Patterns of biofilm structure and formation kinetics among Acinetobacter baumannii clinical isolates with different antibiotic resistance profiles†
E.
Dahdouh
*a,
B.
Orgaz
b,
R.
Gómez-Gil
c,
J.
Mingorance
c,
Z.
Daoud
d,
M.
Suarez
a and
C.
San Jose
b
aFaculty of Veterinary, Department of Animal Health, University Complutense of Madrid, Madrid, Spain. E-mail: elie_dahdouh@hotmail.com
bFaculty of Veterinary, Department of Food Science and Technology, University Complutense of Madrid, Madrid, Spain
cServicio de Microbiología, Hospital Universitario La Paz, IdiPAZ, Madrid, Spain
dFaculty of Medicine and Medical Sciences, Department of Clinical Microbiology, University of Balamand, Amioun, Lebanon
First published on 13th October 2015
Abstract
Acinetobacter baumannii is a ubiquitous organism that has been involved in a wide range of nosocomial infections. Its ability to produce biofilms, among other characteristics, allows it to persist in hospitals for prolonged periods. In this study, in order to check the possible relationship between its resistance to different antibiotics and its ability to form biofilms on inert surfaces, the rate of biofilm formation as well as siderophore production and detection of OmpA and CsuE by PCR were investigated for 12 A. baumannii clinical isolates. The biofilms were cultured at 37 °C on steel coupons immersed in BHI broth and the attached viable cells were counted after 5, 24 and 48 h. Confocal Laser Scanning Microscopy (CLSM) images were obtained for some of the strains that were noted to produce a brown pigment. The biofilm volume and substratum coverage were estimated with an image analysis software program. Our data, though preliminary, show that the quicker biofilm formers were strains susceptible to aminoglycosides, whereas the biofilms providing thicker and more uniform surface coverage were produced by carbapenem-resistant strains, producing a brown pigment with a plausible siderophore role. Further investigation into a wider set of isolates could help better understand the relationship between biofilm formation and various clinical findings.
Introduction
Acinetobacter spp. are Gram-negative bacteria that have diverse roles ranging from bioremediation agents and oil extraction aids to antibiotic-resistant nosocomial pathogens. They possess an outstanding chemical ability to degrade xenobiotic compounds, from alkanes to herbicides and even pharmaceuticals.1,2A. baumannii is known to easily acquire resistance to various antimicrobials, thanks to its wide array of natural resistance mechanisms, such as the production of efflux pumps, down-regulation of porins, production of degrading enzymes and/or modification of the target site of antibiotics.3,4 These resistance mechanisms have converted this ubiquitous microorganism into one of the most concerning and dangerous nosocomial pathogens, contributing to ventilator-associated pneumonia, as well as bloodstream-, burn-, wound- and catheter-related infections. Very frequent antibiotic resistances are found in clinical isolates and mortality rates as high as 20–60% have been registered for associated infection cases.5–7The ability of the clinical isolates of A. baumannii to produce biofilms has often been reported8–10 as well as their involvement in a wide range of genetic expression changes. Regulation of the formation mechanisms, including the quorum sensing ones, is progressively being characterized.11–13 Biofilm formation is considered to provide bacteria with protection against many hazards, ranging from antimicrobial agents to macrophage attacks as well as stress conditions such as desiccation. They assist in the development of antibiotic recalcitrance by different mechanisms.7,14–16 They differ according to the antibiotic, nature and history of the bacteria forming the biofilm and various environmental factors. Biofilms not only provide diffusion limitations due to the charged macromolecular mesh of the matrix, but also act as a protective barrier for embedded cells. Sub-lethal exposure to antimicrobial agents is prone to induce resistance responses such as the production of degradation enzymes. In addition, dissemination of resistance plasmids is favored by the high rate of gene exchange operating at high cell densities. Moreover, slowly dividing cells at deep biofilm layers are scarcely susceptible targets for agents that hinder cell division. Besides, the high cell densities in biofilms give rise, under antimicrobial exposure, to very small subpopulations of persister cells that survive antimicrobial exposure and will eventually repopulate the site.
In short, antibiotic resistance and biofilm formation depend both on genetic diversity and genetic expression. Understanding the connections among these phenomena has, as Badmasti et al.17 recently pointed out, a threading potential to understand A. baumannii persistence in the hospital environment and its colonization of medical equipment. In the present study, 12 clinical isolates from a Spanish hospital have been compared in terms of the kinetics of biofilm formation and the structures formed on abiotic surfaces. The association between these features, antibiotic resistance profiles, the presence of CsuE and OmpA genes, and siderophore and pigment production has also been explored. The association between clinical findings and the ability to attach and form biofilms on abiotic surfaces is important from an infection control perspective where special care could be taken for strains with certain clinical characteristics so as to avoid the colonization of abiotic surfaces and the persistence in the hospital of these isolates for prolonged periods of time.
Results and discussion
Details of the materials and methods used in this study are available in the ESI.†Antibiotic susceptibility and characteristics of A. baumannii clinical strains
The twelve isolates studied here are clinical bloodstream isolates and were deliberately chosen for their heterogeneity in antibiotic susceptibility and clinical history. Their origin, according to the patient's unit assignment and/or treatment, is shown in Table 1. Five were isolated from the general Intensive Care Unit (ICU) and four from the Burn Unit. Strain 30 was obtained from the Hematology/Oncology Unit and strain 59 came from the Internal Medicine Unit. Strain 20 also came from the Internal Medicine Unit, but from a patient not previously admitted to the ICU. Two thirds of the isolates were sampled from patients previously exposed to mechanical ventilation and catheterization, both of which are regarded as risk factors for biofilm formation and subsequent A. baumannii infection.6 75% of the source patients, except those infected by isolates 9, 20, and 38, had undergone antibiotic treatment in the 15 days prior to strain isolation.| Strain | Risk factors | Previous antimicrobial treatment | Disease | Primary infection | Patient outcome |
|---|---|---|---|---|---|
| 3 | MV | Carbapenems | 2nd degree burns on 70% of the body surface | Unknown | Recovered |
| CVC | |||||
| UC | |||||
| 9 | None | None | Rapidly progressive glomerulonephritis | Urinary tract infection | Recovered |
| 12 | MV | Carbapenems | Flame burn on 26% of the body surface | Respiratory infection | Died |
| CVC | Colistin | ||||
| UC | Linezolid | ||||
| 15 | MV | Carbapenems | Amputation of left leg | Respiratory infection and soft tissue infection | Recovered |
| CVC | Colistin | ||||
| UC | Linezolid | ||||
| 20 | None | None | Multifactorial chronic anemia | Soft tissue infection | Recovered |
| Septic shock | |||||
| 26 | MV | Carbapenems | Bilateral eosinophilic | Respiratory infection | Recovered |
| CVC | Linezolid | Pneumonia | |||
| 30 | None | Carbapenems | Severe combined immunodeficiency | Unknown | Recovered |
| Vancomycin | Bone marrow transplant | ||||
| 35 | MV | Carbapenem | Flame burn on 70% of the body surface | Respiratory infection | Died |
| CVC | Colistin | ||||
| UC | Linezolid | ||||
| 38 | MV | None | Intracranial hematoma | Respiratory infection | Recovered |
| CVC | |||||
| UC | |||||
| 45 | MV | Carbapenems | Non-Hodgkin lymphoma | Respiratory infection | Died |
| CVC | Colistin | Allogeneic transplant | |||
| UC | Tigecycline | ||||
| 52 | MV | Carbapenems | Acute lymphoblastic leukemia | Respiratory infection | Died |
| CVC | Colistin | Bone marrow transplant | |||
| UC | Vancomycin | ||||
| 59 | None | Piperacillin | Crohn's disease | Unknown | Recovered |
| Tazobactam |
According to the AST criteria (Table 2), the only entirely antibiotic-susceptible strains were strains 38 and 59. The rest were resistant to β-lactams and quinolones. This was not surprising since A. baumannii is known to have a wide array of intrinsic resistance mechanisms (causing resistance to first and second generation cephalosporins and most third generation cephalosporins) and an outstanding ability to acquire resistance from the environment.5
| Strain | Antimicrobial Agents | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-Lactams | Polymyxin | Aminoglycosides | Tetracycline | Fluoroquinolones | Folic acid synthesis inhibitors | ||||||||||||
| TIC | PIP | A/S | P/T | CTZ | CFP | IMI | MER | COL | G | TO | AK | MIN | CIP | LEV | T/S | ||
| 3 | MIC | >64 | >64 | 16/8 | >64/4 | >32 | 16 | >8 | >8 | ≤0.5 | ≤1 | ≤1 | ≤2 | 2 | >2 | >4 | >4/76 |
| S-I-R | R | R | I | R | R | I | R | R | S | S | S | S | S | R | R | R | |
| 9 | MIC | >64 | >64 | 16/8 | >64/4 | >32 | 32 | >8 | >8 | ≤0.5 | >8 | >8 | >32 | 8 | >2 | >4 | >4/76 |
| S-I-R | R | R | I | R | R | R | R | R | S | R | R | R | I | R | R | R | |
| 12 | MIC | 64 | >64 | 4/2 | >64/4 | 16 | 32 | >8 | >8 | 16 | >8 | >8 | 8 | ≤1 | >2 | >4 | >4/76 |
| S-I-R | I | R | S | R | I | R | R | R | R | R | R | S | S | R | R | R | |
| 15 | MIC | >64 | >64 | 16/8 | >64/4 | >32 | >32 | >8 | >8 | ≤0.5 | >8 | >8 | 16 | ≤1 | >2 | >4 | >4/76 |
| S-I-R | R | R | I | R | R | R | R | R | S | R | R | S | S | R | R | R | |
| 20 | MIC | >64 | >64 | 16/8 | >64/4 | >32 | >32 | >8 | >8 | ≤0.5 | >8 | >8 | >32 | 8 | >2 | >4 | >4/76 |
| S-I-R | R | R | I | R | R | R | R | R | S | R | R | R | I | R | R | R | |
| 26 | MIC | 32 | >64 | 4/2 | >64/4 | >32 | 16 | ≤1 | 1 | ≤0.5 | ≤1 | ≤1 | ≤2 | 2 | >2 | >4 | >4/76 |
| S-I-R | I | R | S | R | R | I | S | S | S | S | S | S | S | R | R | R | |
| 30 | MIC | >64 | >64 | 4 | 64/4 | 16 | 16 | 2 | 2 | ≤0.5 | >8 | 8 | 4 | ≤1 | >2 | >4 | >4/76 |
| S-I-R | R | R | S | I | I | I | S | S | S | R | I | S | S | R | R | R | |
| 35 | MIC | >64 | >64 | >16/8 | >64/4 | >16 | 8 | >8 | 8 | ≤0.5 | >8 | >8 | ≤2 | ≤1 | >2 | >4 | >4/76 |
| S-I-R | R | R | R | R | R | I | R | I | S | R | R | S | S | R | R | R | |
| 38 | MIC | ≤8 | ≤8 | ≤2/1 | ≤4/2 | 4 | 2 | ≤1 | ≤0.25 | ≤0.5 | ≤1 | >8 | ≤2 | ≤1 | ≤0.25 | ≤0.12 | ≤2/38 |
| S-I-R | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| 45 | MIC | >64 | >64 | >16 | >64/4 | >32 | 8 | >8 | 8 | 8 | >8 | 8 | ≤2 | 2 | >2 | >4 | >4/76 |
| S-I-R | R | R | R | R | R | I | R | I | R | R | I | S | S | R | R | R | |
| 52 | MIC | >64 | >64 | >16/8 | >64/4 | >32 | >32 | >8 | >8 | ≤0.5 | 4 | ≤1 | ≤2 | ≤1 | >2 | >4 | >4/76 |
| S-I-R | R | R | I | R | R | R | R | R | S | S | S | S | S | R | R | R | |
| 59 | MIC | ≤8 | ≤8 | ≤2/1 | ≤4/2 | 2 | 2 | ≤1 | 1 | ≤0.5 | ≤1 | ≤1 | ≤2 | ≤1 | ≤0.25 | ≤0.12 | ≤2/38 |
| S-I-R | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
Our data show that eight of the twelve strains were resistant to carbapenems. Protection mechanisms against these agents in A. baumannii strains are a global concern, since they tend to be the most commonly used ones to deal with recalcitrant strains. Carbapenem resistance has been also associated with increased mortality rates among infected individuals.18 Strains 12 and 45 were also resistant to colistin. The use of this antibiotic has re-emerged in the clinical setting as an alternative to deal with carbapenem-resistant A. baumannii isolates.19 Resistance to both carbapenems and colistin leaves the clinician with very few options for therapy and may result in treatment failure. Moreover, previous patient treatments with carbapenems and colistin have been defined as a risk factor for ventilator-associated pneumonia by Inchai et al.20
All strains tested here were still relatively susceptible to aminoglycosides and tetracyclines. The exceptions were strains 9 and 20 which were resistant to aminoglycosides and had an intermediate resistance to tetracyclines. This conclusion however was mainly based on susceptibility to amikacin, as strains 9, 12, 15, 30, 35 and 45 were resistant to gentamicin and tobramycin and strains 30 and 45 were moderately resistant to tobramycin. According to the definitions proposed by Magiorakos et al.,21 strains 9, 12, 15, 20, 35 and 45 would be classified as Extensively Drug Resistant (XDR), strains 3, 26, 30 and 52 as Multi Drug Resistant (MDR), and strains 38 and 59 were susceptible. As is shown further on, all tested strains, including XDR and MDR isolates, were found to produce biofilms, a significant aspect from a public health point of view, as it allows them to persist for long periods of time in hospital settings and gives rise to repeated outbreaks.
Biofilm formation patterns of A. baumannii clinical strains
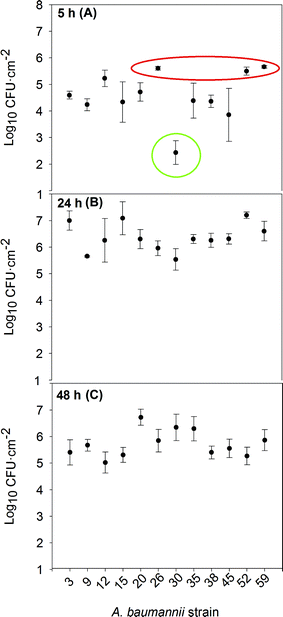
The isolates were tested for their biofilm formation ability on abiotic surfaces, which is linked to the attachment densities of the cells on steel coupons. As shown in Fig. 1, all of the strains were able to attach to the stainless steel coupons at 37 °C, though their development kinetics differed. A quick formation pattern is presumably due to an earlier adherence to the substratum, acting as a bottleneck step for attached cell division and overall inert surface colonization. The speed of attachment and subsequent biofilm formation on abiotic surfaces reflects the bacterium's ability to quickly adhere unto surfaces in the hospital setting and start protecting itself through the formation of biofilms. This, in turn, leads to a longer persistence in the hospital and a possible source of repeated infections and outbreaks.According to the density of the attached cells per surface unit attained after 5 hours of incubation (Fig. 1a), the strains could be classified into three different groups. Group 1 (in red) includes strains 26, 52 and 59: quick biofilm formers, reaching more than 5 log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU cm−2. Strains 26 and 52 came from patients with a respiratory infection, who had stayed at the ICU and undergone mechanical ventilation and catheterization. Strain 59 was isolated from a patient with Crohn's disease. These individuals tend to have an altered intestinal microbiota, allowing/selecting for strongly adherent microbial strains.22 Group 1 strains were susceptible to all aminoglycosides; only strain 52 was resistant to carbapenems. Group 2 (in green) represented the slow biofilm forming mode and consisted only of strain 30, which attained just around 2 log
CFU cm−2. Strains 26 and 52 came from patients with a respiratory infection, who had stayed at the ICU and undergone mechanical ventilation and catheterization. Strain 59 was isolated from a patient with Crohn's disease. These individuals tend to have an altered intestinal microbiota, allowing/selecting for strongly adherent microbial strains.22 Group 1 strains were susceptible to all aminoglycosides; only strain 52 was resistant to carbapenems. Group 2 (in green) represented the slow biofilm forming mode and consisted only of strain 30, which attained just around 2 log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU cm−2 after 5 h. This rather special strain was isolated from an oncology immunodeficient patient not exposed to catheterization; it was sensitive to carbapenems, resistant to gentamicin and moderately resistant to tobramycin. The rest of the strains, attaining intermediate attached cell densities after 5 h, constituted group 3. Except for strain 38, they were all resistant to carbapenems; in addition, strains 12 and 45 were resistant to colistin. Strains 9, 12, 15, 20, 35, and 45 were resistant to at least one aminoglycoside. All XDR isolates belonged to this group 3, with intermediate attachment densities.
CFU cm−2 after 5 h. This rather special strain was isolated from an oncology immunodeficient patient not exposed to catheterization; it was sensitive to carbapenems, resistant to gentamicin and moderately resistant to tobramycin. The rest of the strains, attaining intermediate attached cell densities after 5 h, constituted group 3. Except for strain 38, they were all resistant to carbapenems; in addition, strains 12 and 45 were resistant to colistin. Strains 9, 12, 15, 20, 35, and 45 were resistant to at least one aminoglycoside. All XDR isolates belonged to this group 3, with intermediate attachment densities.
The distribution of antibiotic susceptibility profiles and biofilm forming patterns suggests some association between good susceptibility to aminoglycosides and rapid biofilm formation, as in the case of our group 1. Hoffman et al.23 reported that sub-inhibitory concentrations of aminoglycoside antibiotics induced biofilm formation in Pseudomonas aeruginosa and Escherichia coli. This induction was found to be inhibited by GTP. These authors postulated that biofilm formation could be a specific defensive reaction to the presence of antibiotics, whose molecular basis is linked to alterations in the level of c-di-GMP. Rodriguez-Baño et al.8 observed that previous use of aminoglycoside was associated with A. baumannii biofilm-forming isolates, whereas for treatment in an intensive care unit, ciprofloxacin resistance and isolation from a respiratory sample were associated with non-biofilm forming isolates. He et al.24 have described biofilm formation being induced by levofloxacin and a correlation between biofilm induction and an up-regulation of the transcription of the gene encoding the adeG efflux pump; they suggested a link between low-dose antimicrobial therapy and a high risk of infection caused by biofilm forming A. baumannii strains. The fact that the XDR strains showed intermediate rates of biofilm formation (our group 3) may correspond to an energy allocation tradeoff between taking advantage of the biofilms' physical protection and expressing antibiotic resistance mechanisms.
After 24 h of incubation (Fig. 1b), most of the strains had similar counts of viable biofilm forming cells and beyond 48 h (Fig. 1c) dispersal caused a general decrease in cell numbers. For only two out of the twelve tested strains (strains 20 and 30), the attached cell densities continued to increase between 24 to 48 h, either as a result of a slow biofilm development pattern, or a less active dispersal mechanism.
Biofilm formation on abiotic surfaces has been previously tested in clinical A. baumannii strains9,25 and rather variable scores after 24 h were observed. De Breij et al.26 also reported variable biofilm formation abilities in a collection of A. baumannii strains. Rao et al.27 and Gurung et al.28 found a positive relationship between antibiotic resistance and biofilm formation ability. Orsinger-Jacobsen et al.29 described biofilm formation on steel for 13 strains of this organism; they also observed a wide variation in cell densities and rather scarce matrix formation, as seen by Scanning Electron Microscopy (SEM). The variations these authors observed could be due to the quantitative differences in the expression of specific biofilm phenotype genes or in the Quorum Sensing (QS) mechanisms involved in their regulation, as it may also be in our case.
In our study, the presence of two biofilm formation-related genes was checked in the isolates. One was Outer Membrane Protein A (OmpA), considered to be a virulence factor in this species and required for the production of robust biofilms on abiotic surfaces and for attachment to epithelial cells.11 All tested strains in this study were positive for the gene encoding OmpA.
The other one was the CsuE gene, coding for a protein included in an assembly system of pili, usually involved in biofilm formation and cellular attachment.30 All tested strains except for strains 12 and 59 had the CsuE gene. The absence of the CsuE gene in strain 59, coming from the patient with Crohn's disease, may imply an alternative, though rather effective mechanism, to either build the pili, or do without them; we should note that this strain belonged to the fast-biofilm forming group. Gaddy et al.11 reported that even though the CsuE protein seems essential for the production of pili and the formation of biofilms, some A. baumannii strains without this gene were still able to form biofilms on abiotic surfaces when cultured in certain chemically controlled media. De Breij et al.31 also showed that certain A. baumannii strains not producing pili through the pathway involving the CsuE protein were still able to attach to human epithelial cells.
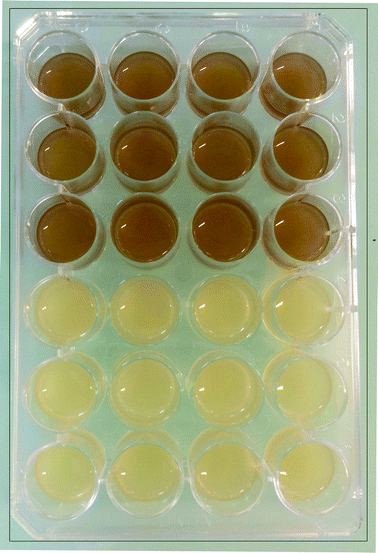
Pigment production and biofilm structure of A. baumannii clinical strains
Strains 3, 26, 45 and 52 were noted to produce a brown pigment when cultured in BHI broth while the rest presented a neutral hue. Pigment production was thus not related to the speed of biofilm formation. The colors displayed by the biofilms of the pigmented strain 52, belonging to group 1, the quick biofilm forming group, and the non-pigmented strain 38 (in group 3, with intermediate biofilm formation speed) are shown in Fig. 2. To check whether pigment production had any relationship with the biofilm features, Confocal Laser Scanning Microscopy (CLSM) images of 24 h biofilms of two pigment-producing strains (52 and 45) and two non-pigmented strains (30 and 38) were analyzed, to estimate the substratum surface coverage and biofilm thickness and volume (Table 3).| A. baumannii strain | Biofilm thickness (μm) | Biovolume (μm3) | Biovolume distribution (%) | |
|---|---|---|---|---|
| Cell | Matrix | |||
| 38 | 6 | 1.6 × 103 | 90 | 10 |
| 30 | 2 | 3.3 × 103 | 51 | 49 |
| 45 | 9 | 112 × 103 | 83 | 17 |
| 52 | 9 | 12.3 × 103 | 73 | 27 |
The pigmented strains developed thicker biofilms than the non-pigmented ones and managed to cover almost the whole surface of the available substratum, showing, at least apparently, better colonization of steel coupons (Fig. 3). The biofilms of strain 45, an XDR organism and an intermediate rate biofilm former, attained the highest biovolume value after 24 h. How much of the extensive antibiotic resistance exhibited by this strain could be due to the protection provided by its dense biofilms is hard to say at this point. The biofilm could have been formed while resistance was being assayed, and besides, the possible previous exposure of each strain to low antibiotic concentrations was not recorded. Strain 52, an MDR organism and a quick biofilm former (Fig. 1) also developed relatively thick biofilms (Table 3). Both pigmented strains 45 and 52, which gave rise to relatively homogeneous biofilms with wide surface coverage (Fig. 3), happened to be resistant to imipenem and meropenem (Table 2).
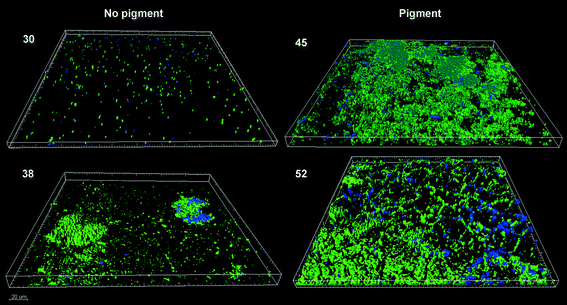 | ||
| Fig. 3 CLSM images (zenital view) of 24 h-biofilms formed by A. baumannii strains. Pigment (+): 45 and 52 and pigment (−): 30 and 38. Cells were in green and EPS in blue. | ||
On the other hand, the non-pigmented strains, which were sensitive to those β-lactam antibiotics, gave rise to small-volume biofilms which did not homogeneously cover the substratum surface. Strain 38, a medium rate biofilm former, fully sensitive to all the antibiotics tested here, produced a heterogeneous biofilm mostly composed of scattered colonies unable to cover the whole surface (Fig. 3). This sort of pattern usually develops from few and separate adhered cells, which later divide at a relatively fast rate, giving rise to cell stacks. Strain 30, the MDR slow biofilm former (single member of group 2), produced a light and thin homogeneous coating (Fig. 3) with a higher proportion of matrix to cells than the others (Table 3).
Vilacoba et al.32 reported an outbreak caused by an XDR indigo-pigmented A. baumannii strain isolated at an acute care hospital unit in Argentina. In that case, there was a link between pigment formation and virulence and/or antibiotic resistance. The specific nature or mechanism of action of the brown pigment observed in this study is not yet established, but one could think that it might be a siderophore. There is a well described need for iron in biofilm development in general,33 and in this species in particular.34 The iron acquisition systems of A. baumannii have been reviewed by Mortensen et al.35 and McConnell et al.6 When all of our isolates were screened for unspecific siderophore production, all turned up to be positive, except for strain 30, the strain which showed very feeble biofilm production. Thus, the features of the pigment-producing strains are compatible with the siderophore role for the brown pigment, but further characterization work is still needed.
Conclusions
Though the number of studied isolates does not allow for more than preliminary conclusions, the tested A. baumannii strains showed rates of biofilm formation that could be grouped into fast, moderate, and slow biofilm forming groups on steel coupons after 5 hours of growth. The fast biofilm forming group seems to be associated with good susceptibility to aminoglycosides. Under CLSM, the pigmented strains that produced more homogenous and voluminous biofilms were resistant to carbapenems, suggesting an interplay between the density of the formed biofilms and the resistance to this class of antibiotics. This also highlights the importance of biofilms in MDR strains that could lead to their persistence in the hospital for prolonged periods of time. No association between the brown pigment noted for certain strains and siderophore production was found. It was however observed that the pigmented strains produced a more voluminous and homogeneous biofilm. All the tested strains were positive for OmpA. One fast biofilm-forming strain, in addition to the one with a moderate rate of biofilm formation, lacked CsuE. Further investigation into the mechanisms of biofilm formation for these strains could be of interest. Finally, investigation of a broader set of clinical isolates may shed clearer light on the interplay between the various clinical and laboratory findings and biofilm formation patterns.References
- H. J. Doughari, P. A. Ndakidemi, I. S. Human and S. Benade, Microbes Environ., 2011, 26, 101–112 CrossRef.
- M. Touchon, J. Cury, E. J. Yoon, L. Krizova, G. C. Cerqueira, C. Murphy, M. Feldgarden, J. Wortman, D. Clermont, T. Lambert, C. Grillot-Courvalin, A. Nemec, P. Courvalin and E. P. Rocha, Genome Biol. Evol., 2014, 6, 2866–2882 CrossRef CAS PubMed.
- F. C. Tenover, Am. J. Med., 2006, 119, S3–S10 CrossRef CAS PubMed.
- L. C. Antunes, P. Visca and K. J. Towner, Pathog. Dis., 2014, 71, 292–301 CrossRef CAS PubMed.
- A. Y. Peleg, H. Seifert and D. L. Paterson, Clin. Microbiol. Rev., 2008, 21, 538–582 CrossRef CAS PubMed.
- M. J. McConnell, L. Actis and J. Pachon, FEMS Microbiol. Rev., 2013, 37, 130–155 CrossRef CAS PubMed.
- Y. Doi, G. L. Murray and A. Y. Peleg, Semin. Respir. Crit. Care Med., 2015, 36, 85–98 CrossRef PubMed.
- J. Rodrıguez-Baño, S. Martı, S. Soto, F. Fernandez-Cuenca, J. M. Cisneros, J. Pachon, A. Pascual, L. Martınez-Martınez, C. McQueary, L. A. Actis, J. Vila and the Spanish Group for the Study of Nosocomial Infections (GEIH), Clin. Microbiol. Infect., 2008, 14, 276–278 CrossRef PubMed.
- H. W. Lee, Y. M. Koh, J. Kim, J. C. Lee, Y. C. Lee, S. Y. Seol, D. T. Cho and J. Kim, Clin. Microbiol. Infect., 2008, 14, 49–54 CrossRef CAS PubMed.
- F. Longo, C. Vuotto and G. Donelli, New Microbiol., 2014, 37, 119–127 CAS.
- J. A. Gaddy, A. P. Tomaras and L. A. Actis, Infect. Immun., 2009, 77, 3150–3160 CrossRef CAS PubMed.
- S. Rumbo-Feal, M. J. Gomez, C. Gayoso, L. Alvarez-Fraga, M. P. Cabral, A. M. Aransay, N. Rodriguez-Ezpeleta, A. Fullaondo, J. Valle, M. Tomas, G. Bou and M. Poza, PLoS One, 2013, 8, e72968 CAS.
- M. L. Liou, P. C. Soo, S. R. Ling, H. Y. Kuo, C. Y. Tang and K. C. Chang, J. Microbiol., Immunol. Infect., 2014, 47, 275–281 CrossRef CAS PubMed.
- O. Ciofu and T. Tolker-Nielsen, in Biofilm Infections, ed. T. Bjarnsholt, C. Moser, P. Jensen and N. Hoiby, Springer Publishing Company, New York, 2011, pp. 215–229 Search PubMed.
- D. Lebeaux, J. M. Ghigo and C. Beloin, Microbiol. Mol. Biol. Rev., 2014, 78, 510–543 CrossRef CAS PubMed.
- I. Olsen, Eur. J. Clin. Microbiol. Infect. Dis., 2015, 34, 877–886 CrossRef CAS PubMed.
- F. Badmasti, S. D. Siadat, S. Bouzari, S. Ajdary and F. Shancheraghi, J. Med. Microbiol., 2015, 64, 538–543 CrossRef PubMed.
- E. V. Lemos, F. P. de la Hoz, T. R. Einarson, W. F. McGhan, F. Quevedo, C. Castaneda and K. Kawai, Clin. Microbiol. Infect., 2014, 20, 416–423 CrossRef CAS PubMed.
- J. Fishbain and A. Y. Peleg, Clin. Infect. Dis., 2010, 51, 79–84 CrossRef PubMed.
- J. Inchai, C. Liwsrisakun, T. Theerakittikul, R. Chaiwarith, W. Khositsakulchai and C. Pothirat, J. Infect. Chemother., 2015, 21, 570–578 CrossRef PubMed.
- A. P. Magiorakos, A. Srinivasan, R. B. Carey, Y. Carmeli, M. E. Falagas, C. G. Giske, S. Harbarth, J. F. Hindler, G. Kahlmeter, B. Olsson-Liljequist, D. L. Paterson, L. B. Rice, J. Stelling, M. J. Struelens, A. Vatopoulos, J. T. Weber and D. L. Monnet, Clin. Microbiol. Infect., 2011, 18, 268–281 CrossRef PubMed.
- B. Chassaing, E. Garénaux, J. Carriere, N. Rolhion, Y. Guérardel, N. Barnich, R. Bonnet and A. Darfeuille-Michaud, J. Bacteriol., 2015, 197, 1451–1465 CrossRef CAS PubMed.
- L. R. Hoffman, D. A. D'Argenio, M. J. MacCoss, Z. Zhang, R. A. Jones and S. I. Miller, Nature, 2005, 436, 1171–1175 CrossRef CAS PubMed.
- X. He, F. Lu, F. Yuan, D. Jiang, P. Zhao, J. Zhu, H. Cheng, J. Cao and G. Lu, Antimicrob. Agents Chemother., 2015, 59, 4817–4825 CrossRef CAS PubMed.
- C. N. McQueary and L. A. Actis, J. Microbiol., 2011, 49, 243–250 CrossRef CAS PubMed.
- A. De Breij, L. Dijkshoorn, E. Lagendijk, J. van der Meer, A. Koster, G. Bloemberg, R. Wolterbeek, P. van den Broek and P. Nibbering, PLoS One, 2010, 5, e10732 Search PubMed.
- R. S. Rao, R. U. Karthika, S. P. Singh, P. Shashikala, R. Kanungo, S. Jayachandran and K. Prashanth, Indian J. Med. Microbiol., 2008, 26, 333–337 CrossRef.
- J. Gurung, A. B. Khyriem, A. Banik, W. V. Lyngdoh, B. Choudhury and P. Bhattacharyya, Indian J. Crit. Care Med., 2013, 17, 214–218 CrossRef CAS PubMed.
- S. J. Orsinger-Jacobsen, S. S. Patel, E. M. Vellozzi, P. Gialanella, L. Nimrichter, K. Miranda and L. R. Martinez, Microbiology, 2013, 159, 2594–2604 CrossRef CAS PubMed.
- J. F. Turton, S. N. Gabriel, C. Valderrey, M. E. Kaufmann and T. L. Pitt, Clin. Microbiol. Infect., 2007, 13, 807–815 CrossRef CAS PubMed.
- A. De Breij, J. Gaddy, J. van der Meer, R. Koning, A. Koster, P. van den Broek, L. Actis, P. Nibbering and L. Disjkshoorn, Res. Microbiol., 2009, 160, 213–218 CrossRef CAS PubMed.
- E. Vilacoba, M. Almuzara, L. Gulone, R. Rodriguez, E. Pallone, R. Bakai and M. S. Ramírez, J. Clin. Microbiol., 2013, 51, 3726–3730 CrossRef CAS PubMed.
- E. Banin, M. L. Vasil and E. P. Greenberg, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 11076–11081 CrossRef CAS PubMed.
- V. Gentile, E. Frangipani, C. Bonchi, F. Minandri, F. Runci and P. Visca, Pathogens, 2014, 3, 704–719 CrossRef CAS PubMed.
- B. L. Mortensen and E. P. Skaar, Front. Cell. Infect. Microbiol., 2013, 3, 95 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5md00377f |
| This journal is © The Royal Society of Chemistry 2016 |