Endless resistance. Endless antibiotics?
Jed F.
Fisher
and
Shahriar
Mobashery
Department of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, IN 46556-5670, USA. E-mail: jfisher1@nd.edu; mobashery@nd.edu
First published on 3rd November 2015
Abstract
The practice of medicine was profoundly transformed by the introduction of the antibiotics (compounds isolated from Nature) and the antibacterials (compounds prepared by synthesis) for the control of bacterial infection. As a result of the extraordinary success of these compounds over decades of time, a timeless biological activity for these compounds has been presumed. This presumption is no longer. The inexorable acquisition of resistance mechanisms by bacteria is retransforming medical practice. Credible answers to this dilemma are far better recognized than they are being implemented. In this perspective we examine (and in key respects, reiterate) the chemical and biological strategies being used to address the challenge of bacterial resistance.
Introduction
Franklin's choice of the inevitability of “death and taxes” was surely not meant as an exclusive list. The modern observer of the diminishing efficacy of the chemotherapeutic armamentarium against bacterial infection would not hesitate to add “resistance” to his list. Bacterial resistance was, is, and forever shall be.1–4 It is found in the hospital, in the microbiome;5 in the animal manure of farms;6,7 in the soil;8–12 in remote caves;13 in the permafrost;14,15 in a centuries-old mummy;16 and in the remote reaches of the oceans.17 Moreover, given the impossibility of removing antibiotics from the environment—that is, removal of the evolutionary pressure—the evolution of the resistome cannot be reversed.18 We are compelled to live with the evolutionary legacy of antibiotics, both as ultimate pollutants in the environment and as persistent genes throughout the microbiome,19 even as we confront the consequences of this legacy. The question is less what we are to do, but how this confrontation must be done. The multi-factorial answers to this question are well known, and have been expressed in a chorus of different voices.20–38 Some of the answers to the question of what to do—such as limiting antibiotics in food sources; improving scientific,39 environmental,40 and clinical stewardship;41–46 and revising clinical trial design and creating post-registration financial incentives to reward the commercial investment in antibacterial discovery and development)47,48—transcend science. The importance and the inseparability of each of these aspects are well recognized. Nonetheless, the slow translation of many of these aspects into the societal and political spheres raises the question whether the full value of the chemical arsenal against bacterial infection—the (isolated from Nature) antibiotics, the semi-synthetic antibiotics, and the synthetic antibacterials—can be preserved. Until this translation occurs, how can society, and how can science, sustain the future of all—both old and new—antibacterial entities? The immediate task for society is the impeccable husbandry of the antibacterials that we already possess, as cogently argued by The Center for Disease Dynamics, Economics, and Policy.49 Nonetheless, their world-wide assessment shows that we remain well short of this standard. The task for science is to contribute to the technologies that will contribute to this stewardship, while simultaneously preparing for the possibility that stewardship will not be enough. The antibacterial future will include the pragmatic resurrection (or repurposing) of existing structures,20,50 identifying advantageous structural permutations of these same structures, and the discovery of new antibacterial strategies and structures. The chemistry, biochemistry, microbiology, and medical aspects of resistance are inseparable. In this brief perspective we exemplify, using for the most part recent discoveries, how scientists are confronting resistance in order to secure an antibiotic future as endless as that of resistance. These discoveries are discussed first from a chemical, and then a biological, perspective.Endless antibiotics: a chemical perspective
The most fundamental chemical perspective is the antibacterial as a composition of matter. The simplest proposal to address the diminishing ability of existing antibacterials is the discovery of better ones. If only this discovery was the simple matter of wishing! The daunting challenge of the discovery of new chemical matter that is efficacious, as a result of the compromise of one of the limited number of validated bacterial targets, is the theme of two separate lamentations from two separate industrial discovery teams.51,52 Is antibacterial discovery so exceptionally challenging? The chemical structures useful against bacterial infection are sourced from two realms. The first realm is the synthetic: structures that are unprecedented, or are only partly precedented, in Nature. The fluoroquinolones are an important example of exceptionally useful antibacterials having (for the most part) origins in the synthetic realm.53,54 The second realm is the antibiotics discovered by Nature, as exemplified by the incredible array of natural products found during the “golden age” of antibiotic isolation and thereafter.55 The interface between the two realms comprises the structures synthetically elaborated from the antibiotics of Nature. This transformation is robustly exemplified by the structure–activity development of the sub-classes (including the penicillins, cephalosporins, carbapenems, monobactams, monocarbams, and monosulfactams) of the β-lactam antibacterials.56 The challenge with respect to the chemistry of antibiotic discovery thus coincides with two inquiries. Is there opportunity for structural development within the “golden age” antibiotic structures? Are there unrecognized compounds with antibacterial activity in the synthetic structures found in chemical libraries, or to be found in Nature?The answer to both questions is “yes”, notwithstanding the daunting difficulty of realizing chemical answers.51,52 A sense of the answer to the first question is given by Scheme 1, where recent structures represent some of the key structural and mechanistic frontiers for the grand classes of the antibacterials of the golden age of discovery (the aminoglycosides, the β-lactams, the tetracyclines, the macrolides, and the quinolones). The focus of the remainder of this review is the answer to the second question: finding new antibacterials from chemical libraries and from Nature.
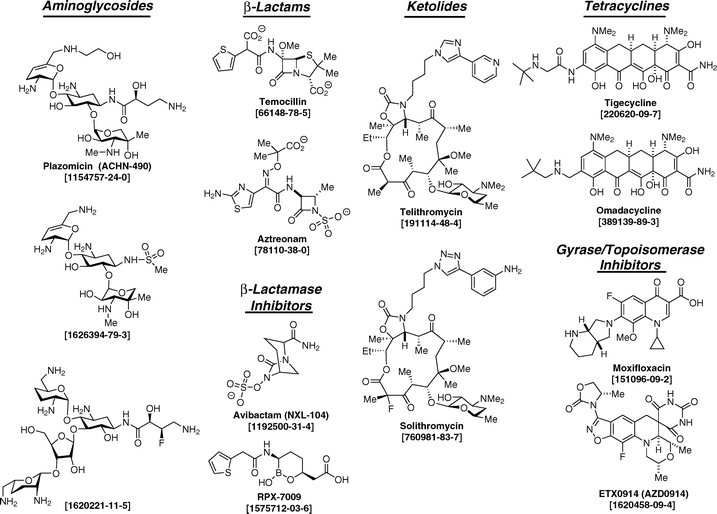 | ||
| Scheme 1 Structure–activity development within the “golden age” classes of the antibiotics continues, and certainly will address the future need for new antibacterials. This Scheme displays structures (identified by their generic name and/or their registry number) that exemplify (but by no means define) the research frontiers for these classes. The Aminoglycoside class (Column 1) is represented by the late-stage clinical candidate plazomicin,57 having broad-spectrum activity, low toxicity, and good evasion of the aminoglycoside-modifying enzymes of resistance.58,59 Structure [1626394-79-3] is an exploratory monosulfonamide-modified derivative of sisomicin with excellent Gram-negative activity and (in mice) exceptionally reduced ototoxicity.60 Structure [1620221-11-5], a second exploratory aminoglycoside, combines selective deletion of alcohol functional groups (to avoid resistance enzyme modification) with a fluoro-dependent reduction of the basicity of the neighboring amine with a consequent reduction in toxicity.61 Among the recent developments in the β-Lactam antibiotics (Column 2) is the reevaluation of older β-lactam structures such as temocillin62 and aztreonam. These latter two structures were perceived at time of their entry into the clinic as undesirably narrow-spectrum. With the proliferation of β-lactamase resistance enzymes over the past two decades, the ability of these older structures to resist hydrolysis by many β-lactamases is now regarded as advantageous. The antibacterial spectrum of aztreonam is further improved upon combination with β-lactamase inhibitors.63 The new β-Lactamase Inhibitor class (Column 2) includes the diazabicyclooctane avibactam.64,65 Other members of the diazabicyclooctane class under active investigation include the MK-7615 structure66 and the OP-0595 structure.67 A second new exploratory β-lactamase inhibitor, the cyclic boronic acid RPX-7009, restores carbapenem activity to bacteria expressing the KPC β-lactamase.68 Macrolides within the new Ketolide class (Column 3) are represented by telithromycin and by the newer fluoro-substituted solithromycin.69 The characterization of the rRNA methylase that confers ketolide resistance to the producing Streptomyces strains will facilitate the structure–activity development of this class, given the probable eventual transition of this activity as a resistance mechanism.70 New structures within the Tetracycline class (Column 4) include the clinically-approved tigecyline69 and the exploratory structure omadacycline. Although the structural difference between the two is subtle, the latter has oral activity.71 A second exploratory class of new tetracyclines is the hexacyclines.72 Resurgent interest in bacterial Gyrase/Topoisomerase Inhibitors (Column 4) is driven both by the proven clinical value of the fluoroquinolones and the consequent resistance development.73 A comparison of the structures of moxifloxacin, a recent generation fluoroquinolone, with that of the new exploratory structure ETX-0914 (having both Gram-positive and Gram-negative activity) shows superficial similarity (both have a modified fluoroquinoline core). Notwithstanding this similarity and the shared target, the mechanistic difference between the two is distinct.74,75 ETX-0914 is a clinical candidate targeting Neisseria gonorrhoeae. It is discussed in this review as an outstanding example of the possible value that synthetic chemical libraries may have for the discovery of new antibacterials. | ||
Part of the reason for the perception of a scarceness of antibacterial matter in chemical libraries is the poor alignment between the desirable chemical properties of drugs that address bacterial, compared to eukaryotic, targets, in such libraries.76–78 Recognition of this difference has led to the design of chemical libraries that are biased toward the generally more polar character of known antibacterials. These efforts have attained promising results.79 Aligning yet further the libraries to coincide with the different antibacterial sub-classes (such as attention to the requirements for optimal inhibition of cell wall targets compared to the ribosome as a target) may improve success.80 Nonetheless—and notwithstanding the reality that successful antibacterial discovery requires more than optimization of the physicochemical properties of the chemical library81—there are antibacterials waiting to be found in chemical libraries. For example, the interplay of library screening, synthesis, and the use of reverse genomics identified the spiropyrimidinetrione inhibitors of the bacterial topoisomerases.82,83 Subsequent structure–activity effort yielded a lead structure (EXT-0914) with excellent in vitro activity, and promising in vivo activity,84 against inter alia Neisseria gonorrhoeae.85–87 Likewise, library screening has had exceptional success in identifying potential antibacterials targeting Mycobacterium tuberculosis, as evidenced by both the imidazopyrimidine88–95 and benzothiazinone96–103 classes. As with any such structure, whether natural or synthetic, it remains to be seen whether it has the robustness (most emphatically, with respect to resistance development) needed to progress. But the structures are there. The new Community for Open Antimicrobial Drug Discovery effort is an opportunity to identify these structures.104,105 This objective of this effort is to expand the chemical diversity of antibacterial (and antifungal) compositions of matter through a free screening program (http://www.co-add.org), requiring submission of only 1 mg of a pure, water- or DMSO-soluble compound for the purpose of screening. The results of the screening are “free” (ownership is retained by the submitters).
Virtual chemical libraries in principle should not have the limitation of (for example) mismatched physical properties, but in fact virtual screening shares this same encumbrance since every virtual structure must have a physical counterpart. The treatment of disease (alas) will never be virtual. Nonetheless, the dramatically improved structural understanding of validated bacterial targets has provided examples of successful virtual screening.106 A recent example of successful virtual screening include the identification of inhibitors of the Penicillin-Binding Proteins having promising in vitro Gram-positive activity.107–109 Other examples include efforts directed toward the cytoskeletal protein FtsZ;110,111 the sortase transpeptidases;112,113 the GlmU uridyltransferase (from a lead identified by high-through put screening);114,115 and the Mur enzymes of peptidoglycan biosynthesis.116–118 The qualification of “promising” for all of these structures (again) refers to the momentous difficulty of taking a structure from in vitro activity to clinical efficacy. But this is a difficulty that is universal for all of drug discovery. A chemist's reflection on the ungainly heterocyclic structures that are approved inhibitors against the kinases of human cancer119 leads to the conclusion that however momentous the difficulty, the difficulty can be surmounted if the resources and incentives are in place. Antibiotic discovery is challenging. But it is not evident that its requirements for resources and incentives are any greater than for any other target, or any other disease. Rather, it is simply that neither the correct resources nor the correct incentives are fully in place.
The conclusion that antibacterial drug discovery is a surmountable challenge also stands on the ingenuity of Nature. If the golden age of antibiotic discovery has passed, a new era of natural product discovery is dawning.120–123 The antibiotics identified in the “golden age” of discovery originated from extraordinary scientific perspicacity. While this requirement has not lessened with time—the challenge of finding and then isolating (or replicating by total synthesis) a natural product, on the mass scale required for antibiotic activity, cannot be understated—the chemist today has an unprecedented ability not just to manipulate secondary biosynthesis, but also to interrogate a vastly greater diversity of bacterial and fungal species.124–137 Many of the genes for secondary metabolism are under epigenetic control.138–140 One can now pair the sequencing of a genome in order to identify the genes dedicated to secondary metabolite biosynthesis, with epigenetic activation of what often are silent biosynthetic pathways. For example, application of this strategy to a filamentous fungus, exploiting histone deacetylase inhibition to alter gene expression, activated 75% of the genes involved in secondary biosynthesis and resulted in the expression of ten secondary metabolites, four of which were new.141 There is no reason to believe that this strategy is not general. It is a strategy that could diversify access to both exploratory antibacterials, such as the nybomycin class of gyrase inhibitors142,143 as well as proven antibiotics such as the glycopeptides.144–149 Nor will it necessarily require a small molecule epigenetic modifier: interspecies communication within the microbial “interactome” has the same ability.150–153 Future natural product discovery will not be limited to the particular proteins encoded by the genes of an organism.124 We now have such a grasp on the modular organization of polyketide assembly that manipulation of the modules is feasible.55,154–156 Future natural-product discovery will not be limited to the genome of a single organism,132,133,157–160 nor to the multimodular synthases they may encode, as evidenced by the emerging ability to reprogram the biosynthetic function of these synthases.161,162 A compelling example of the future possibilities for the discovery of antibiotics that were previously hidden, is the use of microbial co-culture to elicit antibiotic expression by the so-called “dark” or “uncultivatable” bacteria.145,163–166 This strategy culminated in the discovery of the Gram-positive active, cell-wall biosynthesis-targeting depsipeptide, teixobactin.167
The transformation of a new structure from Nature into a clinically useful antibiotic follows in many cases combined empirical synthetic tailoring with property-based and structure-based design.69,168,169 Recent exemplifications (from among many) include structural development of the tetracyclines,69 the glycopeptides,170,171 and the antifolates.172–175 The selection of a clinical candidate among chemical structures with similar in vitro characteristics will be facilitated by an emerging new pharmacological criterion, a long residence time for drug engagement of its target.176–182 Structures that possess this ability achieve an advantageous kinetic selectivity, wherein off-target interactions are minimized. The relevance of this criterion was exemplified recently during the assessment of inhibitors of LpxC, a key enzyme in the biosynthetic pathway to the lipopolysaccharides of the outer membrane of the Gram-negative bacteria. LpxC is a validated antibacterial target. However, its conformational mobility as a protein enables it to accommodate resistance mutations.183,184 Incorporation of kinetic performance, measured as the on-rate for formation and off-rate for breakdown within a series of LpxC inhibitors, yielded a pharmacodynamic model wherein the dose–response curves for these inhibitors in a Pseudomonas aeruginosa animal model of infection was predicted.185 The use of comparative kinetic data in compound evaluation will facilitate the pre-clinical assessment of exploratory antibacterial structures.
Endless antibiotics: a biological perspective
Notwithstanding the fact that antibiotic resistance in Nature coincided with the discovery of the antibiotics, the conceptualization of “resistance” continues to evolve.186–188 This evolution is multi-dimensional. Its directions include not just the mechanism(s) of the resistance against a particular antibacterial, but the spectrum of thought ranging from reflection on the ecological purpose of the antibiotics, to the criteria necessary to attain the multi-agent synergy against infectious disease that the future may demand. Here we offer a concise perspective on the eclectic breadth of the conceptualization of bacterial resistance, with emphasis on (and acknowledgement of) the most recent contributions defining its directions.The emerging methodologies to probe the relationships among bacterial pathways and antibacterial structures will prove transformative for antibacterial discovery.189,190 Both the “Comprehensive Antibiotic Resistance Database” (“CARD”: http://arpcard.mcmaster.ca)191,192 and the NCBI National Database for Antimicrobial Resistant Organisms (http://www.ncbi.nlm.nih.gov/projects/pathogens/) address the bioinformatic aspects of these relationships. One example of a new methodology to complement bioinformatics analysis is the use of sub-μm fluorescence to attain resolution within the dimensions of the bacterial cell. With this resolution, an immediate visual assignment of the antibacterial mechanism by examination of the cytological profile of fluorescent reporters as a result of the presence of the antibacterial.193,194 A second example is the use of imaging mass spectrometry to evaluate specialized metabolite synthesis by Streptomyces coelicolor as the result of interspecies interaction.150,151 This methodology has the promise of improving our understanding of bacterial communication, especially as it relates to antibiotic synthesis, mechanism, and resistance responses. With respect to these mechanistic aspects, it is prudent for us to appreciate how poor is our understanding, even for the “golden age” antibiotics. The venerable class of β-lactam antibiotics exemplifies our ignorance. We are reminded that there is an enormous breadth of structure around the β-lactam core among the sub-families of this class. Individual β-lactam structures show differential affinity for their Penicillin-Binding Protein (PBP) enzyme targets. Each bacterium has a family of PBPs, and each of these PBPs uniquely contributes—some essentially, others much less so—to the growth and shape of the bacterium. Hence, each β-lactam structure uniquely profiles the PBP family of a bacterium.195,196 This uniqueness explains why particular β-lactams are clinically efficacious for infections by particular bacteria, but does not reveal the mechanistic interconnection between PBP inactivation and subsequent bactericidal cell lysis. While recent profiling of the relationship among β-lactam structure, PBP inactivation, and MIC value confirms the importance of selective PBP inactivation, it also identifies particular β-lactams for which the MIC does not coincide with PBP inactivation. The answer to the “bactericidal mechanism of the β-lactams” is remarkably incomplete.197–199
A further context around this incompleteness is our equally rudimentary understanding of the ecological purpose in Nature for the antibiotics. The conception that many natural products produced by microbes are messengers, a molecular realm termed by Davies as the “parvome”,200–204 is now well accepted. But what is their message? The traditional explanation for the antibiotics as defensive molecules to secure and preserve an ecological niche is supported by recent experiments.205,206 Nonetheless, the antibiotic concentrations used in chemotherapy vastly exceed the concentrations attained in ecological niches, and accordingly we must be mindful of understanding the bacterial responses to sub-MIC (sub-lethal) antibiotic exposure.207,208 The proven relationships among quorum sensing,209–212 biofilm formation,213,214 and virulence establish communication as a key role for the antibiotics of Nature.215–217 Antibiotics, even if simplistically conceptualized as weaponry, communicate. Their communication ability is intimate to the complexity of bacterial tolerance218 and persistence219–227 within the diversity of the ecological microbiomes.5,228–231 Chemical communication among bacteria is understandably an evolutionary force for genetic transformation19,208,232–234 and thus is one and the same with resistance.
Here we return to the mysterious depth of the resistome, and the underestimated ability of bacteria to adapt to what we naively might believe to be an even more than decimating assault by the antibacterial concentrations attained during clinical use. We cannot be surprised that while we know that antibiotics are powerful evolutionary forces, we do not understand the relative pressure of these forces in particular ecological niches235–237 and within the universe of resistance mechanisms.238–241 Both new mechanisms for antibacterial invention (such as the discovery that the allosteric regulation of the essential PBP2a enzyme of methicillin-resistant Staphylococcus aureus can be disrupted)109,242–244 and new mechanisms to secure resistance (as just demonstrated for the tetracyclines)245,246 will be found. We face the dilemma that while the micobiome is heterogeneous247 and the natural state for bacteria is a surface-bound community,248 there are compelling arguments to study the behavior of individual bacteria,249,250 even for the determination of the MIC for an antibacterial.251
How is this labyrinth to be confronted for antibacterial discovery? The obvious answer is the use of the antibiotics themselves, as superlative chemical probes, to provide both understanding and opportunity with respect to the confluence of targets and pathways. Two complementary themes exemplify efforts toward such identification. One is the relationship of antibiotic activity to metabolism (defined in the broadest sense). The second is the ability of antibacterial pairs to synergize their respective antibiotic activities. A direct relationship between metabolism and antibiotic activity is well recognized.252–257 Less understood is why the relationship directly correlates for some antibiotics, but indirectly for others.258 The ability to correlate the mechanism of ribosome-directed antibiotics, their antibiotic efficacy, and the rate of growth of E. coli provides promise that an understanding of the key aspects of this relationship, in terms of clinical strategies, will be forthcoming.259 A more demanding question is for which bacteria, and for which circumstances, the generation of reactive-oxygen species260 contributes253,261–265 or does not contribute266–268 to antibacterial efficacy. An answer to this question may contribute (for example) to a mechanistic understanding of how the new antibiotic lysocin acts through interference with the menaquinone of the bacterial membrane;269 and whether the role of glutamate dehydrogenase activity in coordinating FtsZ-dependent cell division in Caulobacter crescentus270 represents a potentially synergistic pathway confluence in bacteria that have FtsZ-dependent bacterial division. Such synergism—collateral sensitivity—is a central theme to the future discovery of antibiotics.271–281 Exploration of this concept with respect to the β-lactam antibiotics in S. aureus has identified synergistic confluence with inhibitors of its cell division pathway,282 of its peptidoglycan biosynthesis pathway,283,284 and of its wall-teichoic acid biosynthesis pathway.285–290 This latter correlation is especially interesting, as the wall-teichoic acids are important contributors to colonization.291,292 Collateral synergy is now also demonstrated between the teichoic acids and the undecaprenyl lipid biosynthesis pathway.205 The undecaprenyl lipids are attractive targets as bacteria have a small pool of these lipids to support cell-wall biosynthesis.293–296 Understanding just how to achieve this sensitivity is incomplete,281,297,298 as evidenced by other studies on the undecaprenyl pathway. Incomplete inhibition of undecaprenyl biosynthesis in B. subtilis induced a stress response that resulted in increased resistance to other cell-wall-active antibiotics.299 Lastly, even when the outcome for the pairing of an antibiotic with a synergistic inhibitor is decisively advantageous in pharmacological models, validating this outcome in the clinic is especially challenging. Not only must safety be proven for the pairing of the both entities, but for optimal efficacy for the structures of the pair and dosing choices for the pair, should correspond to matched pharmacokinetics of the two entities. The current cluster of β-lactam/β-lactamase inhibitors in clinical evaluation for Gram-negative bacterial infection reflects not just their promise of efficacy as a pairing, but the ability to bring to the clinical design the established clinical experience of the β-lactam partner. The task is simpler when one of the entities is known.
Conclusion
The title of this perspective pairs a declaration with a question. The declaration is irrefutable. This perspective has addressed the question, but without separation of the question into its two very different contexts. New antibiotics will be discovered. In this context of the question, our answer is resoundingly positive. Indeed, the basis for this opinion is the central theme of this perspective. There is, however, a second context for this same question. Will these new antibiotics achieve clinical impact? The answer to this question is less positive. The enthusiastic optimism and resoundingly positive answer to the question in the first context, and reserved (even deeply reserved) pessimism for the answer in the second context, is not cognizant dissonance. At this time good progress is being made with respect to the 10 × 20 initiative of the Infectious Diseases Society of America.300,301 New antibiotics (especially new β-lactamase-inhibitor combinations)65,302–310 are reaching the clinic. Yet over this same decade a fundamental transition has occurred with respect to early antibiotic discovery. This task has transitioned from major pharma companies to smaller biotechnology companies, and to academic centers. An excellent example of success from such collaboration is SMT-19969, now in Phase II clinical trials for Clostridium difficile infection.311,312 SMT-19969 is a member of the synthetic, DNA-interacting bis-benzimidazole class of structures. Its mechanism is not gyrase inhibition, as is the case for other members of this class.311 SMT-19969 shows good selectivity for C. difficile. As it is less active against other Gram-positive anaerobes and the Gram-negative anerobes, and only weakly active against both Gram-positive aerobes it has potential as a selective, microbiome-sparing entity. Although the transition of early discovery away from major pharma certainly does not itself mean that antibacterial discovery and development will falter, there is reason for concern. The urgency for the preservation of existing antibacterials, and the discovery of new antibacterials, is such that Nathan has argued for an open discovery initiative embracing academic, industrial, foundational, and governmental collaboration.313 Given the present requirements for the size and scope of clinical evaluation (although these requirements are changing for the better) within the unchanging cost structure for the antibiotic market, there are credible reasons for such a proposal. Absolute ownership of intellectual property is a necessity for all new drug entities. Acquisition of this ownership requires tight integration of the timing of the patent prosecution with clinical development. The financial return on the investment required to bring an antibacterial to market, even using optimistic estimates, is predicted to occur only in the final years of the patent life. This sobering reality is illustrated (Fig. 1). The necessity for a fundamental change in how the necessary investment in the discovery and development of anti-infective drugs is made appears inescapable.28,47,313,314 As we stated emphatically a decade ago, bringing a new drug to market (nor even the discovery of a new antibiotic) is not an instantaneous event.315 Achieving intellectual property ownership while sustaining credible progression to the return on investment, in a development model where early discovery does not tightly transition to clinical development, is a profound challenge. The financial incentives for antibiotic discovery must change if clinically relevant antibiotic discovery is to be sustainable. The interim solutions of reconsidering old antibiotics316,317 or synergistic pairing of existing antibacterials (as we have just discussed) are essential. But neither solution, alone or together, offers the prospect of endless antibiotics.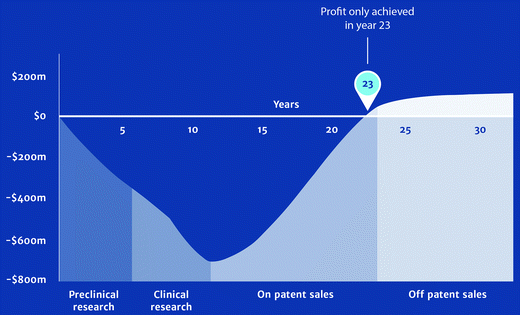 | ||
| Fig. 1 A simulation of the cost to discover and develop an antibacterial (preclinical and clinical research) compared to the return on the cost of the investment (on-patent and off-patent sales). This Figure is taken from p. 11 of the 2015 document “Securing new drugs for future generations: The pipeline of antibiotics” of the Wellcome Trust.48 The factual basis for this figure is provided in the appendix to this document. (Acknowledgement: ‘Review on Antimicrobial Resistance. Securing new drugs for future generations: the pipeline of antibiotics. 2015’). | ||
Acknowledgements
Research in this laboratory is supported by a grant from the National Institutes of Health (AI104987).References
- V. M. D'Costa, C. E. King, L. Kalan, M. Morar, W. W. Sung, C. Schwarz, D. Froese, G. Zazula, F. Calmels, R. Debruyne, G. B. Golding, H. N. Poinar and G. D. Wright, Nature, 2011, 477, 457 CrossRef PubMed.
- G. D. Wright and H. Poinar, Trends Microbiol., 2012, 20, 157 CrossRef CAS PubMed.
- R. L. Finley, P. Collignon, D. G. J. Larsson, S. A. McEwen, X. Z. Li, W. H. Gaze, R. Reid-Smith, M. Timinouni, D. W. Graham and E. Topp, Clin. Infect. Dis., 2013, 57, 704 CrossRef PubMed.
- T. U. Berendonk, C. M. Manaia, C. Merlin, D. Fatta-Kassinos, E. Cytryn, F. Walsh, H. Bürgmann, H. Sørum, M. Norström, M. N. Pons, N. Kreuzinger, P. Huovinen, S. Stefani, T. Schwartz, V. Kisand, F. Baquero and J. L. Martinez, Nat. Rev. Microbiol., 2015, 13, 310 CrossRef CAS PubMed.
- M. O. A. Sommer, G. Dantas and G. M. Church, Science, 2009, 325, 1128 CrossRef CAS PubMed.
- C. Larson, Science, 2015, 347, 704 CrossRef CAS PubMed.
- L. B. Price, B. J. Koch and B. A. Hungate, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 5554 CrossRef CAS PubMed.
- H. K. Allen, J. Donato, H. H. Wang, K. A. Cloud-Hansen, J. Davies and J. Handelsman, Nat. Rev. Microbiol., 2010, 8, 251 CrossRef CAS PubMed.
- K. J. Forsberg, A. Reyes, B. Wang, E. M. Selleck, M. O. A. Sommer and G. Dantas, Science, 2012, 337, 1107 CrossRef CAS PubMed.
- B. S. Griffiths and L. Philippot, FEMS Microbiol. Rev., 2013, 37, 112 CrossRef CAS PubMed.
- J. Nesme, S. Cécillon, T. O. Delmont, J. M. Monier, T. M. Vogel and P. Simonet, Curr. Biol., 2014, 24, 1096 CrossRef CAS PubMed.
- J. Nesme and P. Simonet, Environ. Microbiol., 2015, 17, 913 CrossRef PubMed.
- K. Bhullar, N. Waglechner, A. Pawlowski, K. Koteva, E. D. Banks, M. D. Johnston, H. A. Barton and G. D. Wright, PLoS One, 2012, 7, e34953 CAS.
- M. Petrova, A. Kurakov, N. Shcherbatova and S. Mindlin, Microbiology, 2014, 160, 2253 CrossRef CAS PubMed.
- G. G. Perron, L. Whyte, P. J. Turnbaugh, J. Goordial, W. P. Hanage, G. Dantas and M. M. Desai, PLoS One, 2015, 10, e0069533 Search PubMed.
- T. M. Santiago-Rodriguez, G. Fornaciari, S. Luciani, S. E. Dowd, G. A. Toranzos, I. Marota and R. J. Cano, PLoS One, 2015, 10, e0138135 Search PubMed.
- M. S. Alves, A. Pereira, S. M. Araújo, B. B. Castro, A. C. Correia and I. Henriques, Front. Microbiol., 2014, 5, 426 CrossRef PubMed.
- D. I. Andersson and D. Hughes, Nat. Rev. Microbiol., 2010, 8, 260 CAS.
- M. R. Gillings, Front. Microbiol., 2013, 4, 4 CrossRef PubMed.
- M. A. Fischbach and C. T. Walsh, Science, 2009, 325, 1089 CrossRef CAS PubMed.
- K. Bush, P. Courvalin, G. Dantas, J. Davies, B. Eisenstein, P. Huovinen, G. A. Jacoby, R. Kishony, B. N. Kreiswirth, E. Kutter, S. A. Lerner, S. Levy, K. Lewis, O. Lomovskaya, J. H. Miller, S. Mobashery, L. J. Piddock, S. Projan, C. M. Thomas, A. Tomasz, P. M. Tulkens, T. R. Walsh, J. D. Watson, J. Witkowski, W. Witte, G. Wright, P. Yeh and H. I. Zgurskaya, Nat. Rev. Microbiol., 2011, 9, 894 CrossRef CAS PubMed.
- L. L. Silver, Clin. Microbiol. Rev., 2011, 24, 71 CrossRef CAS PubMed.
- J. G. Bartlett, D. N. Gilbert and B. Spellberg, Clin. Infect. Dis., 2013, 56, 1445 CrossRef CAS PubMed.
- K. Lewis, Nat. Rev. Drug Discovery, 2013, 12, 371 CrossRef CAS PubMed.
- K. M. G. O'Connell, J. T. Hodgkinson, H. F. Sore, M. Welch, G. P. C. Salmond and D. R. Spring, Angew. Chem., Int. Ed., 2013, 52, 10706 CrossRef PubMed.
- G. Dantas and M. O. A. Sommer, Am. Sci., 2014, 102, 42 CrossRef.
- J. F. Fisher, J. W. Johnson and S. Mobashery, in Handbook of Antimicrobial Resistance, ed. M. Götte, A. Berghuis, G. Matlashewski, M. Wainberg and D. Sheppard, Springer Science + Business Media, New York, 2014, ch. 12 Search PubMed.
- M. Metz and D. M. Shlaes, Antimicrob. Agents Chemother., 2014, 58, 4253 CrossRef CAS PubMed.
- C. Nathan and O. Cars, N. Engl. J. Med., 2014, 371, 1761 CrossRef PubMed.
- E. Oldfield and X. Feng, Trends Pharmacol. Sci., 2014, 35, 664 CrossRef CAS PubMed.
- L. J. V. Piddock, Microbiology, 2014, 160, 2366 CrossRef CAS PubMed.
- G. M. Rossolini, F. Arena, P. Pecile and S. Pollini, Curr. Opin. Pharmacol., 2014, 18, 56 CrossRef CAS PubMed.
- S. B. Singh, Bioorg. Med. Chem. Lett., 2014, 24, 3683 CrossRef CAS PubMed.
- B. Spellberg and D. N. Gilbert, Clin. Infect. Dis., 2014, 59(Suppl 2), S71 CrossRef PubMed.
- M. Perros, Science, 2015, 347, 1062 CrossRef CAS PubMed.
- G. Tillotson, Lancet Infect. Dis., 2015, 15, 758 CrossRef PubMed.
- G. D. Wright, ACS Infect. Dis., 2015, 1, 80 CrossRef CAS.
- K. Bush, ACS Infect. Dis., 2015, 1, DOI:10.1021/acsinfecdis.5b00100 , in press.
- L. Bowater, J. Antimicrob. Chemother., 2015, 70, 1925 CrossRef PubMed.
- S. Jechalke, H. Heuer, J. Siemens, W. Amelung and K. Smalla, Trends Microbiol., 2014, 22, 536 CrossRef CAS PubMed.
- E. Charani, J. Cooke and A. Holmes, J. Antimicrob. Chemother., 2010, 65, 2275 CrossRef CAS PubMed.
- D. J. O'Brien and I. M. Gould, Curr. Opin. Infect. Dis., 2013, 26, 352 CrossRef PubMed.
- T. M. File Jr., A. Srinivasan and J. G. Bartlett, Clin. Infect. Dis., 2014, 59(Suppl 3), S93 CrossRef PubMed.
- D. M. Livermore, Int. J. Antimicrob. Agents, 2014, 43, 319 CrossRef CAS PubMed.
- P. D. Tamma, A. Holmes and E. D. Ashley, Curr. Opin. Infect. Dis., 2014, 27, 348 CrossRef CAS PubMed.
- J. P. Metlay, Clin. Infect. Dis., 2015, 60, 1317 Search PubMed.
- D. M. Shlaes, ACS Infect. Dis., 2015, 1, 232 CrossRef CAS.
- J. O'Neill, Review on Antimicrobial Resistance, 2015, http://amr-review.org/sites/default/files/SECURING%20NEW%20DRUGS%20FOR%20FUTURE%20GENERATIONS%20FINAL%20WEB_0.pdf Search PubMed.
- H. Gelband, M. Miller-Petrie, S. Pant, S. Gandra, J. Levinson, D. Barter, A. White and R. Laxminarayan, The Center for Disease Dynamics, Economics, and Policy (Global Antibiotic Resistance Partnership), 2015, http://cddep.org/publications/state_worlds_antibiotics_2015 Search PubMed.
- G. Cox and G. D. Wright, Int. J. Med. Microbiol., 2013, 303, 287 CrossRef CAS PubMed.
- D. J. Payne, M. N. Gwynn, D. J. Holmes and D. L. Pompliano, Nat. Rev. Drug Discovery, 2007, 6, 29 CrossRef CAS PubMed.
- R. Tommasi, D. G. Brown, G. K. Walkup, J. I. Manchester and A. A. Miller, Nat. Rev. Drug Discovery, 2015, 14, 529 CrossRef CAS PubMed.
- G. S. Bisacchi and J. I. Manchester, ACS Infect. Dis., 2015, 1, 4 CrossRef CAS.
- G. S. Bisacchi, J. Med. Chem., 2015, 58, 4874 CrossRef CAS PubMed.
- C. T. Walsh and T. A. Wencewicz, J. Antibiot., 2013, 67, 7 CrossRef PubMed.
- S. A. Testero, J. F. Fisher and S. Mobashery, Burger's Medicinal Chemistry, Drug Discovery and Development, Seventh Edition, 2010, vol. 7, p. 259 Search PubMed.
- I. Karaiskos, M. Souli and H. Giamarellou, Expert Opin. Invest. Drugs, 2015, 24, 1501 CrossRef CAS PubMed.
- B. Becker and M. A. Cooper, ACS Chem. Biol., 2013, 8, 105 CrossRef CAS PubMed.
- M. Y. Fosso, Y. Li and S. Garneau-Tsodikova, MedChemComm, 2014, 5, 1075 RSC.
- M. E. Huth, K. H. Han, K. Sotoudeh, Y. J. Hsieh, T. Effertz, A. A. Vu, S. Verhoeven, M. H. Hsieh, R. Greenhouse, A. G. Cheng and A. J. Ricci, J. Clin. Invest., 2015, 125, 583 Search PubMed.
- J. P. Maianti, H. Kanazawa, P. Dozzo, R. D. Matias, L. A. Feeney, E. S. Armstrong, D. J. Hildebrandt, T. R. Kane, M. J. Gliedt, A. A. Goldblum, M. S. Linsell, J. B. Aggen, J. Kondo and S. Hanessian, ACS Chem. Biol., 2014, 9, 2067 CrossRef CAS PubMed.
- C. G. Giske, Clin. Microbiol. Infect., 2015, 21, 899 CrossRef CAS PubMed.
- H. Li, M. Estabrook, G. A. Jacoby, W. W. Nichols, R. T. Testa and K. Bush, Antimicrob. Agents Chemother., 2015, 59, 1789 CrossRef CAS PubMed.
- S. M. Drawz, K. M. Papp-Wallace and R. A. Bonomo, Antimicrob. Agents Chemother., 2014, 58, 1835 CrossRef PubMed.
- I. Olsen, Eur. J. Clin. Microbiol. Infect. Dis., 2015, 34, 1303 CrossRef CAS PubMed.
- T. A. Blizzard, H. Chen, S. Kim, J. Wu, R. Bodner, C. Gude, J. Imbriglio, K. Young, Y. W. Park, A. Ogawa, S. Raghoobar, N. Hairston, R. E. Painter, D. Wisniewski, G. Scapin, P. Fitzgerald, N. Sharma, J. Lu, S. Ha, J. Hermes and M. L. Hammond, Bioorg. Med. Chem. Lett., 2014, 24, 780 CrossRef CAS PubMed.
- A. Morinaka, Y. Tsutsumi, M. Yamada, K. Suzuki, T. Watanabe, T. Abe, T. Furuuchi, S. Inamura, Y. Sakamaki, N. Mitsuhashi, T. Ida and D. M. Livermore, J. Antimicrob. Chemother., 2015, 70, 2779 CrossRef CAS PubMed.
- S. J. Hecker, K. R. Reddy, M. Totrov, G. C. Hirst, O. Lomovskaya, D. C. Griffith, P. King, R. Tsivkovski, D. Sun, M. Sabet, Z. Tarazi, M. C. Clifton, K. Atkins, A. Raymond, K. T. Potts, J. Abendroth, S. H. Boyer, J. S. Loutit, E. E. Morgan, S. Durso and M. N. Dudley, J. Med. Chem., 2015, 58, 3682 CrossRef CAS PubMed.
- P. M. Wright, I. B. Seiple and A. G. Myers, Angew. Chem., Int. Ed., 2014, 53, 8840 CrossRef CAS PubMed.
- M. M. Almutairi, S. R. Park, S. Rose, D. A. Hansen, N. Vázquez-Laslop, S. Douthwaite, D. H. Sherman and A. S. Mankin, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 12956 CrossRef CAS PubMed.
- L. Honeyman, M. Ismail, M. L. Nelson, B. Bhatia, T. E. Bowser, J. Chen, R. Mechiche, K. Ohemeng, A. K. Verma, E. P. Cannon, A. Macone, S. K. Tanaka and S. Levy, Antimicrob. Agents Chemother., 2015, 59, 7044 CrossRef PubMed.
- C. Sun, D. K. Hunt, C. L. Chen, Y. Deng, M. He, R. B. Clark, C. Fyfe, T. H. Grossman, J. A. Sutcliffe and X.-Y. Xiao, J. Med. Chem., 2015, 58, 4703 CrossRef CAS PubMed.
- K. J. Aldred, R. J. Kerns and N. Osheroff, Biochemistry, 2014, 53, 1565 CrossRef CAS PubMed.
- G. S. Basarab, P. Doig, V. Galullo, G. Kern, A. Kimzey, A. Kutschke, J. P. Newman, M. Morningstar, J. Mueller, L. Otterson, K. Vishwanathan, F. Zhou and M. Gowravaram, J. Med. Chem., 2015, 58, 6264 CrossRef CAS PubMed.
- G. Kern, T. Palmer, D. E. Ehmann, A. B. Shapiro, B. Andrews, G. S. Basarab, P. Doig, J. Fan, N. Gao, S. D. Mills, J. Mueller, S. Sriram, J. Thresher and G. K. Walkup, J. Biol. Chem., 2015, 290, 20984 CrossRef CAS PubMed.
- R. O'Shea and H. E. Moser, J. Med. Chem., 2008, 51, 2871 CrossRef PubMed.
- M. N. Gwynn, A. Portnoy, S. F. Rittenhouse and D. J. Payne, Ann. N. Y. Acad. Sci., 2010, 1213, 5 CrossRef PubMed.
- S. Barelier, O. Eidam, I. Fish, J. Hollander, F. Figaroa, R. Nachane, J. J. Irwin, B. K. Shoichet and G. Siegal, ACS Chem. Biol., 2014, 9, 1528 CrossRef CAS PubMed.
- R. Fleeman, T. M. LaVoi, R. G. Santos, A. Morales, A. Nefzi, G. S. Welmaker, J. L. Medina-Franco, M. A. Giulianotti, R. A. Houghten and L. N. Shaw, J. Med. Chem., 2015, 58, 3340 CrossRef CAS PubMed.
- G. Mugumbate and J. P. Overington, Bioorg. Med. Chem., 2015, 23, 5218 CrossRef CAS PubMed.
- D. G. Brown, T. L. May-Dracka, M. M. Gagnon and R. Tommasi, J. Med. Chem., 2014, 57, 10144 CrossRef CAS PubMed.
- A. A. Miller, G. L. Bundy, J. E. Mott, J. E. Skepner, T. P. Boyle, D. W. Harris, A. E. Hromockyj, K. R. Marotti, G. E. Zurenko, J. B. Munzner, M. T. Sweeney, G. F. Bammert, J. C. Hamel, C. W. Ford, W. Z. Zhong, D. R. Graber, G. E. Martin, F. Han, L. A. Dolak, E. P. Seest, J. C. Ruble, G. M. Kamilar, J. R. Palmer, L. S. Banitt, A. R. Hurd and M. R. Barbachyn, Antimicrob. Agents Chemother., 2008, 52, 2806 CrossRef CAS PubMed.
- J. C. Ruble, A. R. Hurd, T. A. Johnson, D. A. Sherry, M. R. Barbachyn, P. L. Toogood, G. L. Bundy, D. R. Graber and G. M. Kamilar, J. Am. Chem. Soc., 2009, 131, 3991 CrossRef CAS PubMed.
- G. S. Basarab, G. H. Kern, J. McNulty, J. P. Mueller, K. Lawrence, K. Vishwanathan, R. A. Alm, K. Barvian, P. Doig, V. Galullo, H. Gardner, M. Gowravaram, M. Huband, A. Kimzey, M. Morningstar, A. Kutschke, S. D. Lahiri, M. Perros, R. Singh, V. J. A. Schuck, R. Tommasi, G. Walkup and J. V. Newman, Sci. Rep., 2015, 5, 11827 CrossRef CAS PubMed.
- S. Jacobsson, D. Golparian, R. A. Alm, M. Huband, J. Mueller, J. S. Jensen, M. Ohnishi and M. Unemo, Antimicrob. Agents Chemother., 2014, 58, 5585 CrossRef PubMed.
- R. A. Alm, S. D. Lahiri, A. Kutschke, L. G. Otterson, R. E. McLaughlin, J. D. Whiteaker, L. A. Lewis, X. Su, M. D. Huband, H. Gardner and J. P. Mueller, Antimicrob. Agents Chemother., 2015, 59, 1478 CrossRef CAS PubMed.
- M. Unemo, J. Ringlander, C. Wiggins, H. Fredlund, S. Jacobsson and M. Cole, Antimicrob. Agents Chemother., 2015, 59, 5220 CrossRef CAS PubMed.
- G. C. Moraski, L. D. Markley, P. A. Hipskind, H. Boshoff, S. Cho, S. G. Franzblau and M. J. Miller, ACS Med. Chem. Lett., 2011, 2, 466 CrossRef CAS PubMed.
- G. C. Moraski, L. D. Markley, M. Chang, S. Cho, S. G. Franzblau, C. H. Hwang, H. Boshoff and M. J. Miller, Bioorg. Med. Chem., 2012, 20, 2214 CrossRef CAS PubMed.
- G. C. Moraski, L. D. Markley, J. Cramer, P. A. Hipskind, H. Boshoff, M. Bailey, T. Alling, J. Ollinger, T. Parish and M. J. Miller, ACS Med. Chem. Lett., 2013, 4, 675 CrossRef CAS PubMed.
- G. C. Moraski, P. A. Miller, M. A. Bailey, J. Ollinger, T. Parish, H. I. Boshoff, S. Cho, J. R. Anderson, S. Mulugeta, S. G. Franzblau and M. J. Miller, ACS Infect. Dis., 2015, 1, 85 CrossRef CAS PubMed.
- K. A. Abrahams, J. A. Cox, V. L. Spivey, N. J. Loman, M. J. Pallen, C. Constantinidou, R. Fernández, C. Alemparte, M. J. Remuiñán, D. Barros, L. Ballell and G. S. Besra, PLoS One, 2012, 7, e52951 CAS.
- K. Pethe, P. Bifani, J. Jang, S. Kang, S. Park, S. Ahn, J. Jiricek, J. Jung, H. K. Jeon, J. Cechetto, T. Christophe, H. Lee, M. Kempf, M. Jackson, A. J. Lenaerts, H. Pham, V. Jones, M. J. Seo, Y. M. Kim, M. Seo, J. J. Seo, D. Park, Y. Ko, I. Choi, R. Kim, S. Y. Kim, S. Lim, S. A. Yim, J. Nam, H. Kang, H. Kwon, C. T. Oh, Y. Cho, Y. Jang, J. Kim, A. Chua, B. H. Tan, M. B. Nanjundappa, S. P. Rao, W. S. Barnes, R. Wintjens, J. R. Walker, S. Alonso, S. Lee, J. Kim, S. Oh, T. Oh, U. Nehrbass, S. J. Han, Z. No, J. Lee, P. Brodin, S. N. Cho, K. Nam and J. Kim, Nat. Med., 2013, 19, 1157 CrossRef CAS PubMed.
- S. Kang, R. Y. Kim, M. J. Seo, S. Lee, Y. M. Kim, M. Seo, J. J. Seo, Y. Ko, I. Choi, J. Jang, J. Nam, S. Park, H. Kang, H. J. Kim, J. Kim, S. Ahn, K. Pethe, K. Nam, Z. No and J. Kim, J. Med. Chem., 2014, 57, 5293 CrossRef CAS PubMed.
- M. S. Kim, J. Jang, N. B. Ab Rahman, K. Pethe, E. A. Berry and L. S. Huang, J. Biol. Chem., 2015, 290, 14350 CrossRef CAS PubMed.
- V. Makarov, G. Manina, K. Mikusova, U. Möllmann, O. Ryabova, B. Saint-Joanis, N. Dhar, M. R. Pasca, S. Buroni, A. P. Lucarelli, A. Milano, E. De Rossi, M. Belanova, A. Bobovska, P. Dianiskova, J. Kordulakova, C. Sala, E. Fullam, P. Schneider, J. D. McKinney, P. Brodin, T. Christophe, S. Waddell, P. Butcher, J. Albrethsen, I. Rosenkrands, R. Brosch, V. Nandi, S. Bharath, S. Gaonkar, R. K. Shandil, V. Balasubramanian, T. Balganesh, S. Tyagi, J. Grosset, G. Riccardi and S. T. Cole, Science, 2009, 324, 801 CrossRef CAS PubMed.
- J. Neres, F. Pojer, E. Molteni, L. R. Chiarelli, N. Dhar, S. Boy-Röttger, S. Buroni, E. Fullam, G. Degiacomi, A. P. Lucarelli, R. J. Read, G. Zanoni, D. E. Edmondson, E. De Rossi, M. R. Pasca, J. D. McKinney, P. J. Dyson, G. Riccardi, A. Mattevi, S. T. Cole and C. Binda, Sci. Transl. Med., 2012, 4, 150ra121 Search PubMed.
- S. M. Batt, T. Jabeen, V. Bhowruth, L. Quill, P. A. Lund, L. Eggeling, L. J. Alderwick, K. Futterer and G. S. Besra, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 11354 CrossRef CAS PubMed.
- R. Tiwari, G. C. Moraski, V. Krchnak, P. A. Miller, M. Colon-Martinez, E. Herrero, A. G. Oliver and M. J. Miller, J. Am. Chem. Soc., 2013, 135, 3539 CrossRef CAS PubMed.
- S. Grover, L. J. Alderwick, A. K. Mishra, K. Krumbach, J. Marienhagen, L. Eggeling, A. Bhatt and G. S. Besra, J. Biol. Chem., 2014, 289, 6177 CrossRef CAS PubMed.
- K. Mikusová, V. Makarov and J. Neres, Curr. Pharm. Des., 2014, 20, 4379 CrossRef.
- V. Makarov, B. Lechartier, M. Zhang, J. Neres, A. M. van der Sar, S. A. Raadsen, R. C. Hartkoorn, O. B. Ryabova, A. Vocat, L. A. Decosterd, N. Widmer, T. Buclin, W. Bitter, K. Andries, F. Pojer, P. J. Dyson and S. T. Cole, EMBO Mol. Med., 2014, 6, 372 CrossRef CAS PubMed.
- R. Tiwari, P. A. Miller, S. Cho, S. G. Franzblau and M. J. Miller, ACS Med. Chem. Lett., 2015, 6, 128 CrossRef CAS PubMed.
- M. A. T. Blaskovich, J. Zuegg, A. G. Elliott and M. A. Cooper, ACS Infect. Dis., 2015, 1, 285 CrossRef CAS.
- M. A. Cooper, Nat. Rev. Drug Discovery, 2015, 14, 587 CrossRef CAS PubMed.
- A. K. Agarwal and C. W. G. Fishwick, Ann. N. Y. Acad. Sci., 2010, 1213, 20 CrossRef CAS PubMed.
- P. I. O'Daniel, Z. Peng, H. Pi, S. A. Testero, D. Ding, E. Spink, E. Leemans, M. A. Boudreau, T. Yamaguchi, V. A. Schroeder, W. R. Wolter, L. I. Llarrull, W. Song, E. Lastochkin, M. Kumarasiri, N. T. Antunes, M. Espahbodi, K. Lichtenwalter, M. A. Suckow, S. Vakulenko, S. Mobashery and M. Chang, J. Am. Chem. Soc., 2014, 136, 3664 CrossRef PubMed.
- E. Spink, D. Ding, Z. Peng, M. A. Boudreau, E. Leemans, E. Lastochkin, W. Song, K. Lichtenwalter, P. I. O'Daniel, S. A. Testero, H. Pi, V. A. Schroeder, W. R. Wolter, N. T. Antunes, M. A. Suckow, S. Vakulenko, M. Chang and S. Mobashery, J. Med. Chem., 2015, 58, 1380 CrossRef CAS PubMed.
- R. Bouley, M. Kumarasiri, Z. Peng, L. H. Otero, W. Song, M. A. Suckow, V. A. Schroeder, W. R. Wolter, E. Lastochkin, N. T. Antunes, H. Pi, S. Vakulenko, J. A. Hermoso, M. Chang and S. Mobashery, J. Am. Chem. Soc., 2015, 137, 1738 CrossRef CAS PubMed.
- F. Y. Chan, N. Sun, M. A. Neves, P. C. Lam, W. H. Chung, L. K. Wong, H. Y. Chow, D. L. Ma, P. H. Chan, Y. C. Leung, T. H. Chan, R. Abagyan and K. Y. Wong, J. Chem. Inf. Model., 2013, 53, 2131 CrossRef CAS PubMed.
- F. Y. Chan, N. Sun, Y. C. Leung and K. Y. Wong, J. Antibiot., 2015, 68, 253 CrossRef CAS PubMed.
- A. H. Chan, J. Wereszczynski, B. R. Amer, S. W. Yi, M. E. Jung, J. A. McCammon and R. T. Clubb, Chem. Biol. Drug Des., 2013, 82, 418 CAS.
- J. Zhang, H. Liu, K. Zhu, S. Gong, S. Dramsi, Y. T. Wang, J. Li, F. Chen, R. Zhang, L. Zhou, L. Lan, H. Jiang, O. Schneewind, C. Luo and C. G. Yang, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 13517 CrossRef CAS PubMed.
- N. A. Larsen, T. J. Nash, M. Morningstar, A. B. Shapiro, C. Joubran, C. J. Blackett, A. D. Patten, P. A. Boriack-Sjodin and P. Doig, Biochem. J., 2012, 446, 405 CrossRef CAS PubMed.
- P. Doig, P. A. Boriack-Sjodin, J. Dumas, J. Hu, K. Itoh, K. Johnson, S. Kazmirski, T. Kinoshita, S. Kuroda, T. Sato, K. Sugimoto, K. Tohyama, H. Aoi, K. Wakamatsu and H. Wang, Bioorg. Med. Chem., 2014, 22, 6256 CrossRef CAS PubMed.
- T. Tomasic, A. Kovac, G. Klebe, D. Blanot, S. Gobec, D. Kikelj and L. P. Masic, J. Mol. Model., 2012, 18, 1063 CrossRef CAS PubMed.
- M. Hrast, I. Sosič, R. Šink and S. Gobec, Bioorg. Chem., 2014, 55, 2 CrossRef CAS PubMed.
- A. Perdih, M. Hrast, H. Barreteau, S. Gobec, G. Wolber and T. Solmajer, J. Chem. Inf. Model., 2014, 54, 1451 CrossRef CAS PubMed.
- P. Wu, T. E. Nielsen and M. H. Clausen, Trends Pharmacol. Sci., 2015, 36, 422 CrossRef CAS PubMed.
- J. Clardy, M. A. Fischbach and C. T. Walsh, Nat. Biotechnol., 2006, 24, 1541 CrossRef CAS PubMed.
- A. L. Demain and S. Sanchez, J. Antibiot., 2009, 62, 5 CrossRef CAS PubMed.
- J. W.-H. Li and J. C. Vederas, Science, 2009, 325, 161 CrossRef PubMed.
- G. D. Wright, Can. J. Microbiol., 2014, 60, 147 CrossRef CAS PubMed.
- C. Corre and G. L. Challis, Nat. Prod. Rep., 2009, 26, 977 RSC.
- M. A. Fischbach, Curr. Opin. Microbiol., 2009, 12, 520 CrossRef CAS PubMed.
- C. T. Walsh and M. A. Fischbach, J. Am. Chem. Soc., 2010, 132, 2469 CrossRef CAS PubMed.
- C. G. Bologa, O. Ursu, T. I. Oprea, C. E. Melançon III and G. P. Tegos, Curr. Opin. Pharmacol., 2013, 13, 678 CrossRef CAS PubMed.
- H.-W. Liu and T. Begley, Curr. Opin. Chem. Biol., 2013, 17, 529 CrossRef CAS PubMed.
- M. A. Hayashi, F. C. Bizerra and P. I. Da Silva Jr., Front. Microbiol., 2013, 4, 195 Search PubMed.
- P. W. Taylor, Int. J. Antimicrob. Agents, 2013, 42, 195 CrossRef CAS PubMed.
- H. A. Kirst, Expert Opin. Drug Discovery, 2013, 8, 479 CrossRef CAS PubMed.
- Z. Charlop-Powers, J. G. Owen, B. V. Reddy, M. A. Ternei and S. F. Brady, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 3757 CrossRef CAS PubMed.
- Z. Charlop-Powers, A. Milshteyn and S. F. Brady, Curr. Opin. Microbiol., 2014, 19, 70 CrossRef CAS PubMed.
- M. R. Seyedsayamdost and J. Clardy, ACS Synth. Biol., 2014, 3, 745 CrossRef CAS PubMed.
- P. Stallforth and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 3655 CrossRef CAS PubMed.
- S. Antoraz, R. I. Santamaría, M. Díaz, D. Sanz and H. Rodríguez, Front. Microbiol., 2015, 6, 461 Search PubMed.
- E. Kim, B. S. Moore and Y. J. Yoon, Nat. Chem. Biol., 2015, 11, 649 CrossRef CAS PubMed.
- F. L. Cherblanc, R. W. M. Davidson, P. Di Fruscia, N. Srimongkolpithak and M. J. Fuchter, Nat. Prod. Rep., 2013, 30, 605 RSC.
- S. E. Unkles, V. Valiante, D. J. Mattern and A. A. Brakhage, Chem. Biol., 2014, 21, 502 CrossRef CAS PubMed.
- D. J. Mattern, V. Valiante, S. E. Unkles and A. A. Brakhage, Front. Microbiol., 2015, 6, 775 Search PubMed.
- X. M. Mao, W. Xu, D. Li, W. B. Yin, Y. H. Chooi, Y. Q. Li, Y. Tang and Y. Hu, Angew. Chem., Int. Ed., 2015, 54, 7592 CrossRef CAS PubMed.
- K. Hiramatsu, M. Igarashi, Y. Morimoto, T. Baba, M. Umekita and Y. Akamatsu, Int. J. Antimicrob. Agents, 2012, 39, 478 CrossRef CAS PubMed.
- E. I. Parkinson, J. S. Bair, B. A. Nakamura, H. Y. Lee, H. I. Kuttab, E. H. Southgate, S. Lezmi, G. W. Lau and P. J. Hergenrother, Nat. Commun., 2015, 6, 6947 CrossRef CAS PubMed.
- M. N. Thaker, W. Wang, P. Spanogiannopoulos, N. Waglechner, A. M. King, R. Medina and G. D. Wright, Nat. Biotechnol., 2013, 31, 922 CrossRef CAS PubMed.
- M. N. Thaker, N. Waglechner and G. D. Wright, Nat. Protoc., 2014, 9, 1469 CrossRef CAS PubMed.
- G. Yim, M. N. Thaker, K. Koteva and G. Wright, J. Antibiot., 2014, 67, 31 CrossRef CAS PubMed.
- G. Yim, L. Kalan, K. Koteva, M. N. Thaker, N. Waglechner, I. Tang and G. D. Wright, ChemBioChem, 2014, 15, 2613 CrossRef CAS PubMed.
- M. N. Thaker and G. D. Wright, ACS Synth. Biol., 2015, 4, 195 CrossRef CAS PubMed.
- M. S. Butler, K. A. Hansford, M. A. T. Blaskovich, R. Halai and M. A. Cooper, J. Antibiot., 2014, 67, 631 CrossRef CAS PubMed.
- M. F. Traxler, J. D. Watrous, T. Alexandrov, P. C. Dorrestein and R. Kolter, mBio, 2013, 4, e00459 CrossRef PubMed.
- D. A. Hopwood, mBio, 2013, 4, e00612 CrossRef PubMed.
- P. Charusanti, N. L. Fong, H. Nagarajan, A. R. Pereira, H. J. Li, E. A. Abate, Y. Su, W. H. Gerwick and B. O. Palsson, PLoS One, 2012, 7, e33727 CAS.
- M. F. Traxler and R. Kolter, Nat. Prod. Rep., 2015, 32, 956 RSC.
- B. Lowry, C. T. Walsh and C. Khosla, Synlett, 2015, 26, 1008 CrossRef CAS PubMed.
- C. T. Walsh, Nat. Chem. Biol., 2015, 11, 620 CrossRef CAS PubMed.
- P. J. Rutledge and G. L. Challis, Nat. Rev. Microbiol., 2015, 13, 509 CrossRef CAS PubMed.
- J. J. Banik and S. F. Brady, Curr. Opin. Microbiol., 2010, 13, 603 CrossRef CAS PubMed.
- Z. Feng, D. Chakraborty, S. B. Dewell, B. V. Reddy and S. F. Brady, J. Am. Chem. Soc., 2012, 134, 2981 CrossRef CAS PubMed.
- A. Milshteyn, J. S. Schneider and S. F. Brady, Chem. Biol., 2014, 21, 1211 CrossRef CAS PubMed.
- L. J. Cohen, H. S. Kang, J. Chu, Y. H. Huang, E. A. Gordon, B. V. Reddy, M. A. Ternei, J. W. Craig and S. F. Brady, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E4825 CrossRef CAS PubMed.
- S. Kapur, B. Lowry, S. Yuzawa, S. Kenthirapalan, A. Y. Chen, D. E. Cane and C. Khosla, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 4110 CrossRef CAS PubMed.
- C. Khosla, D. Herschlag, D. E. Cane and C. T. Walsh, Biochemistry, 2014, 53, 2875 CrossRef CAS PubMed.
- C. Lok, Nature, 2015, 522, 270 CrossRef CAS PubMed.
- F. von Nussbaum and R. D. Süssmuth, Angew. Chem., Int. Ed., 2015, 54, 6684 CrossRef CAS PubMed.
- P. Hunter, EMBO Rep., 2015, 16, 563 CrossRef CAS PubMed.
- L. J. V. Piddock, J. Antimicrob. Chemother., 2015, 70, 2679 CrossRef CAS PubMed.
- L. L. Ling, T. Schneider, A. J. Peoples, A. L. Spoering, I. Engels, B. P. Conlon, A. Mueller, T. F. Schaberle, D. E. Hughes, S. Epstein, M. Jones, L. Lazarides, V. A. Steadman, D. R. Cohen, C. R. Felix, K. A. Fetterman, W. P. Millett, A. G. Nitti, A. M. Zullo, C. Chen and K. Lewis, Nature, 2015, 517, 455 CrossRef CAS PubMed.
- R. Cain, S. Narramore, M. McPhillie, K. Simmons and C. W. G. Fishwick, Bioorg. Chem., 2014, 55, 69 CrossRef CAS PubMed.
- J. Hong, Chem. – Eur. J., 2014, 20, 10204 CrossRef CAS PubMed.
- A. Okano, A. Nakayama, A. W. Schammel and D. L. Boger, J. Am. Chem. Soc., 2014, 136, 13522 CrossRef CAS PubMed.
- A. Okano, A. Nakayama, K. Wu, E. A. Lindsey, A. W. Schammel, Y. Feng, K. C. Collins and D. L. Boger, J. Am. Chem. Soc., 2015, 137, 3693 CrossRef CAS PubMed.
- W. Zhou, E. W. Scocchera, D. L. Wright and A. C. Anderson, Med. Chem. Commun., 2013, 4, 908 RSC.
- K. M. Lamb, N. G-Dayanandan, D. L. Wright and A. C. Anderson, Biochemistry, 2013, 52, 7318 CrossRef CAS PubMed.
- K. M. Lamb, M. N. Lombardo, J. Alverson, N. D. Priestley, D. L. Wright and A. C. Anderson, Antimicrob. Agents Chemother., 2014, 58, 7484 CrossRef PubMed.
- S. Keshipeddy, S. M. Reeve, A. C. Anderson and D. L. Wright, J. Am. Chem. Soc., 2015, 137, 8983 CrossRef CAS PubMed.
- H. Lu and P. J. Tonge, Curr. Opin. Chem. Biol., 2010, 14, 467 CrossRef CAS PubMed.
- G. Dahl and T. Akerud, Drug Discovery Today, 2013, 18, 697 CrossRef PubMed.
- L. Liu, J. Med. Chem., 2014, 57, 2843 CrossRef PubMed.
- J. M. Bradshaw, J. M. McFarland, V. O. Paavilainen, A. Bisconte, D. Tam, V. T. Phan, S. Romanov, D. Finkle, J. Shu, V. Patel, T. Ton, X. Li, D. G. Loughhead, P. A. Nunn, D. E. Karr, M. E. Gerritsen, J. O. Funk, T. D. Owens, E. Verner, K. A. Brameld, R. J. Hill, D. M. Goldstein and J. Taunton, Nat. Chem. Biol., 2015, 11, 525 CrossRef CAS PubMed.
- R. A. Copeland, Nat. Chem. Biol., 2015, 11, 451 CrossRef CAS PubMed.
- K. P. Cusack, Y. Wang, M. Z. Hoemann, J. Marjanovic, R. G. Heym and A. Vasudevan, Bioorg. Med. Chem. Lett., 2015, 25, 2019 CrossRef CAS PubMed.
- R. Zhang, Nat. Chem. Biol., 2015, 11, 382 CrossRef CAS PubMed.
- C. J. Lee, X. Liang, X. Chen, D. Zeng, S. H. Joo, H. S. Chung, A. W. Barb, S. M. Swanson, R. A. Nicholas, Y. Li, E. J. Toone, C. R. H. Raetz and P. Zhou, Chem. Biol., 2011, 18, 38 CrossRef CAS PubMed.
- C. J. Lee, X. Liang, R. Gopalaswamy, J. Najeeb, E. D. Ark, E. J. Toone and P. Zhou, ACS Chem. Biol., 2014, 9, 237 CrossRef CAS PubMed.
- G. K. Walkup, Z. You, P. L. Ross, E. K. H. Allen, F. Daryaee, M. R. Hale, J. O'Donnell, D. E. Ehmann, V. J. A. Schuck, E. T. Buurman, A. L. Choy, L. Hajec, K. Murphy-Benenato, V. Marone, S. A. Patey, L. A. Grosser, M. Johnstone, S. G. Walker, P. J. Tonge and S. L. Fisher, Nat. Chem. Biol., 2015, 11, 416 CrossRef CAS PubMed.
- J. A. Perry, E. L. Westman and G. D. Wright, Curr. Opin. Microbiol., 2014, 21, 45 CrossRef CAS PubMed.
- J. M. A. Blair, M. A. Webber, A. J. Baylay, D. O. Ogbolu and L. J. V. Piddock, Nat. Rev. Microbiol., 2015, 13, 42 CrossRef CAS PubMed.
- M. J. Culyba, C. Y. Mo and R. M. Kohli, Biochemistry, 2015, 54, 3573 CrossRef CAS PubMed.
- G. D. Wright, Chem. Biol., 2012, 19, 3 CrossRef CAS PubMed.
- M. A. Farha and E. D. Brown, Ann. N. Y. Acad. Sci., 2015, 1354, 54 CrossRef PubMed.
- A. G. McArthur, N. Waglechner, F. Nizam, A. Yan, M. A. Azad, A. J. Baylay, K. Bhullar, M. J. Canova, G. De Pascale, L. Ejim, L. Kalan, A. M. King, K. Koteva, M. Morar, M. R. Mulvey, J. S. O'Brien, A. C. Pawlowski, L. J. V. Piddock, P. Spanogiannopoulos, A. D. Sutherland, I. Tang, P. L. Taylor, M. Thaker, W. Wang, M. Yan, T. Yu and G. D. Wright, Antimicrob. Agents Chemother., 2013, 57, 3348 CrossRef CAS PubMed.
- A. G. McArthur and G. D. Wright, Curr. Opin. Microbiol., 2015, 27, 45 CrossRef PubMed.
- P. Nonejuie, M. Burkart, K. Pogliano and J. Pogliano, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 16169 CrossRef CAS PubMed.
- W. F. Penwell, A. B. Shapiro, R. A. Giacobbe, R.-F. Gu, N. Gao, J. Thresher, R. E. McLaughlin, M. D. Huband, B. L. M. DeJonge, D. E. Ehmann and A. A. Miller, Antimicrob. Agents Chemother., 2015, 59, 1680 CrossRef PubMed.
- O. Kocaoglu and E. E. Carlson, Antimicrob. Agents Chemother., 2015, 59, 2785 CrossRef CAS PubMed.
- O. Kocaoglu, H.-C. T. Tsui, M. E. Winkler and E. E. Carlson, Antimicrob. Agents Chemother., 2015, 59, 3548 CrossRef CAS PubMed.
- Z. Yao, D. Kahne and R. Kishony, Mol. Cell, 2012, 48, 705 CrossRef CAS PubMed.
- H. Cho, T. Uehara and T. G. Bernhardt, Cell, 2014, 159, 1300 CrossRef CAS PubMed.
- T. S. B. Kjeldsen, M. O. A. Sommer and J. E. Olsen, BMC Microbiol., 2015, 15, 63 CrossRef PubMed.
- J. Davies and D. Davies, Microbiol. Mol. Biol. Rev., 2010, 74, 417 CrossRef CAS PubMed.
- J. Davies, Curr. Opin. Chem. Biol., 2011, 15, 5 CrossRef CAS PubMed.
- J. Davies and K. S. Ryan, ACS Chem. Biol., 2012, 7, 252 CrossRef CAS PubMed.
- J. Davies, J. Antibiot., 2013, 66, 361 CrossRef CAS PubMed.
- J. Davies, in Handbook of Antimicrobial Resistance, ed. M. Götte, A. Berghuis, G. Matlashewski, M. Wainberg and D. Sheppard, Springer Science + Business Media, New York, 2014, ch. 1 Search PubMed.
- M. A. Farha, T. L. Czarny, C. L. Myers, L. J. Worrall, S. French, D. G. Conrady, Y. Wang, E. Oldfield, N. C. Strynadka and E. D. Brown, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 11048–11053 CrossRef CAS PubMed.
- D. M. Cornforth and K. R. Foster, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 10827 CrossRef CAS PubMed.
- D. I. Andersson and D. Hughes, Nat. Rev. Microbiol., 2014, 12, 465 CrossRef CAS PubMed.
- F. Baquero and T. M. Coque, mBio, 2014, 5, e02270 CrossRef PubMed.
- T. G. Platt and C. Fuqua, Trends Microbiol., 2010, 18, 383 CrossRef CAS PubMed.
- B. LaSarre and M. J. Federle, Microbiol. Mol. Biol. Rev., 2013, 77, 73 CrossRef CAS PubMed.
- J. P. Gerdt and H. E. Blackwell, ACS Chem. Biol., 2014, 9, 2291 CrossRef CAS PubMed.
- G. Rampioni, L. Leoni and P. Williams, Bioorg. Chem., 2014, 55, 60 CrossRef CAS PubMed.
- M. S. Blackledge, R. J. Worthington and C. Melander, Curr. Opin. Pharmacol., 2013, 13, 699 CrossRef CAS PubMed.
- C. Beloin, S. Renard, J.-M. Ghigo and D. Lebeaux, Curr. Opin. Pharmacol., 2014, 18, 61 CrossRef CAS PubMed.
- R. J. Worthington and C. Melander, Angew. Chem., Int. Ed., 2012, 51, 6314 CrossRef CAS PubMed.
- A. R. Stacy, S. P. Diggle and M. Whiteley, Curr. Opin. Microbiol., 2012, 15, 155 CrossRef PubMed.
- S. Yajima, Tetrahedron Lett., 2014, 55, 2773 CrossRef.
- H. R. Meredith, J. K. Srimani, A. J. Lee, A. J. Lopatkin and L. You, Nat. Chem. Biol., 2015, 11, 182 CrossRef CAS PubMed.
- K. Lewis, Annu. Rev. Microbiol., 2010, 64, 357 CrossRef CAS PubMed.
- N. M. Vega, K. R. Allison, A. S. Khalil and J. J. Collins, Nat. Chem. Biol., 2012, 8, 431 CrossRef CAS PubMed.
- N. Q. Balaban, K. Gerdes, K. Lewis and J. D. McKinney, Nat. Rev. Microbiol., 2013, 11, 587 CrossRef CAS PubMed.
- N. R. Cohen, M. A. Lobritz and J. J. Collins, Cell Host Microbe, 2013, 13, 632 CAS.
- S. Helaine and E. Kugelberg, Trends Microbiol., 2014, 22, 417 CrossRef CAS PubMed.
- E. Maisonneuve and K. Gerdes, Cell, 2014, 157, 539 CrossRef CAS PubMed.
- B. R. Levin, J. Concepción-Acevedo and K. I. Udekwu, Curr. Opin. Microbiol., 2014, 21, 18 CrossRef CAS PubMed.
- M. Ayrapetyan, T. C. Williams and J. D. Oliver, Trends Microbiol., 2015, 23, 7 CrossRef CAS PubMed.
- D. W. Holden, Science, 2015, 347, 30 CrossRef CAS PubMed.
- M. O. A. Sommer and G. Dantas, Curr. Opin. Microbiol., 2011, 14, 556 CrossRef CAS PubMed.
- D. J. Triggle, Biochem. Pharmacol., 2012, 84, 1543 CrossRef CAS PubMed.
- M. A. Riley, S. M. Robinson, C. M. Roy and R. L. Dorit, Future Med. Chem., 2013, 5, 1231 CrossRef CAS PubMed.
- M. K. Waldor, G. Tyson, E. Borenstein, H. Ochman, A. Moeller, B. B. Finlay, H. H. Kong, J. I. Gordon, K. E. Nelson, K. Dabbagh and H. Smith, PLoS Biol., 2015, 13, e1002050 CrossRef PubMed.
- E. Gullberg, S. Cao, O. G. Berg, C. Ilback, L. Sandegren, D. Hughes and D. I. Andersson, PLoS Pathog., 2011, 7, e1002158 CAS.
- J. Blázquez, A. Couce, J. Rodriguez-Beltrán and A. Rodriguez-Rojas, Curr. Opin. Microbiol., 2012, 15, 561 CrossRef PubMed.
- E. Gullberg, L. M. Albrecht, C. Karlsson, L. Sandegren and D. I. Andersson, mBio, 2014, 5, e01918 CrossRef CAS PubMed.
- K. J. Forsberg, S. Patel, M. K. Gibson, C. L. Lauber, R. Knight, N. Fierer and G. Dantas, Nature, 2014, 509, 612 CrossRef CAS PubMed.
- J. A. Perry and G. D. Wright, BioEssays, 2014, 36, 1179 CrossRef PubMed.
- M. O. A. Sommer, Nature, 2014, 509, 567 CrossRef CAS PubMed.
- M. K. Gibson, K. J. Forsberg and G. Dantas, ISME J., 2015, 9, 207 CrossRef CAS PubMed.
- J. L. Martínez, T. M. Coque and F. Baquero, Nat. Rev. Microbiol., 2015, 13, 116 CrossRef PubMed.
- J. L. Martínez, T. M. Coque and F. Baquero, Nat. Rev. Microbiol., 2015, 13, 396 CrossRef PubMed.
- J. Bengtsson-Palme and D. G. J. Larsson, Nat. Rev. Microbiol., 2015, 13, 396 CrossRef CAS PubMed.
- L. H. Otero, A. Rojas-Altuve, L. I. Llarrull, C. Carrasco-López, M. Kumarasiri, E. Lastochkin, J. Fishovitz, M. Dawley, D. Hesek, M. Lee, J. W. Johnson, J. F. Fisher, M. Chang, S. Mobashery and J. A. Hermoso, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 16808 CrossRef PubMed.
- J. Fishovitz, A. Rojas-Altuve, L. H. Otero, M. Dawley, C. Carrasco-Lopez, M. Chang, J. A. Hermoso and S. Mobashery, J. Am. Chem. Soc., 2014, 136, 9814 CrossRef CAS PubMed.
- J. Fishovitz, N. Taghizadeh, J. F. Fisher, M. Chang and S. Mobashery, J. Am. Chem. Soc., 2015, 137, 6500 CrossRef CAS PubMed.
- K. J. Forsberg, S. Patel, T. A. Wencewicz and G. Dantas, Chem. Biol., 2015, 22, 888 CrossRef CAS PubMed.
- D. W. Graham, Chem. Biol., 2015, 22, 805 CrossRef CAS PubMed.
- M. Ackermann, Nat. Rev. Microbiol., 2015, 13, 497 CrossRef CAS PubMed.
- A. Persat, C. D. Nadell, M. K. Kim, F. Ingremeau, A. Siryaporn, K. Drescher, N. S. Wingreen, B. L. Bassler, Z. Gitai and H. A. Stone, Cell, 2015, 161, 988 CrossRef CAS PubMed.
- M. Ackermann and F. Schreiber, Environ. Microbiol., 2015, 17, 2193 CrossRef PubMed.
- Y. Wang, M. Ran, J. Wang, Q. Ouyang and C. Luo, PLoS One, 2015, 10, e0127115 Search PubMed.
- T. Artemova, Y. Gerardin, C. Dudley, N. M. Vega and J. Gore, Mol. Syst. Biol., 2015, 11, 822 CrossRef PubMed.
- K. R. Allison, M. P. Brynildsen and J. J. Collins, Nature, 2011, 473, 216 CrossRef CAS PubMed.
- D. J. Dwyer, P. A. Belenky, J. H. Yang, I. C. MacDonald, J. D. Martell, N. Takahashi, C. T. Y. Chan, M. A. Lobritz, D. Braff, E. G. Schwarz, J. D. Ye, M. Pati, M. Vercruysse, P. S. Ralifo, K. R. Allison, A. S. Khalil, A. Y. Ting, G. C. Walker and J. J. Collins, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, E2100 CrossRef CAS PubMed.
- B. Peng, Y. B. Su, H. Li, Y. Han, C. Guo, Y. M. Tian and X. X. Peng, Cell Metab., 2015, 21, 249 CrossRef CAS PubMed.
- P. Bhargava and J. J. Collins, Cell Metab., 2015, 21, 154 CrossRef CAS PubMed.
- Y. Su, B. Peng, Y. Han, H. Li and X. Peng, J. Proteome Res., 2015, 14, 1612 CrossRef CAS PubMed.
- D. J. Dwyer, J. J. Collins and G. C. Walker, Annu. Rev. Pharmacol. Toxicol., 2015, 55, 313 CrossRef CAS PubMed.
- A. C. Palmer and R. Kishony, Nat. Commun., 2014, 5, 4296 CAS.
- P. Greulich, M. Scott, M. R. Evans and R. J. Allen, Mol. Syst. Biol., 2015, 11, 796 CrossRef PubMed.
- F. C. Fang, Nat. Biotechnol., 2013, 31, 415 CrossRef CAS PubMed.
- M. P. Brynildsen, J. A. Winkler, C. S. Spina, I. C. Macdonald and J. J. Collins, Nat. Biotechnol., 2013, 31, 160 CrossRef CAS PubMed.
- M. Mosel, L. Li, K. Drlica and X. Zhao, Antimicrob. Agents Chemother., 2013, 57, 5755 CrossRef CAS PubMed.
- M. A. Lobritz, P. Belenky, C. B. M. Porter, A. Gutierrez, J. H. Yang, E. G. Schwarz, D. J. Dwyer, A. S. Khalil and J. J. Collins, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 8173 CrossRef CAS PubMed.
- X. Zhao and K. Drlica, Curr. Opin. Microbiol., 2014, 21, 1 CrossRef CAS PubMed.
- X. Zhao, Y. Hong and K. Drlica, J. Antimicrob. Chemother., 2015, 70, 639 CrossRef CAS PubMed.
- I. Keren, Y. Wu, J. Inocencio, L. R. Mulcahy and K. Lewis, Science, 2013, 339, 1213 CrossRef CAS PubMed.
- Y. Liu and J. A. Imlay, Science, 2013, 339, 1210 CrossRef CAS PubMed.
- N. Kuhnert, Angew. Chem., Int. Ed., 2013, 52, 10946 CrossRef CAS PubMed.
- H. Hamamoto, M. Urai, K. Ishii, J. Yasukawa, A. Paudel, M. Murai, T. Kaji, T. Kuranaga, K. Hamase, T. Katsu, J. Su, T. Adachi, R. Uchida, H. Tomoda, M. Yamada, M. Souma, H. Kurihara, M. Inoue and K. Sekimizu, Nat. Chem. Biol., 2015, 11, 127 CrossRef CAS PubMed.
- F. Beaufay, J. Coppine, A. Mayard, G. Laloux, X. De Bolle and R. Hallez, EMBO J., 2015, 34, 1786 CrossRef CAS PubMed.
- T. Roemer, J. Davies, G. Giaever and C. Nislow, Nat. Chem. Biol., 2012, 8, 46 CrossRef CAS PubMed.
- L. Imamovic and M. O. A. Sommer, Sci. Transl. Med., 2013, 5, 204ra132 Search PubMed.
- V. Lázár, G. Pal Singh, R. Spohn, I. Nagy, B. Horváth, M. Hrtyan, R. Busa-Fekete, B. Bogos, O. Méhi, B. Csörgő, G. Pósfai, G. Fekete, B. Szappanos, B. Kégl, B. Papp and C. Pál, Mol. Syst. Biol., 2013, 9, 700 CrossRef PubMed.
- T. Roemer and C. Boone, Nat. Chem. Biol., 2013, 9, 222 CrossRef CAS PubMed.
- C. Munck, H. K. Gumpert, A. I. N. Wallin, H. H. Wang and M. O. A. Sommer, Sci. Transl. Med., 2014, 6, 262ra156 CrossRef PubMed.
- T. Bollenbach, Curr. Opin. Microbiol., 2015, 27, 1 CrossRef CAS PubMed.
- G. Chevereau and T. Bollenbach, Mol. Syst. Biol., 2015, 11, 807 CrossRef PubMed.
- E. E. Gill, O. L. Franco and R. E. W. Hancock, Chem. Biol. Drug Des., 2015, 85, 56 CAS.
- C. Pál, B. Papp and V. Lázár, Trends Microbiol., 2015, 23, 401 CrossRef PubMed.
- K. Pettus, S. Sharpe and J. R. Papp, Antimicrob. Agents Chemother., 2015, 59, 2443 CrossRef CAS PubMed.
- M. Rodriguez de Evgrafov, H. Gumpert, C. Munck, T. T. Thomsen and M. O. A. Sommer, Mol. Biol. Evol., 2015, 32, 1175 CrossRef PubMed.
- C. M. Tan, A. G. Therien, J. Lu, S. H. Lee, A. Caron, C. J. Gill, C. Lebeau-Jacob, L. Benton-Perdomo, J. M. Monteiro, P. M. Pereira, N. L. Elsen, J. Wu, K. Deschamps, M. Petcu, S. Wong, E. Daigneault, S. Kramer, L. Liang, E. Maxwell, D. Claveau, J. Vaillancourt, K. Skorey, J. Tam, H. Wang, T. C. Meredith, S. Sillaots, L. Wang-Jarantow, Y. Ramtohul, E. Langlois, F. Landry, J. C. Reid, G. Parthasarathy, S. Sharma, A. Baryshnikova, K. J. Lumb, M. G. Pinho, S. M. Soisson and T. Roemer, Sci. Transl. Med., 2012, 4, 126ra35 Search PubMed.
- T. Roemer, T. Schneider and M. G. Pinho, Curr. Opin. Microbiol., 2013, 16, 538 CrossRef CAS PubMed.
- P. A. Mann, A. Muller, L. Xiao, P. M. Pereira, C. Yang, S. H. Lee, H. Wang, J. Trzeciak, J. Schneeweis, M. M. Dos Santos, N. Murgolo, X. She, C. Gill, C. J. Balibar, M. Labroli, J. Su, A. Flattery, B. Sherborne, R. Maier, C. M. Tan, T. Black, K. Onder, S. Kargman, F. J. Monsma, M. G. Pinho, T. Schneider and T. Roemer, ACS Chem. Biol., 2013, 8, 2442 CrossRef CAS PubMed.
- J. Campbell, A. K. Singh, J. G. Swoboda, M. S. Gilmore, B. J. Wilkinson and S. Walker, Antimicrob. Agents Chemother., 2012, 56, 1810 CrossRef CAS PubMed.
- S. Brown, J. P. Santa Maria Jr. and S. Walker, Annu. Rev. Microbiol., 2013, 67, 313 CrossRef CAS PubMed.
- H. Wang, C. J. Gill, S. H. Lee, P. Mann, P. Zuck, T. C. Meredith, N. Murgolo, X. She, S. Kales, L. Liang, J. Liu, J. Wu, J. Santa Maria, J. Su, J. Pan, J. Hailey, D. Mcguinness, C. M. Tan, A. Flattery, S. Walker, T. Black and T. Roemer, Chem. Biol., 2013, 20, 272 CrossRef CAS PubMed.
- M. A. Farha, K. Koteva, R. T. Gale, E. W. Sewell, G. D. Wright and E. D. Brown, Bioorg. Med. Chem. Lett., 2014, 24, 905 CrossRef CAS PubMed.
- J. P. Santa Maria Jr., A. Sadaka, S. H. Moussa, S. Brown, Y. J. Zhang, E. J. Rubin, M. S. Gilmore and S. Walker, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 12510 CrossRef PubMed.
- E. W. C. Sewell and E. D. Brown, J. Antibiot., 2014, 67, 43 CrossRef CAS PubMed.
- T. Kohler, C. Weidenmaier and A. Peschel, J. Bacteriol., 2009, 191, 4482 CrossRef CAS PubMed.
- V. Winstel, P. Kühner, F. Salomon, J. Larsen, R. Skov, W. Hoffmann, A. Peschel and C. Weidenmaier, mBio, 2015, 6, e00632 CrossRef PubMed.
- H. Barreteau, S. Magnet, M. El Ghachi, T. Touzé, M. Arthur, D. Mengin-Lecreulx and D. Blanot, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2009, 877, 213 CrossRef CAS PubMed.
- J. W. Johnson, J. F. Fisher and S. Mobashery, Ann. N. Y. Acad. Sci., 2013, 1277, 54 CrossRef CAS PubMed.
- A. W. Kingston, H. Zhao, G. M. Cook and J. D. Helmann, Mol. Microbiol., 2014, 93, 37 CrossRef CAS PubMed.
- G. Manat, S. Roure, R. Auger, A. Bouhss, H. Barreteau, D. Mengin-Lecreulx and T. Touzé, Microb. Drug Resist., 2014, 20, 199 CrossRef CAS PubMed.
- T. Day, S. Huijben and A. F. Read, Trends Microbiol., 2015, 23, 126 CrossRef CAS PubMed.
- A. C. Palmer and R. Kishony, Nat. Rev. Genet., 2013, 14, 243 CrossRef CAS PubMed.
- Y. H. Lee and J. D. Helmann, Antimicrob. Agents Chemother., 2013, 57, 4267 CrossRef CAS PubMed.
- D. N. Gilbert, R. J. Guidos, H. W. Boucher, G. H. Talbot, B. Spellberg, J. E. Edwards, W. M. Scheld, J. S. Bradley and J. G. Bartlett, Clin. Infect. Dis., 2010, 50, 1081 CrossRef PubMed.
- H. W. Boucher, G. H. Talbot, D. K. J. Benjamin, J. Bradley, R. J. Guidos, R. N. Jones, B. E. Murray, R. A. Bonomo and D. Gilbert, Clin. Infect. Dis., 2013, 56, 1685 CrossRef PubMed.
- M. G. P. Page and K. Bush, Curr. Opin. Pharmacol., 2014, 18, 91 CrossRef CAS PubMed.
- P. N. Harris, P. A. Tambyah and D. L. Paterson, Lancet Infect. Dis., 2015, 15, 475 CrossRef CAS PubMed.
- D. T. King, A. M. King, S. M. Lal, G. D. Wright and N. C. J. Strynadka, ACS Infect. Dis., 2015, 1, 175 CrossRef CAS.
- J. H. Lee, J. J. Lee, K. S. Park and S. H. Lee, Lancet Infect. Dis., 2015, 15, 876 CrossRef PubMed.
- J. L. Liscio, M. V. Mahoney and E. B. Hirsch, Int. J. Antimicrob. Agents, 2015, 46, 266 CrossRef CAS PubMed.
- C. Pitart, F. Marco, T. A. Keating, W. W. Nichols and J. Vila, Antimicrob. Agents Chemother., 2015, 59, 3059 CrossRef CAS PubMed.
- S. Shiber, D. Yahav, T. Avni, L. Leibovici and M. Paul, J. Antimicrob. Chemother., 2015, 70, 41 CrossRef CAS PubMed.
- M. L. Winkler, K. M. Papp-Wallace and R. A. Bonomo, J. Antimicrob. Chemother., 2015, 70, 2279 CrossRef CAS PubMed.
- M. L. Winkler, K. M. Papp-Wallace, M. A. Taracila and R. A. Bonomo, Antimicrob. Agents Chemother., 2015, 59, 3700 CrossRef CAS PubMed.
- J. Mann, P. W. Taylor, C. R. Dorgan, P. D. Johnson, F. X. Wilson, R. Vickers, A. G. Dale and S. Neidle, Med. Chem. Commun., 2015, 6, 1420 RSC.
- R. Vickers, N. Robinson, E. Best, R. Echols, G. Tillotson and M. Wilcox, BMC Infect. Dis., 2015, 15, 91 CrossRef PubMed.
- C. Nathan, Nat. Rev. Microbiol., 2015, 13, 651 CrossRef CAS PubMed.
- K. Servick, Science, 2015, 348, 850 CrossRef CAS PubMed.
- J. F. Fisher, S. O. Meroueh and S. Mobashery, Chem. Rev., 2005, 105, 395 CrossRef CAS PubMed.
- C. Pulcini, K. Bush, W. A. Craig, N. Frimodt-Moller, M. L. Grayson, J. W. Mouton, J. Turnidge, S. Harbarth and I. C. Gyssens, Clin. Infect. Dis., 2012, 54, 268 CrossRef PubMed.
- U. Theuretzbacher, F. Van Bambeke, R. Cantón, C. G. Giske, J. W. Mouton, R. L. Nation, M. Paul, J. D. Turnidge and G. Kahlmeter, J. Antimicrob. Chemother., 2015, 70, 2177 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2016 |
