Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synergistic activity of a short lipidated antimicrobial peptide (lipoAMP) and colistin or tobramycin against Pseudomonas aeruginosa from cystic fibrosis patients†‡
Martin G.
de Gier
a,
H.
Bauke Albada
*de,
Michaele
Josten
c,
Rob
Willems
b,
Helen
Leavis
b,
Rosa
van Mansveld
b,
Fernanda L.
Paganelli
b,
Bertie
Dekker
b,
Jan-Willem J.
Lammers
a,
Hans-Georg
Sahl
c and
Nils
Metzler-Nolte
 d
d
aDepartment of Pulmonology, University Medical Center Utrecht, University of Utrecht, Utrecht, The Netherlands
bDepartment of Medical Microbiology, University Medical Center Utrecht, University of Utrecht, Utrecht, The Netherlands
cInstitute of Medical Microbiology, Immunology, and Parasitology, Pharmaceutical Microbiology Section, University of Bonn, Bonn, Germany
dInorganic Chemistry I – Bioinorganic Chemistry, Ruhr University Bochum, Bochum, Germany
eThe Center for Nanoscience and Nanotechnology, Institute of Chemistry, The Hebrew University of Jerusalem, 91904, Jerusalem, Israel. E-mail: h.b.albada@gmail.com
First published on 5th October 2015
Abstract
Declining pulmonary function, ultimately culminating in respiratory failure, is mainly caused by chronic Pseudomonas aeruginosa (P. aeruginosa) infections in patients with cystic fibrosis (CF). Due to its hypermutability, allowing rapid adaptation to the selective constraints in a lung with CF, and the ability to form biofilms, P. aeruginosa is able to colonize and damage the lung by chronic infection. Exacerbations are being treated with a combination of common anti-pseudomonal antibiotics, but (pan)resistance is increasingly reported. Antimicrobial peptides (AMPs) have a broad spectrum of antibacterial activity, and their effectiveness is, still, less affected by induction of resistance. Here, we explore the in vitro applicability of a RWRWRWK(C10) synthetic lipoAMP (named BA250-C10), a lipidated peptide with a C10-lipid chain attached to the C-terminus, as a novel antibacterial agent against P. aeruginosa; and in particular, its ability to inhibit biofilm formation. BA250-C10 was tested for its in vitro antibacterial activity against 20 clinical P. aeruginosa isolates from CF patients, each having a different resistance profile and ability to form biofilms. The modest antibacterial activity of the peptide against most P. aeruginosa strains (16–256 μg mL−1) was significantly increased in the presence of colistin or tobramycin, supported by the results from the checkerboard assay and growth curves. In three biofilm-forming strains, a synergistic effect was observed for BA250-C10 with colistin, but less with tobramycin. This indicates that combinations of lipidated AMPs and colistin may be further pursued as a potential novel treatment strategy against P. aeruginosa infections in CF patients.
Introduction
Pseudomonas aeruginosa (P. aeruginosa) is the most prevalent and significant pulmonary pathogen in patients with cystic fibrosis (CF). Colonization of P. aeruginosa is associated with a faster decline of pulmonary function and overall worsening prognosis.1 A crucial obstacle in antibiotic treatment is the ability of P. aeruginosa to form biofilms and its ability to rapidly adapt2 to the ever-changing physiology within the CF airway.3 Anti-pseudomonal therapies are used in three distinct clinical settings: (i) in delaying onset of chronic colonization, (ii) in chronic maintenance therapy, and (iii) in periodic administration of intensive antibiotic regimens.4 The standard treatment for exacerbation of CF is intravenous therapy with two antibiotics, mainly aimed at decreasing the risk of resistance, but also to decrease dose-related toxicity, to treat polymicrobial infection, and to promote antimicrobial synergism.5 Unfortunately, current antibiotics are becoming less effective in treating chronic Pseudomonas infections due to increasing antibiotic resistance and highly antibiotic-refractory biofilms.6 In the past decade, no new antibiotics have been developed,7 and there are only minor improvements in inhaled anti-pseudomonal antibiotics. New therapeutic options for patients with CF are designed to correct the function of the defective CF transmembrane conductance regulator (CFTR)-modulating protein,8 and clinical effects of this treatment have been shown in different randomized clinical trials.9 However, these treatments will be available only for a selection of CF patients, depending on the type of their genetic defect.8 Therefore, CF patients will continue to suffer from pulmonary infections and new antibacterial therapies and treatment strategies are in continuous demand.10A relatively new class of antibacterial compounds is the large family of host defense peptides (HDPs) and antimicrobial peptides (AMPs).11 Many of these occur naturally as a part of the host defense system; whereas HDPs combine direct broad-spectrum antibiotic activities with modulation of immune responses,12 AMPs have only direct antibacterial activities.13 Whereas naturally occurring HDPs and AMPs hold great promise when it comes to the antimicrobial activity and the ability to inhibit biofilm formation,14 their applicability in a clinical setting is limited due to poor PK/PD profiles.12 In addition, their intricate structure hampers large-scale production and severely encumbers the modulation of their therapeutic profile. Nevertheless, the emergence of resistance against HDPs and AMPs is considered to be less of a problem compared to conventional antibiotics since many AMPs target the bacterial membrane rather than a specific single biomolecule.12,15 Therefore, AMPs are considered as relevant new candidate treatment options in diseases such as CF in which multidrug-resistant organisms cause infections in a hyperinflammatory environment.12,16
Anti-pseudomonal synthetic AMPs (synAMPs) have been developed in recent years.17 In addition, AMPs with specific anti-biofilm properties have been discovered.11b,17a,b,d,18 For example, a dodecameric peptide with the sequence VRLIVAVRIWRR-NH2 was shown to potently inhibit biofilm formation of CF pathogens by blocking a widespread stress response that contributes to biofilm development.19 Short synAMPs can be prepared on a large scale, and can easily be modified to improve proteolytic stability, circulation lifetime, and bacterial specificity or to decrease general toxicity. These make them attractive candidates for clinical applications. En route to that goal, the mode of action of one specific family of short synAMPs, i.e. those that contain the Arg-Trp sequence,20 has been determined.21 The activities against methicillin resistant Staphylococcus aureus (MRSA) of the organometallic derivatives of such peptides are now identical to,22 or even better than, vancomycin without inducing substantial hemolysis and displaying high toxicity in vitro.23 These last two effects are usually problematic for lipidated AMPs. Their effect on planktonic growth and biofilm formation of Escherichia coli was also determined, showing promising results for the former, but limited results for the latter.24 Similarly, N- or C-terminal lipidation of an Arg-Trp hexapeptide, resulting in the so-called lipoAMPs, led to high activity against a broad spectrum of bacterial pathogens, including P. aeruginosa and A. baumannii.25 Even more, their hemolytic activity could be reduced from ~16% to less than 1% when human red blood cells were treated with 250 μg mL−1 of the peptide.26 Moreover, only a few examples have emerged in which the synergy of AMPs with existing antibiotics27 as well as AMPs with anti-pseudomonal antibiotics have been described,17b,18b,28 but a detailed study with a large panel of clinically isolated P. aeruginosa strains and lipoAMPs has not been performed yet.
Here, we now determined the activity of lipoAMPs against CF-related P. aeruginosa strains,29,30 the synergistic activity of the most active lipoAMP with conventional antibiotics,31–33 and their ability to inhibit biofilm formation. We assessed the activity of 12 different lipidated short peptides (i.e. the lipoAMPs) against three CF-related P. aeruginosa isolates. The peptide with the lowest MIC-value was used for further evaluation against a wider panel of clinical P. aeruginosa isolates. Growth curves and checkerboard assays were applied to probe for synergy between the lipoAMP and two commonly applied antibiotics, i.e. colistin and tobramycin, and biofilm interfering capacity was obtained using a polystyrene biofilm assay.
Experimental
All experimental details and procedures are provided in the ESI.†Results
The peptides that were used in this study have been described before.25 Apart from the lipidated peptides, which are identified by the position and length of their lipid chain (i.e. C8 refers to the C(O)C7H15 lipid attached to a C-terminal positioned lysine residue; N8 refers to the same lipid when attached to an N-terminal positioned lysine residue), two ferrocenoyl-derivatized peptides (indicated by ‘Fc’), and one dye-labelled peptide, i.e. BA250-DEC, were also included.Initially, the MIC-values of the 12 lipoAMPs against three clinical isolates of P. aeruginosa were determined (Table 1). The three isolates were chosen for their different susceptibility profiles to standard applied anti-pseudomonal antibiotics: very resistant KD491 and intermediate resistant LES431 and VW1633. LipoAMPs containing either a C- or an N-terminal positioned lipidated lysine residue were tested, as well as the two commonly applied antibiotics ciprofloxacin and polymyxin B. The general activity of these lipoAMPs against the very resistant KD491 was higher than against the less resistant strain LES431.
| LipoAMPa | Clinical isolate | ||
|---|---|---|---|
| VW1633 | LES431 | KD491 | |
| MIC (μg mL−L) | MIC (μg mL−L) | MIC (μg mL−L) | |
| a All peptides were obtained in high purity (>95%) after preparative HPLC and in acceptable yields of 21–46%; HR-MS spectrometry confirmed the identity of the peptides.25 | |||
| BA250-CFc | 32 | >128 | 32 |
| BA250-C6 | 16 | >128 | 32 |
| BA250-C8 | 16 | 128 | 16 |
| BA250-C10 | 16 | 32 | 16 |
| BA250-C12 | 32 | 64 | >128 |
| BA250-C14 | >128 | 128 | >128 |
| BA250-NFc | 32 | >128 | 128 |
| BA250-N6 | 32 | >128 | 64 |
| BA250-N8 | 32 | 64 | 16 |
| BA250-N10 | 64 | 32 | 32 |
| BA250-N12 | >128 | 64 | >128 |
| BA250-N14 | >128 | >128 | >128 |
| BA250-DEC | 64 | >128 | 64 |
| Ciprofloxacin | 6.4 | 6.4 | 1.6 |
| Polymyxin B | 1.6 | 0.8 | 1.6 |
| DMSO | >128 | >128 | >128 |
LipoAMP BA250-C10 was the most promising candidate in our study, with MIC-values of 16–32 μg mL−1 (i.e. 9–18 μM) (Table 1), and the activity of this peptide was further studied against a larger panel of clinical isolates of P. aeruginosa (Table 2). The redox-active Fc-labelled lipoAMP did not display enhanced activity; in fact, the activity of this lipophilic peptide, of which the lipophilicity resembles that of a peptide containing a seven C-atom long lipid, is more or less within the expected range of lipidated AMPs. This indicates that this moiety mostly acts as a lipophilic moiety, potentially as a membrane anchor. Of the 20 clinically isolated P. aeruginosa strains against which activity was determined (Table 2), 6 were international P. aeruginosa isolates34–38 and 14 were obtained from the University Medical Center Utrecht (UMCU).
| Entry | Strain | Ciprofloxacin | Colistin | Tobramycin | Ceftazidime | Tazocin | Meropenem | BA250-C10 | Resistance | Biofilm |
|---|---|---|---|---|---|---|---|---|---|---|
| Notes: Minimal inhibitory concentration (MIC) values are given in μg mL−L; CLSI breakpoints for susceptibility of various strains for specific antibiotics are given in brackets beside the MIC-values: I = intermediate, R = resistant, S = susceptible (S is left out for clarity); the cut-off limits for the respective antibiotics are given below. Resistance is based on the number of antibiotics against which resistance is observed. The origins of the strains are indicated when known: entry-numbers that are marked with an asterisk (*) indicate that these strains were obtained from CF patients treated in the University Medical Center Utrecht; ‘KD’ refers to a child, ‘VW’ to an adult. Entries marked in bold indicate the strains that were used in the subsequent studies. Cut-off limits for the CLSI breakpoints for susceptibility: ciprofloxacin: S ≤ 0.5 μg mL−L and R > 1 μg mL−L – colistin: S ≤ 4 μg mL−L and R > 4 μg mL−L – tobramycin: S ≤ 4 μg mL−L and R > 4 μg mL−L – ceftazidime: S ≤ 8 μg mL−L and R > 8 μg mL−L – tazocin: S ≤ 16 μg mL−L and R > 16 μg mL−L – meropenem: S ≤ 2 μg mL−L and R > 8 μg mL−L.a The ability to form biofilms is measured using the crystal violet assay where ‘++’ indicates high, ‘+’ indicates intermediate, and ‘—’ indicates low tendency for biofilm formation. | ||||||||||
| 1* | D599 | 0.25 | 4 | 0.5 | 1 | 4 | 0.5 | 128 | 0 | + |
| 2 ref. 34 | Pa01 | 0.5–0.25 | 4 | 2 | 1 | 8 | 2 | 256 | 0 | ++ |
| 3 ref. 35 | Clone C | 0.25 | 4 | 1 | 2 | 16 | 2 | 128 | 0 | ++ |
| 4* | VW1501 | 16 (R) | 2 | 4 | 4 | 4 | 0.25 | 128 | 1 | — |
| 5* | kl 1.1 | 4 (R) | 4 | 4 | 2 | 4 | 4 (I) | 128 | 1–2 | — |
| 6* | KD557 | 0.5 | 8 (R) | 1 | 2 | 16–32 (R) | 1–2 | 256 | 2 | + |
| 7* | VW1540 | 2–4 (R) | 4 | 0.125 | 16 (R) | 8 | 1–2 | 64 | 2 | — |
| 8* | VW178 | 1 (R) | 4 | 32 (R) | 2 | 8 | 2 | 32 | 2 | + |
| 9* | VW1633 | 1 (R) | 2 | 0.125 | >256 (R) | >512 (R) | 1 | 32 | 3 | — |
| 10* | VW1485 | 8 (R) | >128 (R) | 16 (R) | 8 | 4 | 0.25 | 256 | 3 | — |
| 11* | VW0247 | 16 (R) | 2 | 4 | 16 (R) | 32 (R) | 0.25 | 64 | 3 | — |
| 12* | kl 3.2 | 2–4 (R) | 8 (R) | 16 (R) | 2 | 4–8 | 4 | 128 | 3 | — |
| 13 ref. 36 | LES431 | 4 (R) | 2 | 2–4 | 256 (R) | 512 (R) | 8 (I) | 64 | 3–4 | — |
| 14* | VW1538 | 8 (R) | 2 | 8 (R) | 8 | 64 (R) | 64 (R) | 64 | 4 | — |
| 15 ref. 37 | MIDLANDS | 4 (R) | 4 | 2 | 64 (R) | 128 (R) | 16 (R) | 64 | 4 | — |
| 16 ref. 37 | LES400 | 2 (R) | 4 | 8 (R) | 32 (R) | 32 (R) | 2 | 64 | 4 | — |
| 17* | VW1471 | 16 (R) | 4 | 8 (R) | 128 (R) | 256 (R) | 32 (R) | 64 | 5 | — |
| 18* | VW313 | 2 (R) | 4 | 16 (R) | >256 (R) | 64 (R) | 16 (R) | 32 | 5 | — |
| 19* | KD491 | 8 (R) | 2 | 8–16 (R) | >256 (R) | >512 (R) | 16 (R) | 32 | 5 | ++ |
| 20 ref. 38 | LESB58 | 8–16 (R) | 32 (R) | 8 (R) | 256 (R) | 512 (R) | 2 | 128 | 5 | + |
The results demonstrated an inverse correlation between the resistance of the P. aeruginosa strains against a number of antibiotics and the MIC-value for BA250-C10 – strains that are more resistant to the more commonly applied antibiotics have lower MIC-values against BA250-C10. For two biofilm-forming P. aeruginosa strains, the MIC-value is 32 μg mL−1 (entries 8 and 19); for the other biofilm-forming P. aeruginosa strains, the MIC-values are 128 and 256 μg mL−1 (entries 2 and 6, respectively). It should be noted that the results displayed in Table 1 were obtained in a different laboratory compared to those displayed in Table 2; this explains the 2-fold difference between the MIC-values of BA250-C10 against VW1633, LES431, and KD491.
Next, synergistic activity of the lipoAMP and colistin and tobramycin was mapped using the checkerboard assay. For this, strains KD491, LESB58, Pa01, and clone C were selected due to their strong tendency to form biofilms. Also, since the activity of BA250-C10 was low when that of colistin and/or tobramycin was high (entries 2 and 3), or when the activity of BA250-C10 was high and that of tobramycin was low (entry 19), synergism in both directions, i.e. of the antibiotics on the activity of lipoAMP or of the lipoAMP on the activity of both antibiotics, was studied. In addition, we determined if synergism could enhance the combined activity of compounds that are poorly active against the multi-resistant strain LESB58 (entry 20).
This study revealed that lipoAMP BA250-C10 showed synergy with colistin in three out of the four tested strains, and with tobramycin in two out of the four tested strains (Table 3). Strong synergy is found for BA250-C10 and colistin against strain KD491 with FIC < 0.5. Whereas the MIC-value of colistin itself is 4 μg mL−1, in the presence of 8 μg mL−1 BA250-C10, the MIC of colistin drops to 1 μg mL−1. Similarly, the MIC-value of BA250-C10 is 32 μg mL−1, but in the presence of 2 μg mL−1 colistin, it drops to 2 μg mL−1. Interestingly, the required amount of the second component is below the MIC-value of that compound. In addition, the two strains that generally display higher levels of resistance, KD491 and LESB58, show very low FIC-indices (i.e. <0.5), which are indicative of a synergistic effect, whereas the two strains that are almost not-resistant against any of the commonly applied antibiotics (see entries 2 and 3 in Table 2), Pa01 and clone C, have higher FIC-indices, i.e. lower synergy.
| KD491 | LESB58 | Pa01 | Clone C | |||||
|---|---|---|---|---|---|---|---|---|
| Colistin | Tobramycin | Colistin | Tobramycin | Colistin | Tobramycin | Colistin | Tobramycin | |
| a Synergism (S) is defined as FIC ≤ 0.5, and indifference (I) as FIC = 0.5–4. | ||||||||
| FIC-index #1 | 0.2625 | 0.5 | 0.375 | 0.5 | 0.3125 | 1 | 0.5 | 0.5 |
| FIC-index #2 | 0.375 | 0.375 | 0.625 | 0.5 | 0.5 | 0.75 | 1 | 1 |
| Effecta | S | S | S/I | S | S | I | S/I | S/I |
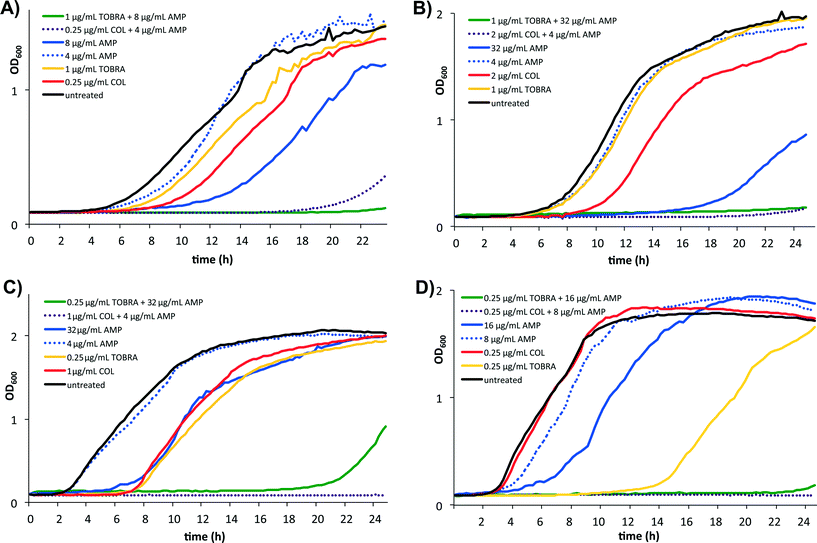
Subsequently, the growth curves of the four strains in the presence of the individual components and of sub-MIC concentrations of the mixtures were generated. The growth curve of KD491 shows normal growth in the presence of 4 μg mL−1 BA250-C10; a prolonged lag phase of 4 hours, with a normal growth rate, is observed in the exponential phase in the presence of 0.25 μg mL−1 colistin (Fig. 1, panel A). However, the combination of 4 μg mL−1 BA250-C10 and 0.25 μg mL−1 colistin almost completely inhibits growth. For the colistin-resistant strain LESB58, there is no growth of LESB58 in the presence of the combination of colistin (2 μg mL−1) and BA250-C10 (4 μg mL−1), even though there is normal growth of LESB58 with 4 μg mL−1 BA250-C10, and a prolonged lag phase with a normal growth rate in the exponential phase in the presence of 2 μg mL−1 colistin (Fig. 1, panel B).
These data confirmed the data of the checkerboard assay that indicated synergy between BA250-C10 and colistin. In Pa01 and clone C, a similar pattern is seen, suggesting synergy between colistin and lipoAMP BA250-C10 during the growth phase of the bacteria (Fig. 1, panels C and D, respectively). In the presence of only BA250-C10 or tobramycin, growth of all strains is delayed and the growth rates in the exponential phase are slower, while the combination of BA250-C10 and tobramycin shows almost complete inhibition of growth.
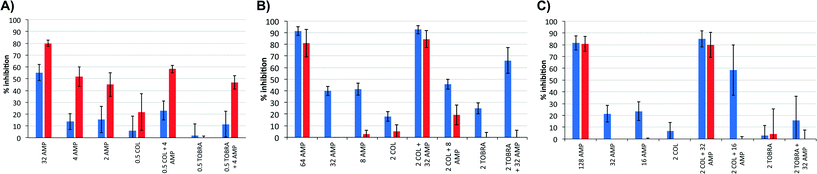
Next, we tested the ability of the isolated lipoAMP in combinations with colistin or tobramycin to inhibit biofilm formation in the polystyrene biofilm assay of KD491, Pa01, and clone C (Fig. 2). For KD491, high concentrations of BA250-C10 were needed to almost fully inhibit biofilm formation: 32 μg mL−1 for 80 ± 3% inhibition (Fig. 2, panel A). However, at 2 and 4 μg mL−1 BA250-C10, significant inhibition of biofilm formation of KD491 was already observed, i.e. 45 ± 11% and 52 ± 9%, respectively. At these concentrations, weak inhibition of planktonic growth was observed. No synergistic activity against biofilm formation between the lipoAMP and colistin or tobramycin was observed against KD491.
For the other two strains, significantly higher concentrations of BA250-C10 were needed in order to achieve substantial inhibition of biofilm formation, i.e. 64 μg mL−1 for 81 ± 11% inhibition of Pa01 and 128 μg mL−1 for 82 ± 6% inhibition of clone C, respectively (Fig. 3, panels B and C, respectively). These concentrations could be lowered to 32 μg mL−1 in the presence of 2 μg mL−1 colistin to achieve a similar level of inhibition of biofilm formation, i.e. 84 ± 8% and 70 ± 11% for Pa01 and clone C, respectively. A concentration of 2 μg mL−1 colistin without the additional compound poorly inhibited biofilm formation, up to 20%. Also, 32 μg mL−1 BA250-C10 without colistin was not able to inhibit biofilm formation in these two strains. Replacing colistin with an equal weight of tobramycin did not lead to biofilm formation inhibition, showing that synergism is strictly limited to colistin. The observation that sub-MIC concentrations of AMPs already lead to observable inhibition of biofilm formation has been described before.18a,39 The differences between the levels of inhibition of planktonic growth and biofilm formation suggest that the AMPs interfere with biofilm formation in a different manner compared to the interference with planktonic growth.
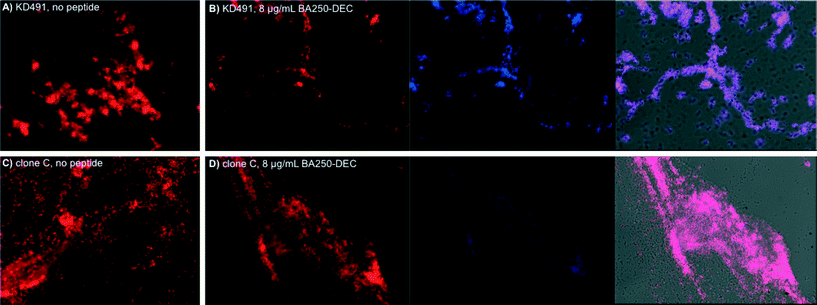
In order to visualize the effect of the lipoAMP on the biofilms that were formed, we performed confocal microscopy studies on the biofilms of KD491 and clone C in the presence or in the absence of the lipoAMP. These two strains were selected since the activity of BA250-C10 against KD491 was distinctly better than against clone C (Table 2, entries 20 and 3, respectively); LESB58 was excluded based on its lower tendency to form biofilms in our assay, and Pa01 was excluded due to its high resistance against the lipoAMPs. Since BA250-C10 cannot be visualized directly with confocal microscopy, we applied the fluorescent peptide BA250-DEC, which contains a fluorescent diethylaminocoumarin moiety (λex = 409 nm, λem = 473 nm) instead of the C10-lipid. The retention time of this dye-labelled peptide is comparable to that of the C10-lipidated peptide, i.e. 19.9 min vs. 20.2 min, respectively (see Fig. 4 for the structures), and the antibacterial activity is 4-fold lower, i.e. 64 μg mL−1 against KD491 (Table 1).
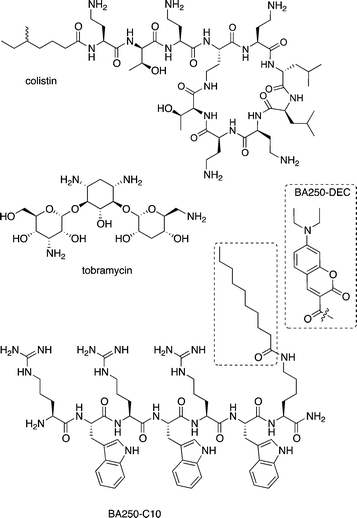 | ||
| Fig. 4 Structural formulas of colistin, tobramycin, and BA250-C10; the structure of the dye in BA250-DEC is shown in the dotted box; it replaced the lipid that is highlighted by the dotted square. | ||
Incubation of P. aeruginosa strains KD491 and clone C revealed that the dye-containing BA250-DEC is also able to inhibit biofilm formation (Fig. 3). Clear difference in biofilm texture is apparent: the biofilm that is formed by KD491 is denser and thicker, whereas that of clone C is more spread-out, containing more isolated cells. For KD491, there is a clear distinction between the biofilms that are formed in the presence or in the absence of the peptide, confirming the inhibition of biofilm formation by the lipoAMP BA250-C10 that was measured in the polystyrene biofilm assay. The lipoAMP more effectively inhibits biofilm formation of KD491 than that of clone C (panels A and B, and panels C and D, Fig. 3, respectively): upon treatment with the lipoAMP, KD491 forms a much thinner biofilm whereas that of clone C was much less altered, which corroborate with the results obtained using the polystyrene biofilm assay.
Co-localization studies reveal a high degree of overlap between the parts of the biofilm that are stained with propidium iodide and those parts that are stained with the peptide. Our results show that the peptide has a high tendency to bind to those areas in the biofilm where bacteria are residing.
Discussion
Patients with cystic fibrosis (CF) are highly dependent on antibiotic treatment since most of these patients endure chronic respiratory infections, causing (slow) degradation of the respiratory tract, which leads to respiratory failure eventually. This accounts for the majority of mortality in CF patients. The main pathogens in a lung with CF are Pseudomonas aeruginosa (>80% of the adult patients), Staphylococcus aureus (30–50%), Haemophilus influenzae, Xenotrophomonas maltophilia (~8%), and Burkholderia cepacia.40 Recently, short Arg-Trp based peptides were discovered that showed broad-spectrum activity against various bacterial pathogens, including P. aeruginosa.25 To explore if such short peptides have the potential to combat P. aeruginosa infections, we tested such lipoAMPs for their direct in vitro anti-pseudomonal activity. The most promising lead compound, i.e. BA250-C10, was further tested for its potential synergy with conventional antibiotics colistin and tobramycin (see Fig. 4 for the structures), and the potential in interfering with biofilm formation.In this study, we demonstrated that the combination of BA250-C10 with one of the conventional anti-pseudomonal antibiotics (colistin or tobramycin) successfully inhibits planktonic growth in a synergetic way. The best synergy was seen in the combination of 2 μg mL−1 BA250-C10 with 2 μg mL−1 colistin. Colistin and tobramycin are frequently used in CF patients intravenously during exacerbations and chronically by nebulization. For both BA250-C10 and colistin, it was shown that they delocalize peripheral membrane proteins,41 hinting at a cooperative activity in weakening the membrane architecture. Such an effect was not observed before for this type of lipoAMP. In addition, for two of the three strains, biofilm formation was inhibited due to the synergistic effect between 2 μg mL−1 colistin and 32 μg mL−1 BA250-C10. With 50% hemolysis at 250 μg mL−1 BA250-C10, which translates to <10% hemolysis at 32 μg mL−1 (assuming a linear correlation between concentration and hemolysis), this amount is still problematic for systemic applications. However, in the case of P. aeruginosa from KD491, only 4 μg mL−1 BA250-C10 is needed to inhibit biofilm formation in the presence of 0.5 μg mL−1 colistin. With this low concentration of lipoAMP, less than 1% hemolysis can be expected, a number that might even be lowered further by performing an L-to-D substitution of certain amino acid residues.26 Although it is too early to investigate the clinical applicability of lipoAMPs like BA250-C10, the current study reveals promising synergy between the lipoAMP and existing antibiotics, both at the level of bacterial growth as well as at the level of biofilm formation inhibition.
Further studies have to focus on the mechanism how BA250-C10 interferes with biofilm formation in KD491, even at low concentrations, and why it only interferes in the biofilm formation in the other two strains at high concentrations. Tuning the lead compound or further testing of different configurations of the parent peptide can reveal a peptide with higher anti-biofilm and immunomodulatory activity. The class of lipoAMPs currently under investigation is particularly interesting as an add-on nebulization therapy for CF patients. Recently, a high throughput screening has been developed for further optimizing peptides to generate novel sequences that possess a variety of biological properties.42
Conclusions
In conclusion, we have demonstrated that the 7-amino acid residue long lipopeptide BA250-C10 has synergistic activity with two conventional anti-pseudomonal antibiotics in inhibiting planktonic growth of four P. aeruginosa strains. Synergism in the inhibition of biofilm formation was shown in three P. aeruginosa strains. For the most resistant biofilm-forming strain, only 2 μg mL−1 BA250-C10 was required to achieve ~50% biofilm formation inhibition; for the less resistant strains, 32 μg mL−1 BA250-C10 and 2 μg mL−1 colistin were needed to obtain near quantitative inhibition. Localization of the lipoAMP in the bacteria was shown using a fluorescently labelled lipoAMP in the confocal microscopy studies. Further studies have to reveal the working mechanism of biofilm interference. Amplification and tuning of the peptide lead compound is relatively easy and is a promising path to obtain peptides with more specific anti-pseudomonal and anti-biofilm properties.Acknowledgements
We thank P. Prochnow and J. E. Bandow (Ruhr University Bochum, Germany) for the initial checkerboard analysis of BA250-C10 with P. aeruginosa type strain DSM50071. For the international P. aeruginosa strains we thank: U. Römling (Karolinska Institutet, Sweden) for clone C; C. Winstanley (University of Liverpool, UK) for LES431, LES400, and LESB58; and S. Molin (Technical University of Denmark, Denmark) for Pa01.References
- J. Emerson, M. Rosenfeld, S. McNamara, B. Ramsey and R. L. Gibson, Pediatr. Pulm., 2002, 34, 91–100 CrossRef PubMed.
- R. L. Marvig, L. M. Sommer, S. Molin and H. K. Johansen, Nat. Genet., 2015, 47, 57–64 CrossRef CAS PubMed.
- (a) L. Jelsbak, H. K. Johansen, A.-L. Frost, R. Thøgersen, L. E. Thomsen, O. Ciofu, L. Yang, J. A. J. Haagensen, N. Høiby and S. Molin, Infect. Immun., 2007, 75, 2214–2224 CrossRef CAS PubMed; (b) D. J. Hasset, T. R. Korfhagen, R. T. Irvin, M. J. Schurr, K. Sauer, G. W. Lau, M. D. Sutton, H. Yu and N. Hoiby, Expert Opin. Ther. Targets, 2010, 14, 117–130 CrossRef PubMed.
- (a) T. S. Cohen and A. Prince, Nat. Med., 2012, 18, 509–519 CrossRef CAS PubMed; (b) R. L. Gibson, J. L. Burns and B. W. Ramsey, Am. J. Respir. Crit. Care Med., 2003, 168, 918–951 CrossRef PubMed; (c) See also: Annual Report 2014, Cystic Fibrosis Foundation, Bethesda, MD (USA), 2014 Search PubMed.
- S. K. Pillai, R. C. Moellering and G. M. Eliopoulos, Antimicrobial combinations, in Antibiotics in Laboratory Medicine, ed. V. Lorian, The Lippincott Williams & Wilkins Co., Philadelphia (PA), USA, 2005, pp. 365–440 Search PubMed.
- See for the case of polymyxins: V. Balaji, S. S. Jeremiah and P. R. Baliga, Indian J. Med. Microbiol., 2011, 29, 230–242 CrossRef CAS PubMed.
- (a) E. E. Gill, O. L. Franco and R. E. W. Hancock, Chem. Biol. Drug Des., 2015, 85, 56–78 CrossRef CAS PubMed; (b) See also: Cystic Fibrosis Foundation: http://www.cff.org/research/DrugDevelopmentPipeline/.
- E. E. Smith, D. G. Buckley, Z. Wu, C. Saenphimmachak, L. R. Hoffman, D. A. D'Argenio, S. I. Miller, B. W. Ramsey, D. P. Speert, S. M. Moskowitz, J. L. Burns, R. Kaul and M. V. Olson, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 8487–8492 CrossRef CAS PubMed.
- S. Patel, I. P. Sinha, K. Dwan, C. Echevarria, M. Schechter and K. W. Southern, Cochrane Database Syst Rev, 2015, 3, CD009841 Search PubMed.
- (a) R. E. W. Hancock and H.-G. Sahl, Curr. Opin. Microbiol., 2013, 16, 519–521 CrossRef PubMed; (b) G. Sharma, S. Rao, A. Bansal, S. Dang, S. Gupta and R. Gabrani, Biologicals, 2014, 42, 1–7 CrossRef CAS PubMed.
- (a) C. D. Fjell, J. A. Hiss, R. E. W. Hancock and G. Schneider, Nat. Rev. Drug Discovery, 2012, 11, 37–51 CAS; (b) A. A. Bahar and D. Ren, Pharmaceuticals, 2013, 6, 1543–1575 CrossRef PubMed; (c) H. Ulm, M. Wilmes, Y. Shai and H.-G. Sahl, Front. Immunol., 2012, 3, 249 Search PubMed.
- A. L. Hilchie, K. Wuerth and R. E. W. Hancock, Nat. Chem. Biol., 2013, 9, 761–768 CrossRef CAS PubMed.
- M. Zasloff, Nature, 2002, 415, 389–395 CrossRef CAS PubMed.
- J. Overhage, A. Campisano, M. Bains, E. C. W. Torfs, B. H. A. Rehm and R. E. W. Hancock, Infect. Immun., 2008, 76, 4176–4182 CrossRef CAS PubMed.
- (a) H. Jenssen, P. Hamill and R. E. W. Hancock, Clin. Microbiol. Rev., 2006, 19, 491–511 CrossRef CAS PubMed; (b) D. A. Steinberg, M. A. Hurst, C. A. Fujii, A. H. Kung, J. F. Ho, F. C. Cheng, D. J. Loury and J. C. Fiddes, Antimicrob. Agents Chemother., 1997, 41, 1738–1742 CAS.
- P. Jorge, A. Lourenco and M. O. Pereira, Biofouling, 2012, 28, 1033–1061 CrossRef CAS PubMed.
- (a) C. de la Fuente-Núñez, V. Korolik, M. Bains, U. Nguyen, E. B. M. Breidenstein, S. Horsman, S. Lewenza, L. Burrows and R. E. W. Hancock, Antimicrob. Agents Chemother., 2012, 56, 2696–2704 CrossRef PubMed; (b) A. Pompilio, V. Crocetta, M. Scocchic, S. Pomponio, V. Di Vincenzo, M. Mardirossian, G. Gherardi, E. Fiscarelli, R. Gennaro and G. Di Bonaventura, BMC Microbiol., 2012, 12, 145 CrossRef CAS PubMed; (c) S. M. Paranjape, T. W. Lauer, R. C. Montelaro, T. A. Mietzner and N. Vij, F1000Research, 2013, 2, 36 Search PubMed; (d) C. Nagant, B. Pitts, K. Nazmi, M. Vandenbranden, J. G. Bolscher, P. S. Stewart and J.-P. Dehaye, Antimicrob. Agents Chemother., 2012, 56, 5698–5708 CrossRef CAS PubMed; (e) B. Deslouches, I. A. Gonzalez, D. DeAlmeida, K. Islam, C. Steele, R. C. Montelaro and T. A. Mietzner, J. Antimicrob. Chemother., 2007, 60, 669–672 CrossRef CAS PubMed.
- (a) C. de la Fuente-Núñez, S. C. Mansour, Z. Wang, L. Jiang, E. B. M. Breidenstein, M. Elliott, F. Reffuveille, D. P. Speert, S. L. Reckseidler-Zenteno, Y. Shen, M. Haapasalo and R. E. W. Hancock, Antibiotics, 2014, 3, 509–526 CrossRef PubMed; (b) F. Reffuveille, C. de la Fuente-Nunez, S. Mansour and R. E. W. Hancock, Antimicrob. Agents Chemother., 2014, 58, 5363–5371 CrossRef PubMed; (c) A. Pompilio, M. Scocchic, S. Pomponio, F. Guida, A. Di Primio, E. Fiscarelli, R. Gennaro and G. Di Bonaventura, Peptides, 2011, 32, 1807–1814 CrossRef CAS PubMed; (d) C. de la Fuente-Núñez, F. Raffuveille, S. C. Mansour, S. L. Reckseidler-Zenteno, D. Hernández, G. Brackman, T. Coenye and R. E. W. Hancock, Chem. Biol., 2015, 22, 196–205 CrossRef PubMed; (e) J.-L. Reymond, M. Bergmann and T. Darbre, Chem. Soc. Rev., 2013, 42, 4814–4822 RSC.
- (a) S. C. Mansour, C. de la Fuente-Núñez and R. E. W. Hancock, J. Pept. Sci., 2015, 21, 323–329 CrossRef CAS PubMed; (b) C. de la Fuente-Núñez, F. Reffuveille, E. F. Haney, S. K. Straus and R. E. W. Hancock, PLoS Pathog., 2014, 10, e1004152 Search PubMed.
- (a) H. Ulvatne, S. Karoliussen, T. Stiberg, O. Rekdal and J. S. Svendsen, J. Antimicrob. Chemother., 2001, 48, 203–208 CrossRef CAS PubMed; (b) A. A. Bahar, Z. Liu, F. Totsingan, C. Buitrago, N. Kallenbach and D. Ren, Appl. Microbiol. Biotechnol., 2015, 99, 8125–8135 CrossRef CAS PubMed.
- M. Wenzel, A. I. Chiriac, A. Otto, D. Zweytick, C. May, C. Schumacher, R. Gust, H. B. Albada, M. Penkova, U. Krämer, R. Erdmann, N. Metzler-Nolte, S. K. Straus, E. Bremer, D. Becher, H. Brötz-Oesterhelt, H.-G. Sahl and J. E. Bandow, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, E1409–E1418 CrossRef CAS PubMed.
- H. B. Albada, A.-I. Chiriac, M. Wenzel, M. Penkova, J. E. Bandow, H.-G. Sahl and N. Metzler-Nolte, Beilstein J. Org. Chem., 2012, 8, 1753–1764 CrossRef CAS PubMed.
- H. B. Albada, P. Prochnow, S. Bobersky, J. E. Bandow and N. Metzler-Nolte, Chem. Sci., 2014, 5, 4453–4459 RSC.
- S. Hou, Z. Liu, A. W. Young, S. L. Mark, N. R. Kallenbach and D. Ren, Appl. Environ. Microbiol., 2010, 76, 1967–1974 CrossRef CAS PubMed.
- H. B. Albada, P. Prochnow, S. Bobersky, S. Langklotz, P. Schriek, J. E. Bandow and N. Metzler-Nolte, ACS Med. Chem. Lett., 2012, 3, 980–984 CrossRef CAS PubMed.
- H. B. Albada, P. Prochnow, S. Bobersky, S. Langklotz, J. E. Bandow and N. Metzler-Nolte, ACS Comb. Sci., 2013, 15, 585–592 CrossRef CAS PubMed.
- (a) Z. Tong, Y. Zhang, J. Ling, J. Ma, L. Huang and L. Zhang, PLoS One, 2014, 9, e89209/1–e89209/9 CAS; (b) Y. Zhang, Y. Liu, Y. Sun, Q. Liu, X. Wang, Z. Li and J. Hao, Curr. Microbiol., 2014, 68, 685–692 CrossRef CAS PubMed; (c) R. Gopal, Y. G. Kim, J. H. Lee, S. K. Lee, J. D. Chae, B. K. Son, C. H. Seo and Y. Park, Antimicrob. Agents Chemother., 2014, 58, 1622–1629 CrossRef PubMed.
- (a) C. Bozkurt-Guzel, P. B. Savage and A. A. Gerceker, Chemotherapy, 2011, 57, 505–510 CrossRef CAS PubMed; (b) M. Berditsch, T. Jäger, N. Strempel, T. Schwartz, J. Overhage and A. S. Ulrich, Antimicrob. Agents Chemother., 2015 DOI:10.1128/AAC.00682-15 , Just Accepted.
- Clinical and Laboratory Standards Institute, Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement, CLSI document M100–S22, 2012 Search PubMed.
- Clinical and Laboratory Standards Institute. Approved standard: M7–A7, Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th edn. Clinical and Laboratory Standards Institute, Wayne, PA (USA), 2006 Search PubMed.
- R. L. White, D. S. Burgess, M. Manduru and J. A. Bosso, Antimicrob. Agents Chemother., 1996, 40, 1914–1918 CAS.
- M. J. Hall, R. F. Middleton and D. Westmacott, J. Antimicrob. Chemother., 1983, 11, 427–433 CrossRef CAS PubMed.
- J. Moody, in Clinical Microbiology Procedures Handbook, Synergism testing: broth microdilution checkerboard and broth macrodilution methods, ed. L. S. Garcia and H. D. Isenberg, ASM Press, Washington, DC (USA), 2nd edn, 2007, ch 5.12, pp. 5.12.1–5.12.23 Search PubMed.
- K. C. Stover, X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory and M. V. Olson, Nature, 2000, 406, 959–964 CrossRef PubMed.
- U. Römling, A. Kader, D. D. Sriramulu, R. Simm and G. Kronvall, Environ. Microbiol., 2005, 7, 1029–1038 CrossRef PubMed.
- (a) P. Salunkhe, C. H. M. Smart, J. A. W. Morgan, S. Panagea, M. J. Walshaw, C. A. Hart, R. Geffers, B. Tümmler and C. Winstanley, J. Bacteriol., 2005, 187, 4908–4920 CrossRef CAS PubMed; (b) M. E. K. Carter, J. L. Fothergill, M. J. Walshaw, K. Rajakumar, A. Kadioglu and C. Winstanley, J. Infect. Dis., 2010, 202, 935–943 CrossRef PubMed.
- C. H. Smart, F. W. Scott, E. A. Wright, M. J. Walshaw, C. A. Hart, T. L. Pitt and C. Winstanley, J. Med. Microbiol., 2006, 55, 1085–1091 CrossRef CAS PubMed.
- (a) I. Kukavica-Ibrulj, A. Bragonzi, M. Paroni, C. Winstanley, F. Sanschagrin, G. A. O'Toole and R. C. Levesque, J. Bacteriol., 2008, 190, 2804–2813 CrossRef CAS PubMed; (b) C. Winstanley, M. G. I. Langille, J. L. Fothergill, I. Kukavica-Ibrulj, C. Paradis-Bleau, F. Sanschagrin, N. R. Thomas, G. L. Winsor, M. A. Quail, N. Lennard, A. Bignell, L. Clarke, K. Seeger, D. Saunders, D. Harris, J. Parkhill, R. E. W. Hancock, F. S. L. Brinkman and R. C. Levexque, Genome Res., 2009, 19, 12–23 CrossRef CAS PubMed; (c) See also ref. 37b.
- (a) P. Lin, Y. Li, K. Dong and Q. Li, Curr. Microbiol., 2015, 71, 170–176 CrossRef CAS PubMed; (b) W. Xu, X. Zhu, T. Tan, W. Li and A. Shan, PLoS One, 2015, 9, e98935 Search PubMed; (c) S. N. Dean, B. M. Bishop and M. L. van Hoek, BMC Microbiol., 2011, 11, 114 CrossRef CAS PubMed.
- (a) H. D. M. Coutinho, V. S. Falcão-Silva and G. F. Gonçalves, Int. Arch. Med., 2008, 1, 24 CrossRef PubMed; (b) See also: Cystic Fibrosis Foundation: Patient Registry Annual Data Report, 2013 Search PubMed.
- P. J. Bergen, C. B. Landersdorfer, J. Zhang, M. Zhao, H. J. Lee, R. L. Nation and J. Li, Diagn. Microbiol. Infect. Dis., 2012, 74, 213–223 CrossRef CAS PubMed.
- E. F. Haney, S. C. Mansour, A. L. Hilchie, C. de la Fuente-Núñez and R. E. W. Hancock, Peptides, 2015, 71, 276–285 CrossRef CAS PubMed.
Footnotes |
| † Author contributions: MGdG, NMN, and HBA designed research, analysed data, and wrote the paper; MJ and HGS conducted the experiments detailed in Table 1; MGdG conducted the synergism experiments (growth inhibition, growth curves and biofilm formation inhibition); RW, HL, JWJL, and BD designed and assisted in the synergistic activity studies; FLP conducted the confocal microscopy studies; and RvM selected the Pseudomonas aeruginosa strains and assisted in the MIC-studies described in Table 2. |
| ‡ Electronic supplementary information (ESI) available. See DOI: 10.1039/c5md00373c |
| This journal is © The Royal Society of Chemistry 2016 |