H,15N HSQC NMR experiments, 15N labelled FecB was prepared using the general isotope-labeling strategy reported previously.41,42 Briefly, E. coli BL21 (DE3) cells were grown in 1 L of M9 medium containing 1 g L−1 15NH4Cl (CIL) as the nitrogen source at 37 °C till the OD at 600 nm reached 0.8. The culture was then induced by adding 0.8 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 3 hrs. Cells were then harvested and suspended in 20 mM Tris, 0.1 M NaCl and 20 mM imidazole (pH 8.0) and lysed by a French Pressure Cell. The cell lysate was centrifuged at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 × g for 45 minutes at 4 °C. Nickel affinity column chromatography was performed using chelating-Sepharose (GE Healthcare), where bound proteins were washed with 20 mM Tris, 100 mM NaCl and 20 mM imidazole (pH 8.0) and FecB was eluted by 20 mM Tris, 100 mM NaCl and 250 mM imidazole (pH 8.0). Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was used to confirm protein homogeneity and purity. The purified protein was then dialyzed against ammonium bicarbonate (4×), subsequently lyophilized and stored at 4 °C. The protein extinction coefficient to determine the protein concentration and the pI were calculated using the ExPaSy ProtParam program.
500 × g for 45 minutes at 4 °C. Nickel affinity column chromatography was performed using chelating-Sepharose (GE Healthcare), where bound proteins were washed with 20 mM Tris, 100 mM NaCl and 20 mM imidazole (pH 8.0) and FecB was eluted by 20 mM Tris, 100 mM NaCl and 250 mM imidazole (pH 8.0). Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was used to confirm protein homogeneity and purity. The purified protein was then dialyzed against ammonium bicarbonate (4×), subsequently lyophilized and stored at 4 °C. The protein extinction coefficient to determine the protein concentration and the pI were calculated using the ExPaSy ProtParam program.
M3+-Cit solution preparation (M3+ = Fe, Ga, Al, In, Sc)
ITC and DSC experiments were performed under two buffer conditions, 20 mM MES pH 5.5 and 20 mM Tris pH 8.0 to get a high concentration of [Fe2(Cit)2]2− and [Fe(Cit)2]5−, respectively, as the predominant species in solution.43 At pH 5.5, 2 mM Fe3+-chloride was mixed with 2 mM Na-citrate to give a final 1 mM [Fe2(Cit)2]2−, whereas at pH 8, 2 mM Fe3+-chloride was mixed with 20 mM Na-citrate to give 2 mM [Fe(Cit)2]5−.43 To make a solution of 1 mM Ga3+-citrate, Ga3+-nitrate was mixed with 2 mM Na-citrate in the respective buffer solution to give a final 1 mM Ga3+-citrate.44 For Al3+-citrate,45 In3+-citrate and Sc3+-citrate 2 mM, the M3+-salt was mixed with 2 mM Na-citrate. For the titration of isocitrate or Fe3+-isocitrate in to FecB, 2 mM Na-isocitrate and a mixture of 2 mM Na-isocitrate and 2 mM Fe3+-chloride, respectively were used.Fluorescence titration
Fluorescence spectroscopy titration experiments were carried out on a Varian Cary Eclipse fluorescence spectrophotometer. FecB was dissolved in 20 mM MES, 100 mM NaCl at pH 5.5 to make a stock solution of 24 μM. This stock solution was diluted to 1 μM with the same buffer and was used for subsequent emission titration experiments. FecB contains seven Trp residues and the protein solution was excited at 295 nm to selectively excite the Trp residues and the emission spectrum was recorded between 310 to 450 nm. Titration experiments were carried out by adding increasing amounts of Fe3+-citrate (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 0–10 μM) at two different temperatures (25 and 15 °C). Ultimately the I0/I ratios for each set of titrations were plotted as a function of [Fe3+-citrate] to determine the linearity of the quenching data (I is the intensity of emission band upon ligand addition whereas I0 represents the emission arising from ligand-free FecB), which can be interpreted as contributions in terms of static and dynamic quenching by ligand addition.46
1, 0–10 μM) at two different temperatures (25 and 15 °C). Ultimately the I0/I ratios for each set of titrations were plotted as a function of [Fe3+-citrate] to determine the linearity of the quenching data (I is the intensity of emission band upon ligand addition whereas I0 represents the emission arising from ligand-free FecB), which can be interpreted as contributions in terms of static and dynamic quenching by ligand addition.46
ITC
ITC experiments were carried out on a Microcal VP-ITC (Malvern) instrument. The desired amount of lyophilised FecB was weighed out and dissolved in either 20 mM MES pH 5.5 or 20 mM Tris pH 8.0 buffers at 25 °C. Protein concentration was calculated based on the extinction coefficient at 280 nm. For all ITC experiments the protein concentration was between 20–30 μM. The heats of dilution were determined in separate experiments and were negligible compared to the actual protein ligand titrations. ITC raw data were fit to a one-site binding model (MicroCal Origin version 7 software) to determine the stoichiometry (N) and association constant (Ka) which were then converted to Kd values using the relation Kd = 1/Ka. ITC experiments were done in duplicate or triplicate.DSC
All DSC experiments described in this paper were carried out on a VP-DSC microcalorimeter (Malvern). The desired amount of the lyophilised protein was dissolved in 20 mM MES, pH 5.5 or 20 mM Tris, pH 8.0. Protein concentration was checked based on absorption at 280 nm using the molar extinction coefficient at 280 nm and was maintained between 10 to 30 μM. In all experiments done in the presence of various ligands, the final [FecB]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M3+-citrate] (where M = Fe3+, Ga3+, Al3+) was kept at 1
[M3+-citrate] (where M = Fe3+, Ga3+, Al3+) was kept at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (for the protocol for preparation of various M-citrate see the M3+-citrate preparation section). In each experiment protein samples were loaded into the DSC instrument immediately after two consecutive buffer-only heating cycles, accounting for the thermal memory compensation of the DSC calorimeter and baseline correction, respectively. The protein samples were heated from 10 to 80 °C at a scan rate of 60 °C h−1 with a filter period of 16 ps and the pressure was kept around 28 psi to keep the sample stable at higher temperatures. A fourth scan was done after cooling the sample after the third one to check for reversibility.
5 (for the protocol for preparation of various M-citrate see the M3+-citrate preparation section). In each experiment protein samples were loaded into the DSC instrument immediately after two consecutive buffer-only heating cycles, accounting for the thermal memory compensation of the DSC calorimeter and baseline correction, respectively. The protein samples were heated from 10 to 80 °C at a scan rate of 60 °C h−1 with a filter period of 16 ps and the pressure was kept around 28 psi to keep the sample stable at higher temperatures. A fourth scan was done after cooling the sample after the third one to check for reversibility.
1H,15N HSQC-NMR spectroscopy
All NMR experiments were performed at 298 K on a Bruker Avance 700 MHz NMR spectrometers equipped with a triple resonance inverse Cryoprobe with a single axis z-gradient. The heteronuclear single quantum coherence Transverse relaxation optimized spectroscopy (HSQC TROSY) experiments of FecB samples were acquired in either 20 mM MES at pH 5.5 or in 20 mM Na-phosphate at pH 7.0 (because of enhanced hydrogen exchange at higher pH values, NMR experiments were not done at pH 8.0). The concentration of 1H,15N-FecB for all titration experiments was kept >400 μM. Samples were prepared with 10% D2O and DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) was used as an internal standard. For all titration experiments the ligand concentrations (metal-free citrate or Ga3+-citrate) were varied from 0–2.5 times of the final protein concentration. All data were processed with the NMRPipe47 software package and analyzed using the program NMRView.Molecular modeling and visualization
We modeled the structure of E. coli FecB (amino acid residue 21–290) using the Swiss-Model program (ESI,† Fig. S3).48 We also calculated the surface charge of FecB (ESI,† Fig. S3b) using the APBS program in PyMol50 (visualization done using the PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC).Results and discussion
As mentioned earlier, currently there is no structural information available for E. coli FecB. The Swiss-Model program selected the HtsA protein (PDB 3EIW), the SA transporter described in the Introduction section as the best template for generating a model for FecB.37 As expected, the model shows FecB as a bilobal protein with the N- and the C-lobes connected by a single α-helical hinge (ESI,† Fig. S3a). The presence of a single α-helical hinge between the N- and the C-lobes of FecB puts it in the Class III PBP, similar to HtsA, FhuD and FepB.20–22,37,49 For Class III periplasmic binding proteins it has been observed that ligand binding does not necessarily involve a large degree of hinge motion.14,21,23 The surface charge calculations show a positive patch in the cleft between the N- and the C-lobes, which can potentially accommodate negatively charged ligands efficiently.FecB binds with several ferric-citrate species
E. coli can grow in diverse biological niches being exposed to a wide range of pH, Fe3+ and citrate concentrations, which in turn can control the speciation of the Fe3+-citrate present in the bio-fluid.40 Hence the possibility that E. coli encounters various species of Fe3+-citrate in the host system is not unreasonable. The OM receptor FecA transporting Fe3+-citrate to the periplasm has been crystallized in the presence of iron-free citrate and [Fe2(Cit2)]2−. The authors also suggested that due to the availability of [Fe(Cit)2]5− in addition to [Fe2(Cit2)]2−in vivo, FecA most likely is able to transport both of these species to the periplasm.35 Hence in our experiments we have explored the possibility of FecB to interact with these two specific species of Fe3+-Cit, [Fe2(Cit2)]2− and [Fe(Cit)2]5−, by adjusting the pH and the concentrations of the individual constituents following a recent report.43 However, we recognize that the speciation thermodynamics and kinetics of Fe3+-citrate in the absence of a binding protein can differ from that in the presence of a protein, such as FecB, which might lead to sequestration of non-major species at those respective pH values.We have carried out initial fluorescence spectroscopy titration experiments (at 15 and 25 °C) with solutions containing FecB, as the protein has seven Trp residues. Under our experimental conditions (20 mM MES, 100 mM NaCl, pH 5.5), the maximum of the emission wavelength is at 335 nm when excited at 295 nm. This indicates an average hydrophobic environment for the Trp.51,52 Addition of increasing amounts of Fe3+-citrate to a solution of FecB gives rise to a concentration dependent quenching of the intrinsic emission band at 335 nm. However, this quenching did not reach saturation even when the final [FecB]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [Fe-Cit] was 1
[Fe-Cit] was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 at both temperatures. As reported earlier, changes in the Trp emission intensity and/or position owing to ligand addition can be correlated to a binding event53–57 and hence the emission titration clearly indicate binding interactions between FecB and Fe3+-citrate at both temperatures tested. Plots of the emission quenching data as I0/I vs. [Fe3+-citrate] at the two temperatures show distinct non-linearity, where the initial additions at both temperatures gave rise to comparable quenching but upon increasing the [Fe3+-citrate] the quenching at higher temperature was much greater than at the lower one (see ESI,† Fig. S4). We interpret these observations as a result of quenching arising from both static and collisional effects.46 The fact that ligand addition leads to static quenching of the intrinsic Trp emission from FecB is an indication of FecB interacting with Fe3+-citrate at both temperatures tested. Further, the fact that the emission maximum did not show any change in position indicates that the average environment of all the Trp remains unchanged upon ligand addition.51,52 However, we did not attempt to determine the Kd value for the binding event using this method owing to the complexity of the need to separate the contributions of static and dynamic quenching from the total quenching data.
10 at both temperatures. As reported earlier, changes in the Trp emission intensity and/or position owing to ligand addition can be correlated to a binding event53–57 and hence the emission titration clearly indicate binding interactions between FecB and Fe3+-citrate at both temperatures tested. Plots of the emission quenching data as I0/I vs. [Fe3+-citrate] at the two temperatures show distinct non-linearity, where the initial additions at both temperatures gave rise to comparable quenching but upon increasing the [Fe3+-citrate] the quenching at higher temperature was much greater than at the lower one (see ESI,† Fig. S4). We interpret these observations as a result of quenching arising from both static and collisional effects.46 The fact that ligand addition leads to static quenching of the intrinsic Trp emission from FecB is an indication of FecB interacting with Fe3+-citrate at both temperatures tested. Further, the fact that the emission maximum did not show any change in position indicates that the average environment of all the Trp remains unchanged upon ligand addition.51,52 However, we did not attempt to determine the Kd value for the binding event using this method owing to the complexity of the need to separate the contributions of static and dynamic quenching from the total quenching data.
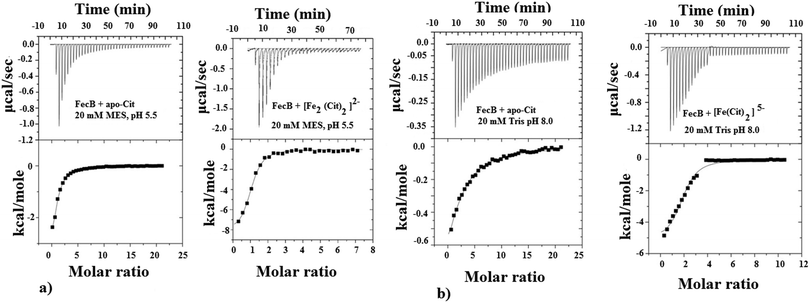
The ability of FecB to interact with iron-free citrate, various Fe3+-citrate species (as controlled by pH and solution Fe3+ and citrate concentrations), various other di-, tri- and poly-carboxylic acids and representative Fe3+-mimics (Ga3+, In3+, Sc3+ and Al3+) and a divalent metal cation (Mg2+) were experimentally explored using ITC and these results are shown in Table 1. As the data in Table 1(a) and (c) show, FecB can bind with iron-free citrate both at pH 5.5 and 8.0 (Fig. 1a and b left panels). However, we also observe that FecB has a greater affinity towards iron-free citrate at pH 5.5 than at pH 8.0. We interpret these differences in the Kd values for iron-free citrate with FecB at the two pH values as being due to the difference in speciation of citrate at these two pH values the major species of citrate (H4Cit) at pH 5.5 is (H2Cit2−) whereas at pH 8.0 is (HCit3−).27 Moreover, the pI of the FecB construct used in these experiments is 7.3, hence in addition to interacting with different major species of iron-free citrate at the respective pH, the protein surface charge is also very different at pH 5.5 than at pH 8.0. Hence these changes in charge for both the ligand and protein can contribute to the weaker binding of iron-free citrate at pH 8.0.
| Ligand | K d (μM) | N |
|---|---|---|
| (a) | ||
| Citrate | 25 ± 5 | 0.7 |
| Isocitrate | 41 ± 3 | 2.0 |
| cis-Aconitic acid | 54 ± 4 | 1.2 |
| Tricarballylic acid | 16 ± 3 | 1.1 |
| Hydroxycitrate | 3 ± 0.5 | 0.7 |
| EGTA | 61 ± 2 | 1.8 |
| Malate | No binding | — |
| Succinate | No binding | — |
| (b) | ||
| [Fe2(Cit)2]2− | 2 ± 0.5 | 0.9 |
| Ga3+-citrate | 1.5 ± 0.3 | 0.9 |
| Al3+-citrate | 2.6 ± 0.5 | 2.7 |
| In3+-citrate | 1.7 ± 0.4 | 1.6 |
| Sc3+-citrate | 72 ± 6 | 1.3 |
| Mg2+-citrate | 53 ± 5 | 0.7 |
| Fe3+-tricarballylic acid | 9 ± 0.7 | 1.3 |
| (c) | ||
| Iron-free citrate | 87 ± 4 | 1.9 |
| [Fe(Cit)2]5− | 6 ± 1 | 1.9 |
| Ga3+-citrate | 1.5 ± 0.5 | 1.6 |
In our ITC experiments we wanted to determine if FecB is capable of binding to [Fe2(Cit2)]2− and [Fe(Cit)2]5−, owing to the previous report that FecA is most likely to transport both these species of Fe3+-citrate.35 In a recent publication,43 the authors described a pH-modulated way to control the speciation of Fe3+-citrate, and we have followed this protocol to obtain [Fe2(Cit2)]2− and [Fe(Cit)2]5−. Our ITC titration experiments show that both of these Fe3+-citrate species show similar binding affinities with FecB (Fig. 1a and b right panels, Table 1b and c). Moreover, comparing the data for iron-free citrate and Fe3+-citrate at pH 5.5 and 8.0 (Table 1a–c), it is evident that FecB can discriminate between the metal-free and metal-loaded species. Hence, our ITC experiments confirm that FecB is able to bind both iron-free and two different species of Fe3+-citrate at two experimental pH values. This is a novel finding, considering that no other Class III type periplasmic binding protein so far has been shown to bind the iron-free form of a siderophore efficiently.54,56–58
In the Citric acid cycle, citrate is converted to oxaloacetate through succinate and malate (dicarboxylic acids) through two other tricarboxylates, cis-aconitic acid and isocitric acid.26 All these di-and tri-carboxylates are potential ligands for Fe3+ and hence we explored the possibility of these compounds acting as ligands with FecB. We also investigated if ethylene glycol tetraacetic acid (EGTA), a non-natural tetra-carboxylic acid can bind FecB. Tricarballylic and hydroxycitric acids, derivatives of citric acid either lacking the alcoholic –OH group or with an additional alcoholic –OH group respectively, were also titrated into solutions of FecB to gather additional information regarding the molecular requirements for the interaction between FecB and citrate.
Table 1a shows that iron-free isocitrate has similar binding affinity towards FecB as compared to iron-free citrate, both of which are di-negative at pH 5.5. cis-Aconitic acid and tricarballylic acid also show similar binding affinity with FecB as compared to iron-free citrate/isocitrate (Table 1a) at pH 5.5. On the other hand succinate and malate, the dicarboxylates, show no binding with FecB (Table 1a). The much larger tetra-carboxylate EGTA shows rather weak binding compared to either iron-free citrate or isocitrate binding to FecB (Table 1a). Finally, hydroxycitrate, having an additional –OH group in comparison to citrate/isocitrate in its structure binds to FecB with much stronger affinity at pH 5.5 (Table 1a). Taken together these binding data indicate that FecB has a distinct preference towards iron-free tricarboxylates, when compared to the di- or tetra-carboxylate compounds tested here.
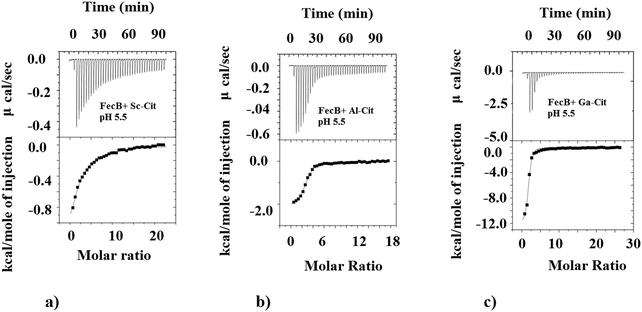
We also investigated the ability of FecB to interact with other M3+-citrate (M3+ = Al3+, Ga3+, In3+, Sc3+) species where M3+ are Fe3+-mimics, and compare this binding ability with representative M2+-citrate (M2+ = Mg2+). The data presented in Table 1b shows the binding data obtained for all these M3+/M2+-citrate with FecB at pH 5.5. As can be observed the Ga3+/Al3+/In3+-citrate species all bind with FecB with very similar affinity as Fe3+-citrate at pH 5.5, whereas Sc3+-citrate binds FecB with a much weaker affinity (Table 1b, Fig. 2). Mg2+-citrate also binds, but it shows weaker binding with FecB (Table 1b). Similar binding affinities for Ga3+ and Al3+-citrate towards FecB compared to Fe3+-citrate at pH 5.5 is perhaps not surprising given the fact that all these M3+ ions have similar ionic radii and form octahedral complexes (Fe3+ (high spin), 64.5 pm; Ga3+, 62 pm; Al3+, 53.5 pm; and In3+, 80 pm). Additionally, a lower binding affinity of FecB towards Mg2+-citrate is also not unexpected owing to the different charge on the central metal ion. However, a simple charge/size argument cannot explain the lower affinity of Sc3+-citrate to FecB. In addition, the fact that the much larger In3+ (in octahedral geometry) binds with FecB with similar affinity to Fe3+-citrate whereas the smaller Sc3+-citrate binds with a lower affinity, indicates a more complex correlation between binding affinity for metal-citrates and FecB than a simple charge/size ratio. However, a recent paper has reported that In3+ can transform from an octahedral to a tetrahedral geometry,59 and it is known that tetra-coordinated In3+ is much smaller in size (62 pm) which resembles Fe3+ ionic radii and could explain the stronger affinity of In3+-citrate towards FecB. The differences in the Kd values between the various M3+-citrate species, where M3+ is always an Fe3+-mimic, can arise for several reasons, including speciation, coordination geometry and net charge on the metal-citrate complex, but with our current data set it is not possible to pinpoint the exact rationale.
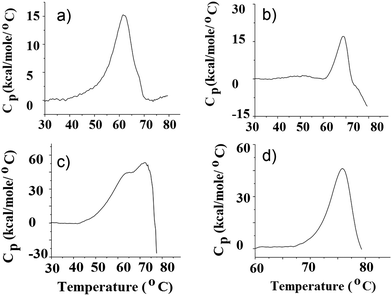
Ligand binding to FecB makes the protein more stable
ITC data provide a direct thermodynamic account of the binding interactions between ligands and proteins, but it does not speak of the inherent stability of the different protein–ligand complexes formed or compare these liganded-forms to the ligand-free protein from a protein folding stability point of view. Hence we carried out DSC experiments with FecB in the absence and presence of different ligand species at pH 5.5 and 8.0 to investigate which protein–ligand complex gives rise to the most stable folded structure. Table 2 summarizes the results of the DSC experiments and Fig. 3 shows DSC data for FecB in the presence of various ligands at both pH values. As can be seen from Table 2, at pH 5.5 ligand-free FecB has the lowest thermal stability (Tm 60 °C), both the N- and C-lobe of the protein melting at the same temperature, possibly in a cooperative manner. Addition of iron-free citrate or Fe3+-citrate (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, protein
5, protein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ligand) to FecB in separate DSC experiments, shows higher thermal stability for the protein ligand assembly (Tm 62 and 68 °C respectively). This higher thermal stability of liganded FecB vs. ligand-free FecB could be due to additional interactions forming between the N- and the C-lobe of FecB upon ligand binding. Further the greater thermal stability of FecB in the presence of Fe3+-citrate vs. iron-free citrate indicates that Fe3+-citrate binding leads to the formation of a more stable ligand-FecB conformation. Comparison of the Tm values obtained for Fe3+-citrate and Fe3+-isocitrate shows that both of these ligands impart the same thermal stability to the protein ligand assembly (Table 2) and hence indicate that FecB does not have the ability to discriminate between the two iron-free isomers of citrate.
ligand) to FecB in separate DSC experiments, shows higher thermal stability for the protein ligand assembly (Tm 62 and 68 °C respectively). This higher thermal stability of liganded FecB vs. ligand-free FecB could be due to additional interactions forming between the N- and the C-lobe of FecB upon ligand binding. Further the greater thermal stability of FecB in the presence of Fe3+-citrate vs. iron-free citrate indicates that Fe3+-citrate binding leads to the formation of a more stable ligand-FecB conformation. Comparison of the Tm values obtained for Fe3+-citrate and Fe3+-isocitrate shows that both of these ligands impart the same thermal stability to the protein ligand assembly (Table 2) and hence indicate that FecB does not have the ability to discriminate between the two iron-free isomers of citrate.
| Species | pH | [Protein]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [Ligand] [Ligand] |
Melting temperature (°C) |
|---|---|---|---|
| apo-FecB | 5.5 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
60.0 |
| FecB + iron-free citrate | 5.5 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
62.0 |
| FecB + [Fe2(Cit)2]2− | 5.5 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
68.0 |
| FecB + Fe3+-isocitrate | 5.5 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
68.0 |
| FecB + Ga3+-citrate | 5.5 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
70.0 |
| FecB + Al3+-citrate | 5.5 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
65.0 |
| FecB + Mg2+-citrate | 5.5 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
63.0 |
| apo-FecB | 8.0 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
63.0, 70.0 |
| FecB + [Fe(Cit)2]5− | 8.0 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
76.0 |
As can be seen from Table 1b at pH 5.5 Fe3+-citrate, Al3+-citrate and Ga3+-citrate all have similar low μM binding with FecB, and using ITC data alone it is not possible to get any information regarding the individual stabilities of the folded conformation of protein–ligand assembly. In the DSC experiments we observe that a solution containing Ga3+-citrate and FecB (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), has the highest thermal stability whereas Al3+-citrate bound to FecB has the lowest thermal stability in this series (Table 2). In other words, at pH 5.5, a complex between Ga3+-citrate and FecB forms the most stable liganded conformation, followed by Fe3+-citrate, whereas Al3+-citrate forms the least thermally stable liganded conformation.
1), has the highest thermal stability whereas Al3+-citrate bound to FecB has the lowest thermal stability in this series (Table 2). In other words, at pH 5.5, a complex between Ga3+-citrate and FecB forms the most stable liganded conformation, followed by Fe3+-citrate, whereas Al3+-citrate forms the least thermally stable liganded conformation.
The experimental outcome at pH 8.0 for ligand-free FecB is entirely different than that obtained for the same experiment at pH 5.5. At pH 8.0, the DSC plot for ligand-free FecB shows two peaks, at 63 and 70 °C (Table 2 and Fig. 4) indicating that the two lobes of the protein at this pH unfold separately. Interestingly the two lobes of FecB once again “melt” at the same temperature, possibly cooperatively, in the presence of Fe3+-citrate at pH 8.0. This can again be interpreted as the ligand being able to form additional interactions between the two lobes of the protein thereby creating a more stable liganded conformation of FecB at this pH (Table 2).
Based on the melting temperatures of FecB in the presence of a 5-times excess of Fe3+-citrate at both pH values, it appears that the liganded form at pH 8.0 is significantly more stable than at pH 5.0 (Table 2). It is also important to mention here that all DSC experiments reported here are irreversible, indicating that once the protein or the protein–ligand complex is thermally denatured, cooling down cannot produce a functionally folded protein structure.
Addition of citrate to 15N FecB causes changes of amino acid environment
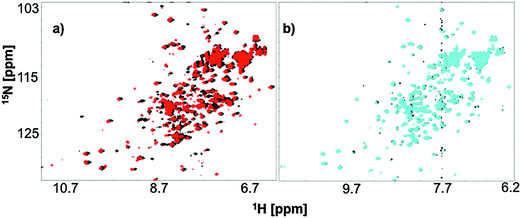
Backbone amide proton HSQC signals provide structural information for a protein on a residue level, which can be affected by the state of ligation, pH and many other factors. In our fingerprint HSQC NMR experiments we titrated in either iron-free citrate or Ga3+-citrate to a solution of 15N FecB at pH 5.5 or pH 7.0 to observe possible structural changes arising from ligand-binding. Diamagnetic Ga3+ was used as a Fe3+-mimic to avoid paramagnetic broadening to carry out the NMR experiments.FecB amide proton signals showed shifts upon addition of metal-free citrate at both pH 5.5 and 7.0 indicating a change of amino acid exposure, which might arise due to binding interaction (Fig. 4). However, as can be seen from Fig. 4, these shifts were more significant at pH 5.5 than at pH 7.0. This information is in accordance with the ITC data, which show that FecB can bind with iron-free citrate much stronger at pH 5.5 than at pH 8.0 (Table 1a and b).
At both pH values tested (5.5 and 7.0) when a 5 time excess of Ga3+-citrate (with respect to the protein concentration) was added to FecB, the signal from the back-bone amide protons showed significant shifts when compared to no-ligand spectra as well as spectra arising from addition of metal-free citrate (ESI,† Fig. S5). This can indicate that at both experimental pH values addition of 5 times Ga3+-citrate brings about large structural changes when compared to the metal-free citrate titration. Further, when the HSQC spectra of the liganded form at these two pH values are overlayed, it is observed that the structural changes brought about by Ga3+-citrate at both pH values are similar (ESI,† Fig. S5). This latter information can be interpreted as that the final liganded form of FecB in the presence of excess Ga3+-citrate is independent of the pH. In other words, at both pH 5.5 and pH 7.0 the conformation of FecB bound to Ga3+-citrate is comparable.
Conclusions
The Fec system in E. coli has been reported several decades ago,32,33 and the protein database UniProt contains several sequences for FecB proteins (the periplasmic component of the fec system) from various organisms.60 However to date there is no comprehensive study to quantitatively analyze the binding of FecB with a variety of ligands. In this work we report that E. coli FecB can bind various tricarboxylates in iron-free and iron-loaded form. We also show that the protein can discriminate between representative M3+- and M2+-citrate species. Our experiments clearly show that at both pH values tested (5.5 and 8.0) FecB can bind both iron-free and iron-loaded citrate although it binds with iron-loaded citrate with a higher affinity at both pH values. Moreover, the capacity of FecB to bind iron-free citrate could be demonstrated in all our experiments (ITC, DSC and HSQC-NMR). Moreover our work demonstrates that FecB can bind with a wide range of tricarboxylates. So far only two other putative periplasmic binding proteins, FpvC and FpvF, found in Pseudomonas aeruginosa and associated with the pyoverdine-I uptake system, have been reported to bind their respective apo-siderophore.61 Moreover, the E. coli OM receptor FecA has been shown to bind iron-free citrate, although it has been argued that it may not be transported in the periplasm through FecA.35,62 Another report has also shown that the TonB-dependent OM transporters, FhuA (ferrichrome transporter) from E. coli and FptA (pyochelin transporter) from P. aeruginosa can bind to their respective apo-siderophores both under in vivo and in vitro conditions.63 A previous report also shows that at least in a mouse model E. coli can grow under both aerobic and anaerobic conditions,64,65 and under anaerobic conditions E. coli might use citrate as an energy and carbon source.64,65 Taken together we note that the ability of E. coli to grow in citrate medium under anaerobic conditions,64 and the fact that FecB can bind iron-free citrate, might indicate a possible role for FecB in transporting iron-free citrate. In addition, E. coli also possesses a gene for the TctC protein which is part of a tripartite tricarboxylate transporter system,66,67 which can also take up citrate under anaerobic conditions. Interestingly, the TctC protein is a Class II PBP and hence it is obviously structurally distinct from FecB, so these two systems are not closely related.68 Clearly further in vivo experiments are required to conclusively determine whether a possible role for FecB in transporting iron-free citrate is a reasonable proposition.[Fe2(Cit)2]2− and [Fe(Cit)2]5− show similar affinity towards FecB in our ITC experiments reported here. A previous report has shown the crystal structure of FecA bound to [Fe2(Cit)2]2− and these authors have also suggested the likelihood of FecA to transport [Fe(Cit)2]5−in vivo.35 Taken together this information indicates that these two species of Fe3+-citrate can both be transported through FecA and can then be sequestered in the periplasm by FecB.
Finally our HSQC-NMR experiments indicate that a structural change occurs in the protein backbone following the addition Ga3+-citrate to a solution of FecB. The most important information obtained from the fingerprint NMR titration experiments is the fact that the final liganded conformation of FecB, irrespective of the pH tested, is the same. Recognition of liganded FecB on the IM permease-ATPase transporter FecC/D involves docking of FecB on the latter. Previous workers have shown that conserved amino acids on FecB in a specific orientation facilitate this recognition/docking and ligand transport.36 Hence forming a liganded species that has a folded structure that is not dependent on the pH to facilitate this docking interaction is reasonable. The structural change that occurs concomitant with ligand binding is further substantiated through our DSC experiments where all liganded forms of FecB show greater stability than the ligand-free proteins. We propose that the structural change that gives rise to greater stability of the protein–ligand complex arises from a small rotation around the hinge region of FecB producing a conformation that can accommodate the ligand between the two lobes.
Acknowledgements
This research project was supported by a CRIO grant from Alberta Innovates Health Solutions (AIHS). SB was the recipient of a postdoctoral fellowship award from AIHS. HJV currently holds the Lance Armstrong Chair in Molecular Cancer Research.References
- R. Crichton, Inorganic Biochemistry of Iron Metabolism: From Molecular Mechanisms to Clinical Consequences, John Wiley & Sons Ltd, Chichester, 2nd edn, 2001 Search PubMed.
- M. L. Guerinot, Annu. Rev. Microbiol., 1994, 48, 743 CrossRef CAS PubMed.
- F. Archibald, FEMS Microbiol. Lett., 1983, 19, 29 CrossRef CAS.
- K. N. Raymond and E. A. Dertz, in Iron transport in bacteria, ed. J. H. Crosa, A. R Mey and S. M. Payne, ASM Press, Washington DC, 2004, pp. 3–17 Search PubMed.
- B. R. Byers and J. E. L. Arceneaux, Met. Ions Biol. Syst., 1998, 35, 37 CAS.
- B. C. Chu, A. Garcia-Herrero, T. H. Johanson, K. D. Krewulak, C. K. Lau, R. S. Peacock, Z. Slavinskaya and H. J. Vogel, BioMetals, 2010, 23, 601 CrossRef CAS PubMed.
- A. L. Crumbliss and J. M. Harrington, Adv. Inorg. Chem., 2009, 61, 179 CrossRef CAS.
- M. Sandy and A. Butler, Chem. Rev., 2009, 109, 4580 CrossRef CAS PubMed.
- A. B. Schryvers and L. J. Morris, Infect. Immun., 1988, 56, 1144 CAS.
- C. N. Cornelissen, G. D. Biswas, J. Tsai, D. K. Paruchuri, S. A. Thompson and P. F. Sparling, J. Bacteriol., 1992, 174, 5788 CAS.
- G. D. Biswas and P. F. Sparling, Infect. Immun., 1995, 63, 2958 CAS.
- T. A. Mietzner, S. B. Tencza, P. Adhikari, K. G. Vaughan and A. J. Nowalk, Curr. Top. Microbiol. Immunol., 1998, 225, 114 Search PubMed.
- K. Postle and R. A. Larsen, BioMetals, 2007, 20, 453 CrossRef CAS PubMed.
- R. P. Berntsson, S. H. Smits, L. Schmitt, D. J. Slotboom and B. Poolman, FEBS Lett., 2010, 584, 2606 CrossRef CAS PubMed.
- B. C. H. Chu and H. J. Vogel, Biol. Chem., 2011, 392, 39 CrossRef CAS PubMed.
- M. Miethke, Metallomics, 2013, 5, 15 RSC.
- R. Tam and M. H. Saier, Microbiol. Rev., 1993, 57, 320 CAS.
- M. A. Dwyer and H. W. Hellinga, Curr. Opin. Struct. Biol., 2004, 14, 495 CrossRef CAS PubMed.
- F. A. Quiocho, Curr. Opin. Struct. Biol., 1991, 1, 922 CrossRef CAS.
- K. D. Krewulak and H. J. Vogel, Biochim. Biophys. Acta, 2008, 1778, 1781 CrossRef CAS PubMed.
- W. W. Ho, H. Li, S. Eakanumkul, Y. Tong, A. Wilks, M. Guo and T. L. Poulos, J. Biol. Chem., 2007, 282, 35796 CrossRef CAS PubMed.
- F. A. Quiocho and P. Ledvina, Mol. Microbiol., 1996, 20, 17 CrossRef CAS PubMed.
- M. T. Sebulsky, B. H. Shilton, C. D. Speziali and D. E. Heinrichs, J. Biol. Chem., 2003, 278, 49890 CrossRef CAS PubMed.
- G. F.-L. Ames, C. Prody and S. Kustu, J. Bacteriol., 1984, 160, 1181 CAS.
- V. Braun, The iron transport systems of Escherichia coli, in The enzymes of biological membranes, ed. A. N. Martonosi, Plenum Publishing Corp., New York, 1984, vol. 3, pp. 617–652 Search PubMed.
- F. L. Breusch, Science, 1943, 97, 490 CAS.
- A. M. N. Silva, X. Kong and R. C. Hider, BioMetals, 2009, 22, 771 CrossRef CAS PubMed.
- K. Hotta, C. Y. Kim, D. T. Fox and A. T. Koppisch, Microbiology, 2010, 156, 1918 CrossRef CAS PubMed.
- R. J. Abergel, M. K. Wilson, J. E. Arceneaux, T. M. Hoette, R. Strong, B. R. Byers and K. N. Raymond, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 18499 CrossRef CAS PubMed.
- V. V. Homann, K. J. Edwards, E. A. Webb and A. Butler, BioMetals, 2009, 22, 565 CrossRef CAS PubMed.
- F. C. Beasley and D. E. Heinrichs, J. Inorg. Biochem., 2010, 104, 282 CrossRef CAS PubMed.
- U. Pressler, H. Staudenmaier, L. Zimmermann and V. Braun, J. Bacteriol., 1988, 170, 2716 CAS.
- C. Härle, I. Kim, A. Angerer and V. Braun, EMBO J., 1995, 14, 1430 Search PubMed.
- A. D. Ferguson, R. Chakraborty, B. S. Smith, L. Esser, D. van der Helm and J. Deisenhofer, Science, 2002, 295, 1658 CrossRef PubMed.
- W. W. Yue, S. Grizot and S. K. Buchanan, J. Mol. Biol., 2003, 332, 353 CrossRef CAS PubMed.
- V. Braun and C. Herrmann, J. Bacteriol., 2007, 189, 6913 CrossRef CAS PubMed.
- J. C. Grigg, J. D. Cooper, J. Cheung, D. E. Heinrichs and M. E. Murphy, J. Biol. Chem., 2010, 285, 11162 CrossRef CAS PubMed.
- A. M Silva, X. Kong, M. C. Parkin, R. Cammack and R. C. Hider, Dalton Trans., 2009, 8616 RSC.
- I. Gautier-Luneau, C. Merle, D. Phanon, C. Lebrun, F. Biaso, G. Serratrice and J. L. Pierre, Chem. – Eur. J., 2005, 11, 2207 CrossRef CAS PubMed.
- P. Brissot, M. Ropert, C. L. Lan and O. Loréal, Biochim. Biophys. Acta, 2012, 1820, 403 CrossRef CAS PubMed.
- V. Tugarinov, V. Kanelis and L. E. Kay, Nat. Protoc., 2006, 1, 749 CrossRef CAS PubMed.
- H. Huang and H. J. Vogel, Methods Mol. Biol., 2013, 963, 99 CAS.
- T. Fukushima, A. K. Sia, B. E. Allred, R. Nichiporuk, Z. Zhou, U. N. Andersen and K. N. Raymond, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 16829 CrossRef CAS PubMed.
- G. E. Jackson and M. J. Byrne, J. Nucl. Med., 1996, 37, 379 CAS.
- H. Hotta, Q. Wang, M. Fukuda, S. Aizawa, T. Umemura, K. Sekizawa and K. Tsunoda, Anal. Sci., 2008, 24, 795 CrossRef CAS PubMed.
- J. S. Kadadevarmath, G. H. Malimath, R. M. Melavanki and N. R. Patil, Spectrochim. Acta, Part A, 2014, 117, 630 CrossRef CAS PubMed.
- F. Delaglio, S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer and A. Bax, J. Biomol. NMR, 1995, 6, 277 CrossRef CAS PubMed.
- T. Schwede, J. Kopp, N. Guex and M. C. Peitsch, Nucleic Acids Res., 2003, 31, 3381 CrossRef CAS PubMed.
- B. C. Chu, R. Otten, K. D. Krewulak, F. A. Mulder and H. J. Vogel, J. Biol. Chem., 2014, 289, 29219 CrossRef CAS PubMed.
- T. J. Dolinsky, J. E. Nielsen, J. A. McCammon and N. A. Baker, Nucleic Acids Res., 2004, 32, W665 CrossRef CAS PubMed.
- Y. K. Reshetnyak and E. A. Burstein, Biophys. J., 2001, 81, 1710 CrossRef CAS PubMed.
- E. G. Matveeva, C. Morisseau, M. H. Goodrow, C. Mullin and B. D. Hammock, Curr. Pharm. Biotechnol., 2009, 10, 589 CAS.
- W. W. Ho, H. Li, S. Eakanumkul, Y. Tong, A. Wilks, M. Guo and T. L. Poulos, J. Biol. Chem., 2007, 282, 35796 CrossRef CAS PubMed.
- M. T. Sebulsky, B. H. Shilton, C. D. Speziali and D. E. Heinrichs, J. Biol. Chem., 2003, 278, 49890 CrossRef CAS PubMed.
- S. Banerjee, C. J. Parker Siburt, S. Mistry, J. Noto, P. DeArmond, M. C. Fitzgerald, L. A. Lambert, C. N. Cornelissen and A. L. Crumbliss, Metallomics, 2012, 4, 361 RSC.
- M. R. Rohrbach, V. Braun and W Köster, J. Bacteriol., 1995, 177, 7186 CAS.
- C. Sprencel, Z. Cao, Z. Qi, D. C. Scott, M. A. Montague, N. Ivanoff, J. Xu, K. N. Raymond, S. M. C. Newton and P. E. Klebba, J. Bacteriol., 2000, 182, 5359 CrossRef CAS PubMed.
- S. Banerjee, A. J. Weerasinghe, C. J. P. Siburt, R. T. Kreulen, S. K. Armstrong, T. J. Brickman, L. A. Lambert and A. L. Crumbliss, Biochemistry, 2014, 53, 3952 CrossRef CAS PubMed.
- H. Narita, M. Tanaka, H. Shiwaku, Y. Okamoto, S. Suzuki, A. Ikeda-Ohno and T. Yaitab, Dalton Trans., 2014, 43, 1630 RSC.
- UniProt: http://www.uniprot.org/uniprot/?query=FecB&sort=score.
- K. Brillet, K. Ruffenach, F. Adams, H. Journet, L. Gasser, V. Hoegy, F. Guillon, L. Hannauer, M. Page and A. I. Shalk, ACS Chem. Biol., 2012, 7, 2036 CrossRef CAS PubMed.
- I. J. Schalk, W. W. Yue and S. K. Buchanan, Mol. Microbiol., 2004, 54, 14 CrossRef CAS PubMed.
- F. Hoegy, H. Celia, G. L. Mislin, M. Vincent, J. Gallay and I. J. Schalk, J. Biol. Chem., 2005, 280, 20222 CrossRef CAS PubMed.
- S. A. Jones, F. Z. Chowdhury, A. J. Fabich, A. Anderson, D. M. Schreiner, A. L. House, S. M. Autieri, M. P. Leatham, J. Lins, J. M. Jorgensen, P. S. Cohen and T. Conway, Infect. Immun., 2007, 75, 4891 CrossRef CAS PubMed.
- H. E. Swim and L. O. Krampitz, J. Biol. Chem., 1954, 67, 419 CAS.
- B. Winnen, R. N. Hvorup and M. H. Saier Jr, Res. Microbiol., 2003, 154, 457 CrossRef CAS PubMed.
- R. Antoine, F. J. Dubuisson, H. Drobecq, E. Willery, S. Lesjean and C. Locht, J. Bacteriol., 2003, 185, 1470 CrossRef CAS PubMed.
- I. Huvent, H. Belrhali, R. Antoine, C. B. C. Locht, F. J. Dubuisson and V. Villeret, J. Mol. Biol., 2006, 356, 1014 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5mt00218d |
| This journal is © The Royal Society of Chemistry 2016 |