Open Access Article
Open Access ArticleNovel p38α MAP kinase inhibitors identified from yoctoReactor DNA-encoded small molecule library†‡
L. K.
Petersen
a,
P.
Blakskjær
a,
A.
Chaikuad
b,
A. B.
Christensen
a,
J.
Dietvorst
a,
J.
Holmkvist
a,
S.
Knapp
bc,
M.
Kořínek
d,
L. K.
Larsen
a,
A. E.
Pedersen
e,
S.
Röhm
c,
F. A.
Sløk
a and
N. J. V.
Hansen
*a
aVipergen ApS, Gammel Kongevej 23A, DK-1610 Copenhagen V, Denmark. E-mail: nha@vipergen.com
bStructural Genomic Consortium, University of Oxford, Old Road Campus Research Building, Roosevelt Drive, Headington, Oxford, OX3 7DQ, UK
cInstitute for Pharmaceutical Chemistry and Buchmann Institute for life sciences, Johann Wolfgang Goethe-University, Max-von-Laue-Str. 9, D-60438 Frankfurt am Main, Germany
dAPIGENEX s.r.o., Poděbradská 173/5, 190 00 Prague, Czech Republic
eThe Faculty of Health and Medical Sciences, Department of Immunology and Microbiology, University of Copenhagen, Blegdamsvej 3B, DK-2200 Copenhagen N, Denmark
First published on 22nd June 2016
Abstract
A highly specific and potent (7 nM cellular IC50) inhibitor of p38α kinase was identified directly from a 12.6 million membered DNA-encoded small molecule library. This was achieved using the high fidelity yoctoReactor technology (yR) for preparing the DNA-encoded library, and a homogeneous screening technique – the binder trap enrichment technology (BTE). Although structurally atypical to other kinase blockers, this inhibitor was found by X-ray crystallography to interact with the ATP binding site and provide strong distortion of the P-loop. Remarkably, it assumed an alternative binding mode as it lacks key features of known kinase inhibitors such as typical hinge binding motifs. Interestingly, the inhibitor bound assuming a canonical type-II (‘DFG-out’) binding mode by forming hinge hydrogen bonds with the backbone, showed excellent shape complementarity, and formed a number of specific polar interactions. Moreover, the crystal structure showed, that although buried in the p38α active site, the original DNA attachment point of the compound was accessible through a channel created by the distorted P-loop conformation. This study demonstrates the usability of DNA-encoded library technologies for identifying novel chemical matter with alternative binding modes to provide a good starting point for drug development.
Introduction
Traditional drug discovery is often based on high-throughput screening (HTS), wherein a large number (up to a few millions) of small molecules are tested individually for their ability to modulate the target of interest. As a viable alternative, DNA-encoded small molecule libraries have been developed1 and offer the power to synthesize and screen million to billion membered collections of compounds. Several technologies including the yR technology have recently been reviewed.2The advent of DNA-encoded libraries signifies a new era of combinatorial libraries and high-throughput screening. The first era in the 1990s of combinatorial chemistry based libraries and HTS did not meet the high expectation of accelerating the drug discovery process.3 However, important lessons were learned including the importance of physical/chemical properties of the compounds, purity of synthesized compounds, and assay fidelity. Technologies based on DNA encoding utilize the powerful means to record and store information and in principle control systems on a single molecule level. Further advantages are low protein and library consumption, single tube format, low requirements for instrumentation, and no requirement for pre-set assay conditions for screening. However, the most important driver for its increasing popularity is that it is tabbing into the unprecedented development in DNA sequencing.4
Historically, the identification of target binding compounds from DNA-encoded libraries have in most cases involved immobilization of the target protein to matrices such as sepharose or paramagnetic beads, either via a chemical crosslinking to the functionalized surface, or to immobilized streptavidin. The target is exposed to the library to allow binding to occur, either before or after the target is immobilized to the matrix. The matrix is then washed multiple times to reduce non- and low-affinity binders. The remaining material is finally eluted, and the DNA is PCR amplified and analysed by DNA sequencing. This approach has in many cases been successful, but features some challenges inherent to heterogeneous assays, such as target denaturation associated with binding to a surface, background originating from binding of library molecules to the matrix, as well as the necessity of the washing steps, which are difficult to control during the procedure. Therefore, multiple rounds of selection are normally called for in order to raise the signal above the background, necessitating the use of high amounts of library as well as rigorous data analysis routines to distinguish binders to the target from binders to the matrix.
The challenges related to immobilization of the target to a matrix have led to the development of homogeneous assays, where the target interacts with the library in solution and no washing steps are required. In some of these techniques, the attachment of nucleic acids onto the protein facilitates or stabilizes intramolecular duplex formation with the nucleic acid on the library molecules, and enables the detection. This principle has been exploited employing primer extension5,6 or, DNA ligation by an internally encoded ribozyme,7 thus encoding the binding event in PCR-amplifiable nucleic acid.
In yet another approach, the binding event between the target protein and a DNA-linked small molecule compound tethers a photo-reactive complementary oligonucleotide to the target. The oligonucleotide is subsequently cross-linked which stabilizes the DNA duplex, and protects the compound encoding DNA strand against nuclease degradation, ultimately enabling detection.8 These promising approaches were all shown to be functional in model studies. It will be interesting to follow their performance for de novo discoveries from complex DNA-encoded libraries.
Here is utilized a recently developed homogeneous screening assay, the binder trap enrichment (BTE), which traps binding complexes consisting of a target protein and a library member in minuscule water droplets in a water-in-oil emulsion. The underlying mathematical principle is simple as many more droplets than target protein molecules are formed. If a library member is bound to the target, it will consistently end up in a droplet together with a target molecule, whereas a non-bound library member will only do so by chance. Consequently, a binder will be observed as a frequent event in a background of the random low frequent event.
Mitogen-activated-protein kinases (MAP kinases) are crucial in transducing extracellular signals that regulate a variety of cellular responses such as proliferation, gene expression, differentiation, cell survival, and apoptosis.9 Many MAP kinase inhibitors including p38 inhibitors have been developed, but unlike inhibitors for proteins that regulate MAP kinase signalling, these have not been approved for clinical use.10 p38 MAP kinases are one of three families of MAP kinases and p38α (MAPK14) is particularly involved in the regulation of pro-inflammatory cytokines11 such as TNF-α, IL-1 and IL-6 whereas the function of the three other isoforms, p38β (MAPK11), p38γ (MAPK12) and p38δ (MAPK13) have not been in the focus of drug development.12 p38 MAP kinase is activated by cellular stress factors such as inflammatory cytokines, lipopolysaccharides (LPS), ultraviolet light, and various growth factors.13 p38α have previously been used as target for interrogation of DNA encoded libraries; in one case, a known binding motif was incorporated in a library of triazines14 and in another case a smaller library of macrocycles was used.15 The findings were based on split-pool and DNA-templated library synthesis, respectively.
Using p38α as an interesting drug target, we here demonstrate feasibility of BTE to screen a DNA-encoded yoctoReactor library, which resulted in the identification of selective single digit nanomolar inhibitors of p38α directly from a naïve library of 12.6 million compounds in a single round of selection.
Results and discussion
Library synthesis
The DNA-encoded small molecule library used in the present study was Lib022 – a yR library sequentially assembled within a DNA junction as previously described16 (see ESI‡). The overall library product comprised small molecules covalently linked to double stranded DNA with each library member uniquely encoded by the DNA sequence. Each molecule was made from three building blocks – each encoded by specific codons in the library DNA. A tight integration of chemical synthesis and encoding was achieved as only products from successful DNA directed chemical reactions were part of the final library, due to multiple purification steps. Thus, further background subtractions or other measures to improve library quality were not required.17 Final library size was 12.6 million building block combinations with average molecular weight of 485 Da and average clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P 2.0, thus within drug-like size and lipophilicity (see ESI‡). A good alignment in physical/chemical descriptors between this library and a recent extensive analysis of drug candidates in clinical trials was observed.18
P 2.0, thus within drug-like size and lipophilicity (see ESI‡). A good alignment in physical/chemical descriptors between this library and a recent extensive analysis of drug candidates in clinical trials was observed.18
The principle of binder trap enrichment
For the BTE method, the protein target was conjugated to DNA followed by in-solution binding to a DNA-encoded small-molecule library. Target DNA and library DNA were designed in such a way that they were capable of ligating and each contained a priming site for PCR amplification of the ligation product.In the first step of the BTE, equilibrium binding between the library members and the protein target was established. Then, the binding mixture was diluted which disturbed the equilibrium, and the kinetics was now dominated by off-rates. Next, a water-in-oil emulsion was formed by combining the aqueous solution with an oil-phase and shaking for one minute (Fig. 1, steps A–C).20,21 Thus, a successful binding event between a library member and target protein causes consistent entrapment of both within the same droplet (Fig. 1, step C). Next, the target and library DNA were ligated inside the droplet to record the co-trapping event (Fig. 1, step D). Then, the emulsion was disrupted by organic extractions22 and the material recovered. The DNA was amplified by PCR where only ligation products comprise two PCR priming sites and were thus amplified exponentially (Fig. 1, step E). Finally, the DNA was subjected to DNA sequencing, and the DNA codes decoded to compounds, which were counted. Compounds observed many times are target binders (hits) whereas compounds observed only a few times are dominantly results of the random co-trapping events (background).
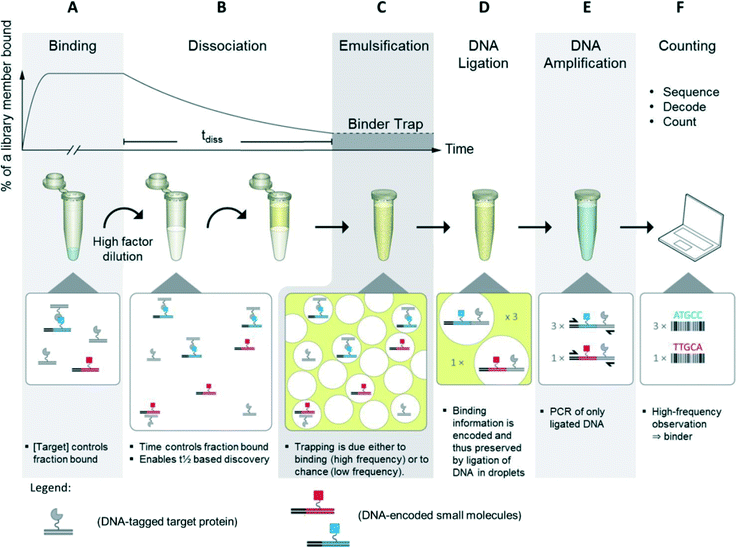 | ||
| Fig. 1 Binder trap enrichment (BTE) technique. A) Equilibrium established between DNA encoded library and DNA tagged protein. B) Dissociation phase triggered by dilution of binding mixture into high volume. (C) After tdiss, the emulsion is formed by shaking water and oil. Binders and protein target, i.e. also target DNA and library DNA, are now trapped within droplets. D) Target DNA and library DNA are now ligated. E) DNA ligation product is isolated and amplified by PCR. F) PCR product is sequenced, decoded to compound, and counted.19 | ||
A mathematical threshold was applied to separate hits from background. This was conveniently done as the number of observations based on random entrapment represents a linear decay line on a logarithmic scale. Having the number of emulsion droplets in great excess over the number of protein target molecules ensured that random trapping of non-target-bound library members with a protein target molecule became very unlikely. Although random entrapment does occur, this leads only to low number of observations.
p38α screening
p38α was screened using Lib022 and BTE. Subsequent DNA sequencing provided about 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 reads which were decoded into compounds and counted.
000 reads which were decoded into compounds and counted.
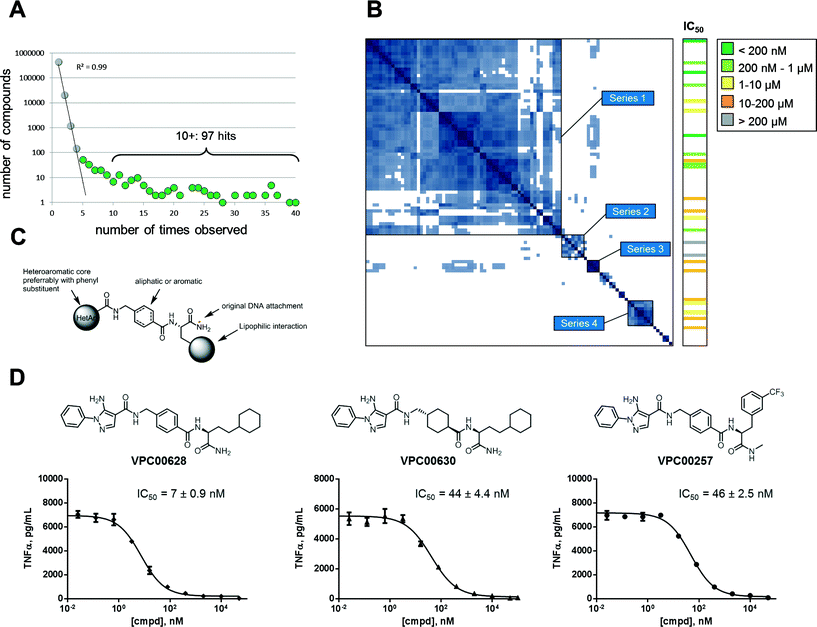
The signal plot of the decoded and counted DNA sequencing data (Fig. 2A) shows the distribution between number of compounds (y-axis) and how many times each were observed (x-axis). When going from low to high number of observations, the curve has two phases: an initial linear decay phase, and a later tail phase. The initial linear decay phase (up to compounds observed 4 times) is dominated by the random co-trapping events, and is thus discarded (light grey spheres). Compounds observed five or more times are likely results of target binding (green spheres) and thus provide 236 primary hits. For simplicity reasons, the 97 most abundant hits (observed ten or more time) were further analyzed. The actual number of observations of a library member in DNA sequencing does not necessarily correlate with potency. At least three factors control how often it is observed: 1) initial frequency in the library, 2) binding affinity to target protein, and 3) the off-rate of complex with target protein.
To cluster the hits into related series, a pairwise calculation of Tanimoto similarities was performed.23 This 97 × 97 symmetrical matrix was sorted using best neighbour method, and several series were observed in the similarity-based heat map as islands along matrix diagonal (Fig. 2B). As indicated, series 1 (62 hits) comprised the main fraction of hits followed by series 2–4 (7, 4, and 8 hits respectively). The remaining 16 hits were singletons or only minor groups with three or fewer members. The hits in series 1, 2, and 4 could be generalized into the structure in Fig. 2C; the building block closest to the original DNA attachment was an amino acid carrying a lipophilic side chain, the second building block was a cyclic amino acid – aliphatic or aromatic – with a 1,4 orientation.
The third building block was various heteroaromatics with pyrazoles dominating (series 1) followed by pyridines (series 2) and thiazoles (series 4). Series 3 hits showed same distal pyrazole building blocks as series 1 but were distinguished by linkage via a diamine to a sulfonamide containing building block closest to DNA attachment.
To validate hits identified by DNA sequencing, twenty four compounds were chosen for resynthesis (off-DNA) from both series and singletons. Assaying compounds enzymatically revealed that the majority (22 of 24) were indeed inhibitors of p38α. This validated the observed signal and demonstrated a low false positive rate (see Table S1‡ for structures and deduced biochemical IC50 values). Fig. 2B (right column) shows the relationship between hit series identified from DNA sequencing and enzyme inhibition: most potent inhibitors were obtained from major series 1, but singletons were also proven effective. Most potent inhibitor (VPC00628) showed 17 observations in signal plot from DNA sequencing underlining the limited correlation between number of observations and potency.
To demonstrate capability of cell penetration and biological function, a set of inhibitors were further subjected to cellular assay. TNF-α secretion in human monocytic cell line (THP-1) was shown to be completely suppressed by inhibitors in nanomolar concentrations. Three example inhibitors are shown in Fig. 2D together with dose response curves and deduced IC50. The most potent (VPC00628) with 7 nM cellular IC50 was selected for further investigations.
VPC00628 in complex with p38α reveals an alternative type-II binding mode
To provide insights into the binding mode of VPC00628, we determined the crystal structure of the p38α-VPC00628 complex. Crystals of the complex diffracted well and were refined at 1.5 Å resolution (Table S2‡). The inhibitor was well defined by electron density and was found to interact with the ATP binding site (Fig. 3A and B). VPC00628 lack key features of kinase inhibitors such as typical hinge binding motifs. Interestingly, the inhibitor bound assuming a canonical type-II (‘DFG-out’) binding mode forming a hinge hydrogen bonds with the backbone of M109 (Fig. 3A and C). VPC00628 showed excellent shape complementarity and formed a number of specific polar interactions: first, the pyrazole nitrogen formed a hydrogen bond with the amide backbone of M109, while the VPC00628 amide adjacent to the pyrazole interacted directly and through a water molecule with the T106 gatekeeper and the carbonyl backbone of the P-loop Y35. Second, the cyclohexane decoration protruded deep into the hydrophobic pocket created by the ‘DFG-out’ conformation and third, the terminal carboxyamide of the inhibitor formed hydrogen bonds with protein backbone (F169 and G170). Interestingly, the P-loop of the kinase was observed to be highly distorted, positioning the sidechain of Y35 at the tip towards the pyrazole phenyl substituent of the inhibitor and the sidechain of the DFG motif phenylalanine (F169) resulting in aromatic stacking interactions. Structural comparison with other type-II inhibitors including BIRB 796 and its allosteric precursor24 and sorafenib25 showed that although the ability of p38α to accommodate type-II inhibitors is well documented, the distorted P-loop in the VPC00628 complex represented an unusual feature of this complex (Fig. 3C). This structural alteration could be due to the induced aromatic stacking of the P-loop Y35 and the DFG F169 through the pyrazole phenyl substituent of the inhibitor, which led to an opening of the so-called P-loop pocket formed between the P-loop and αC. This pocket has been targeted by the selective ERK1/2 inhibitor SCH772984,26 and here likely served to provide a channel for the linker that originally tethered the compound to the library DNA. An interesting observation was made that both VPC00628 and the inhibitors “Allosteric 1” and BIRB 796 comprise a lipophilic pyrazole moiety, yet an inverse binding mode of VPC00628 was observed.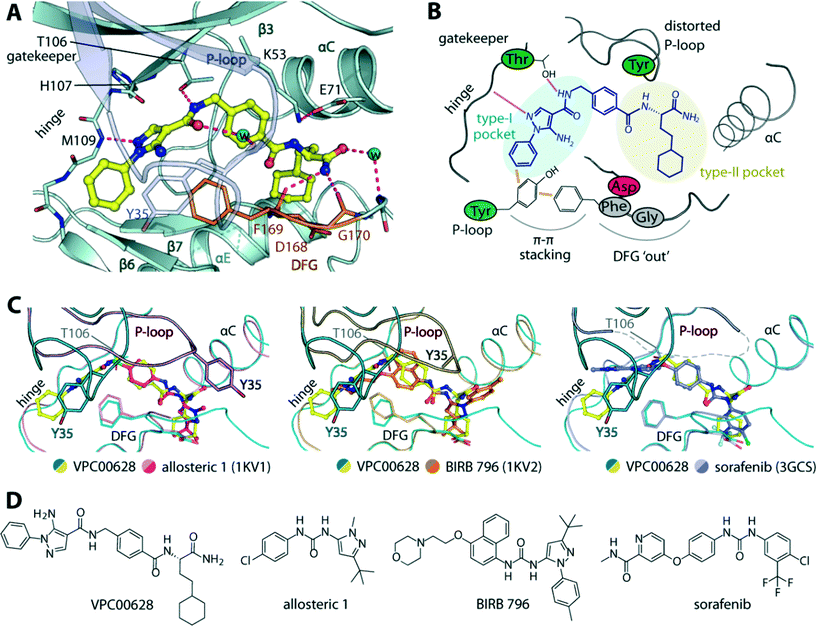 | ||
| Fig. 3 Crystal structure of p38α in complex with VPC00628. A) Detailed interactions of the inhibitor within the kinase. Potential hydrogen bonds are shown in magenta dashed lines, and water molecules shown in cyan spheres. B) Schematic illustration of the key interactions and structural alterations upon accommodation of the inhibitor within the kinase type-I and type-II pockets. C) Structural comparison between the binding modes of VPC00628 and other type-II inhibitors (PDB IDs shown in brackets), revealing high similarity of overall p38α pocket except the P-loop conformation which highly distorted upon the accommodation of VPC00628. D) Chemical structures of the compared inhibitors. PDB accession code “5LAR”. | ||
VPC00628 highly selective inhibitor of p38a
Based on the alternative binding mode and exploitation of new interactions described above for VPC00628, an interesting selectivity profile was to be expected for this novel inhibitor. Indeed, when profiled against 99 human kinases as part of KinomeScan selection,27 VPC00628 was found to be a selective inhibitor of only p38α and p38β with only weak additional activity on the tyrosine kinase KIT (Fig. 4).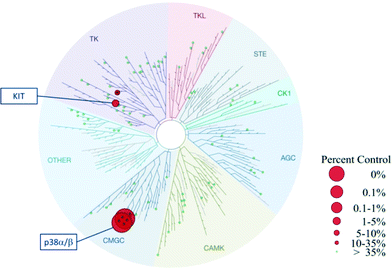 | ||
| Fig. 4 TREEspot™ interaction map for VPC00628 inhibitor was tested at fixed concentration (2 μM) against 99 kinases.27 Red spheres: kinases inhibited to <35% of control; green spheres: >35% kinase activity remains at 2 μM. | ||
Conclusions
Feasibility of a yoctoReactor-generated DNA-encoded small molecule library and binder trap enrichment screening technology was successfully demonstrated on p38α as an interesting drug target. The presented library (Lib022) was not specifically designed for kinases, yet novel chemical matter was identified. As DNA-encoding allows for control of large compound collections in a single tube, libraries can be expanded to be generally applicable across target classes.This is interesting in respect to the ongoing focusing and filtration of classical compound collections, whereby compounds with unexpected chemical characteristics may be removed prior to HTS screening to both 1) improve the expected hit-rate and 2) to screen fewer compounds and thereby decrease costs. In contrast, by using DNA-encoded libraries, the cost and consumption of goods is low and vast compound collections can be screened in a single tube format once the library is prepared. From libraries like Lib022, we have successfully identified modulators of enzymes, protein–protein interaction targets, and epigenetic targets. Due to the built-in diversity of the library, no preconceived target knowledge is required. Secondly, by exploiting a low false-positive rate and a highly reproducible selection technique in BTE, compounds with interesting properties were identified using a highly efficient approach. In the present case, with synthesis and assay of only 24 primary hits from a total library size of 12.6 million compounds, the work load to access new chemical matter was drastically reduced. X-ray crystallography proofed an alternative binding mode of the inhibitor and good correlation between biochemical and cellular inhibitory data was verified.
In conclusion, it was shown that a strong and selective inhibitor was identified from a naïve combinatorial DNA-encoded small molecule library in a cost-effective manner. In the process, no optimization of compound properties was performed and a novel binding mode to the kinase was observed. Based on the present work, new molecular modifications can be envisioned. As the library members are all highly modular, optimizing compounds for further development is readily achieved.
Experimental
Preparation of activated target DNA
A DNA duplex with a 3′ GA overhang and a blunt end consisting of a 5′ phosphorylated 98 nt top-strand and a 5′ C12 amino modified 100 nt bottom-strand was prepared. A COOH moiety was installed at the amino modification by treating 60 nmoles DNA in 100 μL water with 200 μL HEPBS buffer (pH 9.0, 1 M), 200 μL 100 mM suberic acid in NMP, 300 μL NMP and 200 μL 0.5 M 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride (DMTMM). The mixture was incubated at 25 °C for 1 h, quenched for 30 min by addition of 200 μL 2 M lithium hydroxide, neutralized with 400 μL 1 M MOPS buffer (pH 5.2), and ethanol precipitated. For each protein conjugation, 300 pmoles of target DNA were activated by converting the carboxylic acid moiety to the succinimide ester by incubation for 30 min at RT with 5 mM EDC and 10 mM sulfo-NHS in 100 mM MOPS, pH 6.0 in a total volume of 100 μL. The activated DNA was precipitated and washed in 2-propanol before further use.Conjugation of protein to target DNA
10 μl (5 μg) target protein (active p38α, Jena Bioscience) was dialyzed against 10 mM MOPS (pH 8.0), 50 mM NaCl, 0.1% Tween 20, 5% glycerol. The precipitated and activated target DNA was dissolved in 6 μL of the dialyzed protein solution, and 0.7 μl 1 M MOPS (pH 8.0) added. The conjugation reaction was incubated at 4 °C for 16 h and was terminated by adding 0.7 μl 1 M Tris-HCl (pH 8.0) and incubated at 4 °C for 30 min and PAGE purified.BTE
The equilibrium binding was performed by incubating 10 nM p38α conjugated to DNA and 6 × 1011 molecules per μL Lib022 in 10 mM Tris-HCl (pH 7.5), 50 mM NaCl and 0.1% Triton X-100. After 1 h of incubation, 0.12 μl of the mixture was transferred to a tube containing 0.6 ml aqueous buffer (50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 0.1% Triton X-100, 0.15 μM BSA, 9.5 mM KCl, 5 v/v% glycerol, 200 μM EDTA, 1 mM DTT, 2 mM ATP, and 10 nM blocking DNA). T4 DNA ligase was added to a final concentration of 1.1 μM. 900 μL oil phase20 was added. Two minutes after the dilution the water-in-oil emulsion was formed by shaking at 6400 rpm for 60 s. (Precellys24, Bertin Technologies). In parallel, an induction emulsion was prepared containing the aqueous buffer supplemented with 405 mM MgCl2. The main emulsion was supplemented with 50 μL induction emulsion, and incubated at 16 °C overnight with gentle rotation. The T4 DNA ligase was inactivated by incubation at 65 °C for 45 min. The emulsion was broken22 by adding 350 μL 1-butanol, 112.5 μL 2-propanol, and 37.5 μL ethanol, and incubation for 1 h with gentle rotation. Phase separation was achieved by centrifugation, and the organic phase was discarded. The DNA was recovered by a PCR clean-up kit (Machery-Nagel), and eluted in 50 μL 5 mM Tris-HCl (pH 7.5), 0.5 mM EDTA and 0.05% Triton X-100. The DNA was amplified in two consecutive PCRs and subsequently sequenced by ion torrent high-throughput sequencing (Proton, Life Technologies). The DNA sequencing data was analyzed by a hidden Markov model for decoding into compound, and counted.16Biochemical assay of p38α inhibition
The ADP Quest assay from DiscoverX Technologies was used according to manufactures instructions.Cellular assay for TNF-α inhibition
The Human Monocytic Cell Line, THP-1 was stimulated with LPS with or without inhibitors and secreted TNF-α was then analysed. Inhibitors were dissolved in dimethyl sulfoxide and pre-dispensed into round bottom 96-well cell culture plates. The starting concentration was 50–100 μM and initial dilutions were made in dimethyl sulfoxide. After the addition of 200 μl of cell suspension the final dimethyl sulfoxide concentration was 0.5% in all wells. The cell suspensions and compound dilutions were combined and incubated for 30 min at 37 °C in a 5% CO2 humidified atmosphere, before the addition of 500 ng ml−1 LPS (or medium for non-LPS control samples). After the addition of LPS, plates were incubated for 2 h, followed by centrifugation to pellet cells. Cell supernatants were stored at −20 °C until analysis for TNF-α content. TNF-α levels were determined by a commercial kit (from Meso Scale Discovery) following the manufacturer's directions. The percentage inhibition was calculated for each inhibitor concentration tested, and an IC50 curve was plotted using Graphpad Prism software.Protein expression, purification, crystallization and structure determination
Recombinant p38α with an N-terminal His6-tag was co-expressed with λ-phosphatase in E. coli Rosetta.28 The protein was purified using Ni-affinity chromatography, and subsequently treated with TEV protease. The cleaved protein was further purified using size-exclusion chromatography in 20 mM Tris, pH 7.5, 100 mM NaCl and 5 mM DTT. The pure protein at ∼13 mg ml−1 was incubated with 1 mM VPC00628, and the complex was crystallized using the reservoir solution containing 15% PEG smears (PEG 2000, 3350, 4000, and 5000 MME), 0.1 M MES pH 6.2.29 Diffraction data collected at Diamond Light Source beamline I04 were processed and scaled with Mosflm30 and Scala,31 respectively. Initial structure solution was obtained by molecular replacement using Phaser32 and the coordinates of p38α-TAB1 complex.28 The p38α-VPC00628 complex was rebuilt in Coot33 and refined using REFMAC.34 Data collection and refinement statistics are summarized in Table S2.‡ Accession code for Protein Data Bank is “5LAR”.Acknowledgements
A. C. is grateful for financial support by the large EU collaborative network PRIMES.Notes and references
- R. E. Kleiner, C. E. Dumelin and D. R. Liu, Chem. Soc. Rev., 2011, 40, 5707–5717 RSC; L. Mannocci, M. Leimbacher, M. Wichert, J. Scheuermann and D. Neri, Chem. Commun., 2011, 47, 12747–12753 RSC.
- P. Blakskjær, T. Heitner and N. J. V. Hansen, Curr. Opin. Chem. Biol., 2015, 26, 62–71 CrossRef PubMed.
- D. B. Lowe, Nat. Chem., 2014, 6, 851–852 CrossRef CAS PubMed.
- E. R. Mardis, Nature, 2011, 470, 198–203 CrossRef CAS PubMed.
- L. M. McGregor, D. J. Gorin, C. E. Dumelin and D. R. Liu, J. Am. Chem. Soc., 2010, 132, 15522–15524 CrossRef CAS PubMed.
- L. M. McGregor, T. Jain and D. R. Liu, J. Am. Chem. Soc., 2014, 136, 3264–3270 CrossRef CAS PubMed.
- B. Katzman, M. Vyazmensky, O. Press, M. Volokita and S. Engel, Biotechnol. J., 2015, 10, 1–7 Search PubMed.
- P. Zhao, Z. Chen, Y. Li, D. Sun, Y. Gao, Y. Huang and X. Li, Angew. Chem., Int. Ed., 2014, 53, 10056–10059 CrossRef CAS PubMed.
- Reviewed in: G. Sabio and R. J. Davis, Semin. Immunol., 2014, 26(3), 237–245 CrossRef CAS PubMed.
- K. Burkhard and P. Shapiro, Methods Mol. Biol., 2010, 661, 107–122 CrossRef CAS PubMed.
- J. Zhang, B. Shen and A. Lin, Trends Pharmacol. Sci., 2007, 28(6), 286–295 CrossRef CAS PubMed.
- K. K. Hale, D. Trollinger, M. Rihanek and C. L. Manthey, J. Immunol., 1999, 162(7), 4246–4252 CAS.
- Z. Chen, T. B. Gibson, F. Robinson, L. Silvestro, G. Pearson, B. Xu, A. Wright, C. Vanderbilt and M. H. Cobb, Chem. Rev., 2001, 101(8), 2449–2476 CrossRef CAS PubMed.
- M. A. Clark, R. A. Acharya, C. C. Arico-Muendel, S. L. Belyanskaya, D. R. Benjamin, N. R. Carlson, P. A. Centrella, C. H. Chiu, S. P. Creaser, J. W. Cuozzo, C. P. Davie, Y. Ding, G. J. Franklin, K. D. Franzen, M. L. Gefter, S. P. Hale, N. J. Hansen, D. I. Israel, J. Jiang, M. J. Kavarana, M. S. Kelley, C. S. Kollmann, F. Li, K. Lind, S. Mataruse, P. F. Medeiros, J. A. Messer, P. Myers, H. O'Keefe, M. C. Oliff, C. E. Rise, A. L. Satz, S. R. Skinner, J. L. Svendsen, L. Tang, K. van Vloten, R. W. Wagner, G. Yao, B. Zhao and B. A. Morgan, Nat. Chem. Biol., 2009, 5(9), 647–654 CrossRef CAS PubMed.
- R. E. Kleiner, C. Dumelin, G. Tiu, K. Sakurai and D. R. Liu, J. Am. Chem. Soc., 2010, 132, 11779–11791 CrossRef CAS PubMed.
- M. H. Hansen, P. Blakskjær, L. K. Petersen, T. H. Hansen, J. W. Højfeldt, K. V. Gothelf and N. J. V. Hansen, J. Am. Chem. Soc., 2009, 131, 1322–1327 CrossRef CAS PubMed.
- R. M. Franzini, S. Biendl, G. Mikutis, F. Samain, S. Scheuermann and D. Neri, ACS Comb. Sci., 2015, 17(7), 393–398 CrossRef CAS PubMed.
- M. J. Waring, J. Arrowsmith, A. R. Leach, P. D. Leeson, S. Mandrell, R. M. Owen, G. Pairaudeau, W. D. Pennie, S. D. Pickett, J. Wang, O. Wallace and A. Weir, Nat. Rev. Drug Discovery, 2015, 14, 475–486 CrossRef CAS PubMed.
- Reproduced from ref. 2 with permission from Elsevier.
- D. J. Turner and M. E. Hurles, Nat. Protoc., 2009, 4(12), 1771–1783 CrossRef CAS PubMed.
- F. Diehl, M. Li, Y. He, K. W. Kinzler, B. Vogelstein and D. Dressman, Nat. Methods, 2006, 3(7), 551–559 CrossRef CAS PubMed.
- T. Schütze, F. Rubelt, J. Repkow, N. Greiner, V. A. Erdmann, H. Lehrach, Z. Konthur and J. Glökler, Anal. Biochem., 2011, 410, 155–157 CrossRef PubMed.
- Software used for the calculations: ChemAxon JChem for Excel ver. 14.http://www.chemaxon.com Search PubMed.
- C. Pargellis, L. Tong, L. Churchill, P. F. Cirillo, T. Gilmore, A. G. Graham, P. M. Grob, E. R. Hickey, N. Moss, S. Pav and J. Regan, Nat. Struct. Biol., 2002, 9(4), 268–272 CrossRef CAS PubMed.
- J. R. Simard, M. Getlik, C. Grütter, V. Pawar, S. Wulfert, M. Rabiller and D. Rauh, J. Am. Chem. Soc., 2009, 131(37), 13286–13296 CrossRef CAS PubMed.
- A. Chaikuad, E. M. Tacconi, J. Zimmer, Y. Liang, N. S. Gray, M. Tarsounas and S. Knapp, Nat. Chem. Biol., 2014, 10(10), 853–860 CrossRef CAS PubMed.
- http://www.discoverx.com .
- G. F. De Nicola, E. D. Martin, A. Chaikuad, R. Bassi, J. Clark, L. Martino, S. Verma, P. Sicard, R. Tata, R. A. Atkinson, S. Knapp, M. R. Conte and M. S. Marber, Nat. Struct. Mol. Biol., 2013, 20(10), 1182–1190 Search PubMed.
- A. Chaikuad, S. Knapp and F. von Delft, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2015, 71, 1627–1639 CrossRef CAS PubMed.
- T. G. Battye, L. Kontogiannis, O. Johnson, H. R. Powell and A. G. Leslie, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 271–281 CrossRef CAS PubMed.
- P. R. Evans, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2006, 62, 72–82 CrossRef PubMed.
- A. J. McCoy, R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni and R. J. Read, J. Appl. Crystallogr., 2007, 40, 658–674 CrossRef CAS PubMed.
- P. Emsley, B. Lohkamp, W. G. Scott and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 486–501 CrossRef CAS PubMed.
- G. N. Murshudov, P. Skubak, A. A. Lebedev, N. S. Pannu, R. A. Steiner, R. A. Nicholls, M. D. Winn, F. Long and A. A. Vagin, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 355–367 CrossRef CAS PubMed.
Footnotes |
| † The authors declare no competing interests. |
| ‡ Electronic supplementary information (ESI) available: Synthesis and analysis of yoctoReactor library, hit re-synthesis and assay data, and X-ray data. See DOI: 10.1039/c6md00241b |
| This journal is © The Royal Society of Chemistry 2016 |