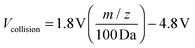Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Glycomic profiling of targeted serum haptoglobin for gastric cancer using nano LC/MS and LC/MS/MS†
Sung Hyeon
Lee
a,
Seunghyup
Jeong
bc,
Jua
Lee
bc,
In Seok
Yeo
 d,
Myung Jin
Oh
bc,
Unyong
Kim
bc,
Sumin
Kim
bc,
Su Hee
Kim
a,
Seung-Yeol
Park
e,
Jae-Han
Kim
f,
Se Hoon
Park
g,
Jung Hoe
Kim‡
*d and
Hyun Joo
An‡
*bc
d,
Myung Jin
Oh
bc,
Unyong
Kim
bc,
Sumin
Kim
bc,
Su Hee
Kim
a,
Seung-Yeol
Park
e,
Jae-Han
Kim
f,
Se Hoon
Park
g,
Jung Hoe
Kim‡
*d and
Hyun Joo
An‡
*bc
aGLYCAN Co., Ltd., Healthcare Innovation Park, 172 Dolma-ro, Bundang-gu, Seongnam 13605, Korea
bAsia-pacific Glycomics Reference Site, Daejeon, Korea
cGraduate School of Analytical Science and Technology, College of Engineering II, Chungnam National University, 99 Daehak-ro, Yuseong-gu, Daejeon 305-764, Republic of Korea. E-mail: hjan@cnu.ac.kr; Fax: +82-42-821-8551
dDepartment of Biological Sciences, Korea Advanced Institute of Science and Technology, Daehak-ro, Yuseong-gu, Daejeon 305-701, Republic of Korea. E-mail: kimjh@kaist.ac.kr; Fax: +82-42-350-2690
eDivision of Rheumatology, Immunology, and Allergy, Brigham and Women's Hospital, and the Department of Medicine, Harvard Medical School, Boston, Massachusetts 02115, USA
fDepartment of Food and Nutrition, Chungnam National University, Daejeon, Korea
gDivision of Hematology-Oncology, Department of Medicine, Sungkyunkwan University Samsung Medical Center, Seoul, Korea
First published on 28th September 2016
Abstract
Gastric cancer has one of the highest cancer mortality rates worldwide, largely because of difficulties in early-stage detection. Aberrant glycosylation in serum proteins is associated with many human diseases including inflammation and various types of cancer. Serum-based global glycan profiling using mass spectrometry has been explored and has already led to several potential glycan markers for several disease states. However, localization of the aberrant glycosylation is desirable in order to improve the specificity and sensitivity for clinical use. Here, we combined protein-specific immunoaffinity purification, glycan release, and MS analysis to examine haptoglobin glycosylation of gastric cancer patients for glyco-markers. Age- and sex-matched 60 serum samples (30 cancer patients and 30 healthy controls) were used to profile and quantify haptoglobin N-glycans. A T-test based statistical analysis was performed to identify potential glyco-markers for gastric cancer. Interestingly, abundances of several tri- and tetra-antennary fucosylated N-glycans were increased in gastric cancer patients. Additionally, structural analysis via LC/MS/MS indicated that the fucosylated complex type N-glycans were primarily decorated with antenna fucose, which can be categorized as sialyl-Lea or sialyl-Lex type structures. This platform demonstrates quantitative, structure-specific profiling of haptoglobin glycosylation for the purposes of biomarker discovery for gastric cancer.
Introduction
Gastric cancer is one of the most commonly occurring cancers and has the highest overall mortality rate worldwide because it has no specific symptoms in its early stage.1 Reported causative factors of gastric cancer include Helicobacter pylori infection, cigarette smoking, and diet.2 Serum markers such as a pepsinogen I/II ratio, carbohydrate antigen 19-9 (CA19-9), and carcinoembryonic antigen (CEA) have been widely used for gastric disorders in clinics.3,4 However, the serum-based tests are limited in the diagnosis of gastric cancer due to their lack of sensitivity and specificity. For example, the ratio of serum pepsinogen I/II usually indicates levels of atrophic gastritis as well as gastric cancer, which could be changeable according to the infection of H. pylori.5 CA19-9 and CEA are applicable only for the recurrence of gastric cancer, not for the initial diagnosis.6 Therefore, many researchers are now on the hunt for new types of biomarkers for gastric cancer diagnostic tests.7–10Glycosylation, which plays a central role in many biological processes is of particular interest because about 50% of all proteins are glycosylated.11,12 Glycans are synthesized by sequential processes involving more than 200 glycosyltransferases following covalent attachment of glycans to a carrier molecule (e.g., polypeptide, lipid, or other organic compound).13 Glycosylation changes have been reported in a wide variety of human diseases, including immune disorders and cancers.14–16 While general profiling of serum N-glycome has already revealed potential biomarkers for various types of cancer,17–19 a targeted glycoproteomic approach is needed for cancer diagnosis with high sensitivity and high specificity.20–24 For example, AFP-L3 (core fucose of alpha-fetoprotein) has been discovered as a specific glycosyl residue for hepatocellular carcinoma (HCC) with increased sensitivity in early stage HCC patients compared to AFP, which is used in clinical practice.25–27
Haptoglobin consisting of two α-subunits and two β-subunits is a major acute phase glycoprotein comprising of 0.4–2.6% total blood proteins.28 This glycoprotein binds hemoglobin released from erythrocytes and recovers an oxidative activity degrading hemoglobin–haptoglobin complex in the liver.29 It also has catalytic and antibacterial capacities in controlling the acute-phase response and plays a role as a natural antagonist for receptor–ligand activation in the immune system.30,31 Haptoglobin is a highly sialylated glycoprotein with four N-glycosylation sites at Asn 184, 207, 211, and 241.32,33 It has gained considerable attention because of its potential as a signature molecule to display aberrant glycosylation in inflammatory disorders (e.g., liver cirrhosis, pancreatitis) and various types of cancer (e.g., of colon, lung, liver, prostate and pancreas).34–45
In this study, we developed a targeted glycoproteomic approach using chip-based nano LC-QTOF MS and MS/MS following antibody-assisted targeted purification to discover glycan signatures of serum haptoglobin for gastric cancer. Initially, we performed lectin blotting of whole serum glycoproteins. Thus, haptoglobin from cancer serum samples displayed significantly higher levels of fucosylation and branching, indicating that this glycoprotein was associated with gastric cancer. Haptoglobin was selected for more detailed examination, and was thus purified from healthy control (n = 30) and gastric cancer patient (n = 30) serum samples using immunoaffinity chromatography. Haptoglobin glycans were enzymatically released, enriched, and profiled via PGC nanoLC/MS and nanoLC/MS/MS. T-test based statistical analysis was performed to identify the potential glycan signatures for gastric cancer. MS profiling of haptoglobin N-glycans was consistent with lectin blotting results. We could further obtain a specific structure of fucosylated molecules that are potential glyco-markers for gastric cancer via LC/MS/MS. The current study demonstrates that glycomic profiling of targeted serum haptoglobin via LC/MS and LC/MS/MS may be used as a powerful platform to monitor the specific glycosylation associated with gastric cancer.
Experimental
Materials and reagents
Four biotinylated lectins were purchased from Vector Laboratories (Burlingame, CA, USA). Commercial human serum and ExtrAvidin Peroxidase were purchased from Sigma (St. Louis, MO, USA). The Rabbit anti-human Hp antibody (Ab) was obtained from Dako (Carpinteria, CA, USA). Goat anti-rabbit IgG conjugated with horseradish peroxidase (HRP) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). CNBr-activated Sepharose 4B was acquired from Amersham Bioscience (Minneapolis, MN, USA). Peptide N-glycosidase F was purchased from New England Biolabs (Ipswich, Suffolk, UK). Graphitized carbon cartridges were purchased from Thermo Fisher Scientific (Runcorn, Cheshire, UK). MS calibrant solution (ESI-TOF Calibrant Mix G1969-85000) was procured from Agilent Technologies (Santa Clara, CA, USA). All other reagents were of analytical grade or higher.Serum samples from gastric cancer patients and normal subjects
Serum samples of patients with gastric cancer stage IV (n = 30) and healthy control (n = 30) were used (Table S1, ESI†). To explore the target glycoprotein for gastric cancer, a small set of serum samples were used (gastric cancer patients each at stage I–IV (n = 4) and a healthy control (n = 2)). The research design was approved by the Ethics Committees of the participating hospitals, Samsung Medical Hospital, Seoul, Republic of Korea (IRB# SMC2015-07-146-001) and Korea Advanced Institute of Science and Technology, Daejeon, Republic of Korea (IRB# KH2010-15). To determine the stage and type of cancer, the patients were examined and diagnosed on the basis of a biopsy by pathologists. Informed consent forms were obtained from all of the subjects. Serum samples were kept at an −80 °C freezer until processing.Lectin blotting and western blotting
Serum samples of 0.5 μL was subjected to electrophoresis on 10% and 12.5% SDS-PAGE gels, transferred to a PVDF membrane (Millipore), and blotted with each of the four biotinylated lectins displayed in Fig. 1C. The membrane was blocked with 5% BSA in T-TBS (TBS [140 mM NaCl, 10 mM Tris-HCl, pH 8.0]/0.05% Tween 20) for 1 hour at RT, rinsed 3 times with T-TBS for 5 minutes, incubated with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000–5000 diluted biotinylated lectin in T-TBS overnight in a cold room, rinsed 3 times with T-TBS, incubated with 1
1000–5000 diluted biotinylated lectin in T-TBS overnight in a cold room, rinsed 3 times with T-TBS, incubated with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000 diluted ExtrAvidin-Peroxidase for 1 hour at RT, rinsed 3 times with T-TBS, and developed using an ECL Supersignal kit (Pierce ECL Western Blotting Substrate, Thermo Science; Rockford, IL, USA). For loading quantification of haptoglobin, the same blots after lectin blotting were subjected to reblotting with anti-Hp Ab. The membrane was incubated with a stripping buffer (Candor Bioscience GmbH; Weissensberg, Germany) at RT for 1 hour, rinsed 5 times with T-TBS, blocked with 5% BSA for 1 hour, immunoblotted with 1
3000 diluted ExtrAvidin-Peroxidase for 1 hour at RT, rinsed 3 times with T-TBS, and developed using an ECL Supersignal kit (Pierce ECL Western Blotting Substrate, Thermo Science; Rockford, IL, USA). For loading quantification of haptoglobin, the same blots after lectin blotting were subjected to reblotting with anti-Hp Ab. The membrane was incubated with a stripping buffer (Candor Bioscience GmbH; Weissensberg, Germany) at RT for 1 hour, rinsed 5 times with T-TBS, blocked with 5% BSA for 1 hour, immunoblotted with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 diluted rabbit anti-Hp Ab overnight in a cold room, rinsed 3 times with T-TBS, incubated with 1
000 diluted rabbit anti-Hp Ab overnight in a cold room, rinsed 3 times with T-TBS, incubated with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 diluted goat anti-rabbit IgG-HRP for 1 hour, rinsed 3 times with T-TBS, and developed with ECL solution.
5000 diluted goat anti-rabbit IgG-HRP for 1 hour, rinsed 3 times with T-TBS, and developed with ECL solution.
In-gel protein digestion
The protein bands of interest were excised and digested in-gel with sequencing-grade modified trypsin (Promega; Madison, WI, USA) as previously described.46 Briefly, the gel band was placed in a polypropylene (Eppendorf) tube, and washed 5 times with 150 μL of acetonitrile/25 mM ammonium bicarbonate (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), pH 7.8. The gel slices were dried in a SpeedVac concentrator and rehydrated in 30 μL of 25 mM ammonium bicarbonate, pH 7.8, containing 20 ng trypsin. The liquid was incubated for 20 hours at 37 °C and then transferred to a new tube. The tryptic peptides remaining in the gel matrix were extracted for 40 minutes at 30 °C with 20 μL of 50% aqueous acetonitrile containing 0.1% formic acid. The combined supernatants were evaporated in a SpeedVac and dissolved in 8 μL of 5% aqueous acetonitrile solution containing 0.1% formic acid for undergoing MS analysis.
1), pH 7.8. The gel slices were dried in a SpeedVac concentrator and rehydrated in 30 μL of 25 mM ammonium bicarbonate, pH 7.8, containing 20 ng trypsin. The liquid was incubated for 20 hours at 37 °C and then transferred to a new tube. The tryptic peptides remaining in the gel matrix were extracted for 40 minutes at 30 °C with 20 μL of 50% aqueous acetonitrile containing 0.1% formic acid. The combined supernatants were evaporated in a SpeedVac and dissolved in 8 μL of 5% aqueous acetonitrile solution containing 0.1% formic acid for undergoing MS analysis.
Identification of proteins via LC-MS/MS
Tryptic peptides were analyzed using reversed-phase capillary HPLC coupled to a Finnigan LCQ ion trap mass spectrometer. A 0.1 × 20 mm trapping column and 0.075 × 130 mm resolving column were packed with Vydac 218 MS low trifluoroacetic acid C18 beads (diameter 5 μm, pore size 300 Å; Vydac, Hesperia, CA, USA). The peptides were captured by the trapping column for 10 minutes with 5% aqueous acetonitrile containing 0.1% formic acid in water (v/v), and the captured peptides were then eluted with a gradient of 5–80% acetonitrile containing 0.1% formic acid for 50 minutes at a flow rate of 0.2 μL min−1. For MS/MS, the full mass scan range mode was m/z = 450–2000 Da. The charge states of the ions were determined on zoom scans, and the product ion spectra were generated in the MS/MS mode with a relative collision energy of 55%. The MS/MS spectra were processed using the TurboSEQUEST software program (Thermoquest, San Jose, CA). The acquired peak list files were compared with the National Center for Biotechnology Information (NCBI) database and the Microsatellite Search and Building Database (MSDB) using the MASCOT program (http://www.matrixscience.com).Haptoglobin purification from the serum samples
Haptoglobin purification was performed using an anti-haptoglobin antibody-conjugated column.42,47 In brief, 450 μL serum from each subject was diluted in 4.5 mL phosphate-buffered saline (PBS: 10 mM phosphate buffer, 2.7 mM potassium chloride, 137 mM sodium chloride, pH 7.4), and then applied to the anti-haptoglobin affinity column. Unbound components were removed by washing the column with 30 mL PBS, haptoglobin was then eluted with elution buffer (0.1 M glycine, 0.5 M NaCl, pH 2.8) and fractionated into a tube containing neutralization buffer (1.0 M Tris-HCl, pH 9.0). The eluent was concentrated, the detergent was removed using a centrifugal filter (MWCO 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, Amicon Ultra, Millipore; Billerica, MA, USA), and assayed for haptoglobin quantification using a Quant-iT Assay Kit (Invitrogen; Carlsbad, CA, USA). To check the existence and purity of the haptoglobin, the eluent was randomly applied to a 12.5% SDS-PAGE with Coomassie Brilliant Blue staining. Each sample was lyophilized and kept at −80 °C until analysis.
000, Amicon Ultra, Millipore; Billerica, MA, USA), and assayed for haptoglobin quantification using a Quant-iT Assay Kit (Invitrogen; Carlsbad, CA, USA). To check the existence and purity of the haptoglobin, the eluent was randomly applied to a 12.5% SDS-PAGE with Coomassie Brilliant Blue staining. Each sample was lyophilized and kept at −80 °C until analysis.
Enzymatic release and enrichment of N-glycan via solid-phase extraction
Serum haptoglobin was denatured via rapid thermal cycling (25–100 °C) in an aqueous solution of 100 mM ammonium bicarbonate and 5 mM dithiothreitol. After cooling, 2.0 μL (or 1000 U) of peptide N-glycosidase F (PNGaseF) was added to release the N-glycans, and the mixture was incubated in a 37 °C water bath for 16 hours. Then, graphitized carbon cartridges were washed with 80% acetonitrile/0.10% trifluoroacetic acid (v/v) and conditioned with water. The released N-glycans were loaded onto the cartridges and washed with pure water to remove any salt and buffer residues. N-glycans were eluted through the sequential addition of 10% and 20% acetonitrile (v/v) (neutral fraction) followed by 40% acetonitrile/0.05% trifluoroacetic acid in water (v/v) (acidic fraction). The samples were dried under a vacuum prior to MS analysis.Chip-based nano-LC/MS and MS/MS analysis
All fractions were combined and 2.0 μL (corresponding to 400 ng haptoglobin) were injected by an autosampler onto a porous graphitized carbon (PGC) nano-LC chip (Agilent). A rapid glycan elution gradient was applied to the analytical column at 0.4 μL min−1 using solutions of (A) 3.0% acetonitrile/0.3% formic acid in water (v/v), and (B) 90.0% acetonitrile/0.3% formic acid in water (v/v), ramping from 6% to 100% B over the course of 40 minutes. The remaining non-glycan compounds were flushed out with 100% B followed by re-equilibration in the final 10 minutes. MS spectra were acquired in a positive ion mode over a mass range of m/z 500–2000 with an acquisition time of 1.6 seconds per spectrum. MS/MS spectra were acquired in the positive ion mode over a mass range of m/z 100–3000 with an acquisition time of 1.6 seconds per spectrum. After the MS scan, precursor ions were selected automatically for MS/MS analysis using the acquisition software based on ion abundance and charge stage (z = 2 or 3), and isolated in the quadrupole with a mass bandpass full width at a half maximum of 1.3 m/z. Collison energies for CID fragmentation were calculated for each precursor compound based on the following equation:Here, Vcollision is the voltage applied across the collision cell in order to accelerate and fragment the precursor.
N-glycan identification and data processing
Raw LC/MS data was filtered with a signal-to-noise ratio of 5.0 and analyzed into a series of extracted ion chromatograms using the Molecular Feature Extractor algorithm included in the MassHunter Qualitative Analysis software (version B.07.00 SP1, Agilent Technologies). Deconvoluted masses of each ECC peak were compared against theoretical glycan masses using a mass error tolerance of 20 ppm. As all the samples originated from human serum, only the glycan compositions consisting of hexose (Hex), N-acetylhexosamine (HexNAc), fucose (Fuc), and N-acetylneuraminic acid (NeuAc) were considered. Using Hierarchical Clustering Explorer (ver.3.5), the Pearson correlation coefficient of haptoglobin derived from commercial sera was calculated. As a statistical test, individual T-test analysis using MS Excel 2013 (Microsoft) and GraphPad Prism (ver.5) was used to examine the quantitative variables of glycan expression between the control and gastric cancer groups.48–50p-Values were applied with 2-tailed analysis and the difference between the two groups was considered to be statistically significant as the p-values were <0.001. The sample variables were measured using mean ± standard error.Results and discussion
Serum haptoglobin is a well-known glycoprotein, having altered glycosylation in various diseases and cancers including human gastrointestinal (GI) cancers (e.g., colonic, pancreatic cancer).43–45 To explore the relationships between haptoglobin glycosylation and gastric cancer, N-glycans of serum haptoglobin were profiled using the streamlined method depicted in Fig. S1 (ESI†). The main workflow includes several steps: (i) purification of serum haptoglobin via immunoaffinity chromatography in order to collect only the target glycoprotein in the serum samples; (ii) PNGaseF treatment to selectively release N-glycans; (iii) desalting in order to prevent any ion suppression during MS analysis, and enrichment of N-glycans via solid-phase extraction using a graphitized carbon cartridge; (iv) N-glycan analysis via PGC nano-LC/MS and LC/MS/MS for detailed structural elucidation; and (v) the discovery of specific glycan signatures for gastric cancer.Glycosylation screening and purification of serum haptoglobin
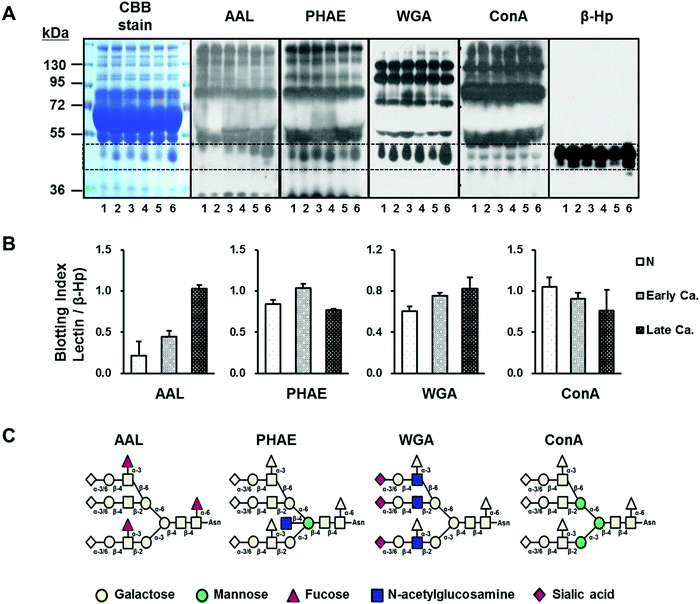
Serum glycoproteins from gastric cancer patients were initially profiled via lectin blotting for overarching glycosylation changes with small sample sets (patient, n = 4; control n = 2) (Fig. 1). Four lectins recognizing each particular monosaccharide residues were used in this study (Fig. 1C). Initial lectin blotting of whole serum glycoproteins revealed that although the overall protein abundances were similar in the control and cancer serum samples, specific proteins from the cancer serum samples were highly expressed in specific lectins. Indeed, one band with a molecular weight of ∼45 kDa (the dotted box in Fig. 1A), which later proved to be the β-chain of haptoglobin, was significantly higher upon AAL lectin signals in the cancer patients vs. the controls. The AAL lectin recognizes fucose residues which are linked to GlcNAc by α1-3/4/6 glycosidic bonds associated with Lewis blood group antigens.51 Through further protein identification using LC-MS/MS following in-gel digested by trypsin, the band having ∼45 kDa to display glycosylation changes in gastric cancer was revealed as the β-chain of haptoglobin with significant hits in a peptide database search (Table S2, ESI†). Additionally, we could confirm the expression of the haptoglobin β-chain in the gastric cancer and control sera by stripping and reblotting the membrane with anti-haptoglobin antibody. Based on the lectin specificity and protein identification, our finding indicates that aberrant glycosylation in serum haptoglobin was related to gastric cancer, which displayed significantly higher levels of fucosylated N-glycans.Glycosylation of haptoglobin was further targeted for more in-depth examination using larger samples from gastric cancer patients. Serum haptoglobin derived from healthy controls (n = 30) and gastric cancer patients (n = 30) were purified via antibody-immobilized affinity chromatography (Fig. S2, ESI†) and further subjected to N-glycan profiling. As shown in Table S1 (ESI†), stage IV gastric cancer patients with adenocarcinoma types were selectively examined for the discovery of evident glycan signatures compared with healthy control. Age and sex in both groups were also matched. In parallel, haptoglobin derived from commercial sera (n = 7) was also prepared to assure the sample preparation including immunoaffinity purification, glycan release, and MS analysis. Haptoglobin 50 μg was used for the subsequent glycan sample preparation including glycan release and enrichment.
LC/MS-based N-glycan profiling of serum haptoglobin
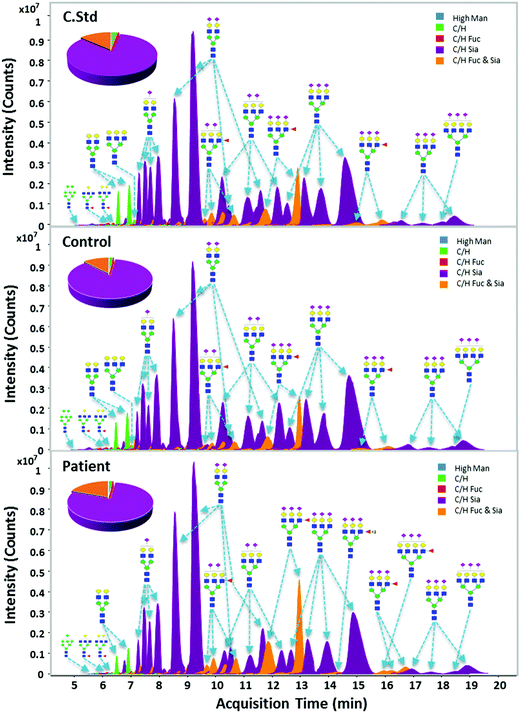
Fig. 2 shows representative extracted compound chromatograms (ECCs) of haptoglobin N-glycans found in different serum samples including the commercial standard, healthy control, and gastric cancer patient. Purified haptoglobin derived from commercial sera was analyzed with the clinical samples to validate the sample preparation and MS analysis. Commercial haptoglobin glycans analyzed using chip-based nano-LC/MS exhibited extremely high reproducibility with a Pearson correlation of R > 0.99 on whole method replicates (Fig. S3, ESI†). N-glycans released from serum haptoglobin were separated by their structure and polarity using PGC chip nanoLC prior to MS detection. The flow rate of 0.3 μL min−1, solutions of (A) 3.0% acetonitrile/0.1% formic acid in water (v/v), and (B) 90.0% acetonitrile/0.1% formic acid in water (v/v) have been generally used for N-glycan analysis with a PGC chip.52 However, this condition is not suitable for serum haptoglobin, which mainly contains highly sialylated N-glycans. For better separation, chromatographic conditions including the flow rate, concentration of formic acid, and LC gradient, have been appropriately optimized. (See the ESI† for details). In the case of N-glycans of haptoglobin in our nanoLC/MS analysis, smaller, and simpler glycans were eluted earlier, while larger, more complex glycans were eluted later. In other words, high mannose glycans are eluted earlier followed by neutral complex glycans and sialylated complex glycans.Native glycans were reproducibly profiled and characterized via chip-based nano-LC/MS and -LC/MS/MS. Using established human glycan structure/composition libraries, glycan signals were rapidly identified and sorted into biologically relevant classes and categories, such as high-mannose glycans, hybrid glycans, fucosylated or sialylated complex glycans, etc.53 Structure-sensitive N-glycan profiling identified hundreds of glycan peaks per haptoglobin, including multiple isomers for most compositions. The ECCs in all serum samples (7 commercial standards, 30 healthy controls, and 30 cancer patients) represent a qualitatively similar chromatographic distribution (Fig. 2). Most haptoglobin exhibited high levels (95.7 to 96.8% relative abundance) of bi- and tri-antennary sialylation, an established hallmark of haptoglobin glycosylation. Interestingly, 5 major compositions quantitatively occupied over 80% of the total haptoglobin glycans. All observed glycan signals were normalized through total ion intensity for a more direct comparison. The top 35 glycans corresponding to 99% of the total glycan quantity were selected for further analysis of glycosylation changes between the gastric cancer and control groups (Table S3, ESI†).
Glycan changes of haptoglobin in patients with gastric cancer
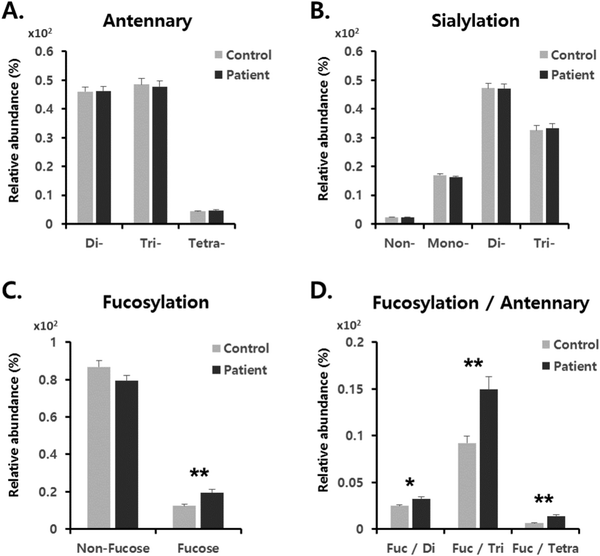
From the perspective of glycan branching and decoration related to cancer,15 N-glycans of serum haptoglobin were grouped together by the following criteria: (1) branching; (2) sialylation; and (3) fucosylation (Fig. 3). The degree of glycan branching (from bi- to tetra-antennae) as well as sialylation (from non- to three-NeuAc) showed no difference in haptoglobin N-glycans between the gastric cancer and healthy control (Fig. 3A and B). On the other hand, as shown in Fig. 3C, relative abundances of fucosylated glycans possessing mainly one fucose reside were increased in the gastric cancer patients (12.2% to 19.5%), while non-fucosylated N-glycans were reduced in the gastric cancer patients (86.5% to 79.0%). Accordingly, we decided to focus on fucosylation and considered a synergistic effect of haptoglobin glycosylation in patients with gastric cancer. Interestingly, abundances of fucosylation in accordance with antenna were significantly increased (1.63 fold in tri-: p = 0.00002, and 2.24 fold in tetra-antennary: p = 0.00004) as shown in Fig. 3D. This MS-based exploration of haptoglobin N-glycan clearly supports the preliminary serum glycan screening data showing increased binding to AAL lectin in gastric cancer patients (Fig. 1). Therefore, these results indicate that the increase of fucosylation with highly branched glycans of serum haptoglobin is the key signature associated with gastric cancer.For a more direct comparison of glycan expressions between the gastric cancer and control groups, an individual T-test was performed using 35 major N-glycans selected from commercial haptoglobin glycans having 100% frequency (presence). In particular, eleven N-glycan compositions were found to increase in abundance in patients with gastric cancer (Table 1). They were only-sialylated and/or fucosylated & sialylated N-glycans, which have mostly branched structures (up to four antenna) and decorated with fucosylation and sialylation. These alterations of the N-glycans were in good agreement with the general feature of malignant cells synthesizing larger N-glycan chains.15,54 Taking into consideration our results (Fig. 1, 3 and Table 1), especially the fucosylation of serum haptoglobin, might be an important key related to gastric cancer. Fucosylation is an important modification notably associated with cancer55 and can be catalyzed by several fucosyltransferases (FUTs) in the Gogi apparatus.51 For example, an increase of α1-3/4 fucosylation reflecting the activation of FUT3-7 has been associated with colon cancer progression.43,56 Furthermore, increased core fucosylation catalyzed by α1-6 FUT8 has been demonstrated in AFP, which is used as a hepatocellular carcinoma (HCC) biomarker, and the fucosylated AFP (AFP-L3) is nowadays used for HCC risk assessment.25 These previous reports support our finding that enhanced fucosylation is related to gastric cancer (Fig. 3).
| Glycan composition | Healthy controls | Cancer patients | T-test | |||||
|---|---|---|---|---|---|---|---|---|
| Hex | HexNAc | Fuc | NeuAc | Avg (%) | Ste | Avg (%) | Ste | (p < 0.001) |
| Complex/hybrid (only-sialylated) | ||||||||
| 4 | 3 | 0 | 1 | 0.196 | 0.011 | 0.241 | 0.018 | 5.6 × 10−4 |
| 6 | 4 | 0 | 1 | 0.274 | 0.014 | 0.320 | 0.020 | 6.0 × 10−4 |
| Complex/hybrid (sialylated & fucosylated) | ||||||||
| 5 | 4 | 1 | 1 | 0.675 | 0.037 | 0.955 | 0.082 | 7.0 × 10−5 |
| 5 | 4 | 1 | 2 | 1.459 | 0.078 | 1.885 | 0.166 | 7.7 × 10−4 |
| 6 | 5 | 1 | 1 | 0.220 | 0.020 | 0.326 | 0.033 | 4.3 × 10−4 |
| 6 | 5 | 1 | 2 | 1.869 | 0.188 | 3.320 | 0.336 | 2.1 × 10−5 |
| 6 | 5 | 1 | 3 | 5.983 | 0.510 | 9.898 | 0.924 | 1.8 × 10−5 |
| 7 | 5 | 2 | 2 | 0.128 | 0.014 | 0.221 | 0.020 | 1.3 × 10−5 |
| 7 | 6 | 1 | 1 | 0.063 | 0.007 | 0.114 | 0.014 | 2.1 × 10−4 |
| 7 | 6 | 1 | 2 | 0.237 | 0.024 | 0.469 | 0.056 | 3.9 × 10−5 |
| 7 | 6 | 1 | 3 | 0.319 | 0.043 | 0.835 | 0.120 | 3.1 × 10−5 |
Characteristic structural features of haptoglobin N-glycans
To obtain structural information of haptoglobin N-glycans, tandem MS via collision-induced dissociation (CID) was conducted. A representative MS/MS spectrum of tri-sialylated mono-fucosylated tri-antennary N-glycan consisting of Hex6HexNAc5Fuc1NeuAc3 (m/z 1009.36 [M + 3H]3+) is shown in Fig. 4. This is the most abundant species in the fucosylated & sialylated N-glycans and shows a stronger significance in differentiating gastric cancer. Generally, specific fragment ions representing Hex (m/z 163.06), HexNAc (m/z 204.08), NeuAc (m/z 292.1), and HexHexNAc (m/z 366.13) were used as the reporter ions of N-glycan due to their common observation in every type of N-glycans. Finally, energy-resolved fragment peaks (m/z 350.12, 512.19, and 1057.39) are specified for core vs. antennal fucosylation. The fragment ions possessing antenna fucose such as m/z 512.19 (Hex1HexNAc1Fuc1) and m/z 350.12 (HexNAc1Fuc1) had relatively high intensities compared with m/z 1057.39 (Hex3HexNAc2Fuc1) containing core fucose which was elusive in MS/MS spectrum, indicating the dominating presence of fucose residues attached to the outer arm GlcNAc (antennary fucosylation). To determine the location of fucose in other glycans with different antennary structures, additional tandem MS was performed on di-sialylated mono fucosylated bi-antennary N-glycan (Hex5HexNAc4Fuc1NeuAc2, m/z 1185.4277 [M + 2H]2+) and tri-sialylated mono fucosylated tetra-antennary N-glycan (Hex7HexNAc6Fuc1NeuAc3, m/z 1131.0742 [M + 3H]3+), respectively. Ion fragments for antennary fucosylation corresponding to m/z 512.19, not for core fucosylation, were clearly observed in both the bi- and tetra-antennary glycan structures (Fig. S4, ESI†). Attachment of a Fuc residue to the outer arm GlcNAc could result in the formation of a Lewis epitope.51 Interestingly, through fragment ion analysis of the haptoglobin N-glycan precursor, we could unearth the SLea or SLex epitope-specific signature ion corresponding to Hex1HexNAc1Fuc1NeuAc1 (m/z 803.29). Increased levels of an SLex epitope from the gastric cancer patients57 and enhanced expression levels of Sialyl Lewis antigen-related molecules (e.g., ST3Gal3 and FUT4 mRNA) in carcinoma tissues of gastric cancer patients were reported in previous research.58 These results support our findings of increased fucosylation on highly branched N-glycans in gastric cancer patient samples, which could reflect the increase of SLea or SLex glycan epitopes.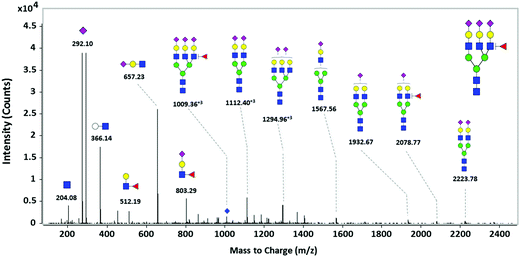 | ||
| Fig. 4 Structural elucidation of tri-sialylated, mono-fucosylated, tri-antennary glycan (Hex6HexNAc5Fuc1NeuAc3, m/z = 1009.363+3) of serum haptoglobin using targeted CID MS/MS. | ||
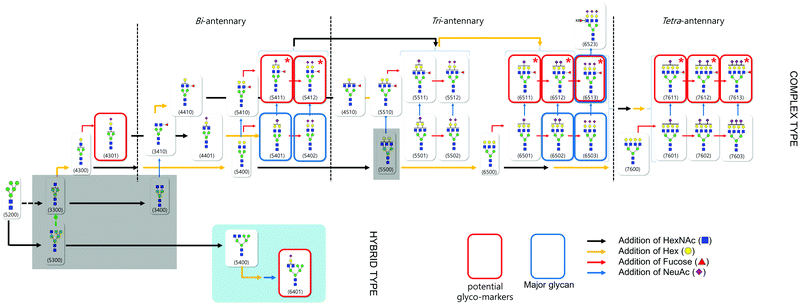
N-glycan signatures of haptoglobin in gastric cancer: a biosynthetic approach
Based on compositional and structural analysis as well as quantitative data of haptoglobin N-glycan, we proposed a ‘glycome map’ covering hypothetical biosynthetic pathways in order to understand the global features of haptoglobin glycosylation (Fig. 5). From a clinical perspective, the creation of a comprehensive map incorporating glycan structures and their biological connectivity would not only provide a useful platform to identify a glyco-marker panel but also open up the possibility of elucidating the molecular mechanism underlying the phenomenon. The glycan components accounting for 99% of the total glycan abundances on serum haptoglobin were used in the map (Table S3, ESI†). Basically, the pathway is constructed depending on known glycobiology.59 Glycans that might be the bridge between two serial glycosylation steps, mainly in the case of branching extension, is additionally depicted to follow a mainstream pathway (grey area). To be more specific, after splitting the pathway into complex and hybrid types at the key cross point Man5GlcNAc3, GlcNAc and galactose (Hex) are sequentially added to yield di-, tri, tetra-antennary (black and yellow arrows, respectively), followed by decoration with sialylation and fucosylation (blue and red arrows, respectively). High contents of complex type glycan (especially sialylated) in serum haptoglobin were observed in both the cancer patient and control groups, while high mannose and hybrid type glycans were relatively lacking in both groups. In particular, ten glycans with marked differences in abundance between the control and cancer groups (p < 0.001, Table 1), whose obvious structures were elucidated using tandem MS, were highlighted in the red boxes. Interestingly, their pathway clearly reveals again that highly branched and decorated glycosylation is a universal feature of serum haptoglobin of patients with gastric cancer. Indeed, ten potential glyco-markers commonly bearing sialylation (and fucosylation) are the set of multiple biosynthetically-related family of glycans.In particular, among the potential glyco-markers, several fucosylated & sialylated complex type glycans originating from three different antennary backbones (di-, tri-, tetra-) may be commonly related to the in vivo synthesis pathway of SLea or SLex (marked with red star). The sequential biosynthetic processes of SLea or SLex are representatively shown in Fig. S5 (ESI†). Briefly, β1,3- and β1,4-galactosyltransferases (β3Gal-Ts and β4Gal-Ts) inceptively synthesize α1→3 disaccharides and α1→4 disaccharides (type 1 and 2 chains). ST3Gal3, a subfamily of salyltransferase (ST), is additionally required to decorate type 1 chains with sialic acid (NeuAc), whereas ST3Gal4 and ST3Gal6 are responsible for type 2 chains decorated by sialylation (blue arrow). Sequential addition of α1,4- and α1,3-linked fucose to type 1 and 2 chains by fucosyltransferase (FUTs) finalizes the biosynthesis of SLea or SLex, respectively (red arrow).58 SLea and SLex are major sialylated and fucosylated antigens associated with cancer and they play a role as carbohydrate ligands for selectin which is a mediator for the extravasation of cancer cells.60,61 Increases in SLea and SLex content in malignant cancer have been correlated with a poor prognosis in cancer patients with colon, breast as well as gastric cancer.62–65 Cancer-associated activation of glycosyltransferase is assumed to influence the expression of determinants. Certain fucosyltransferases are up-regulated in cancer cells, and are responsible for the final step in the synthesis of SLea and SLex.66,67 Note that SLea or SLex is synthesized through the stepwise pathway from branching to sialylation and fucosylation. Taken together, we can suggest that fucosylation by activation of fucosyltransferase is the final crucial glycosylation step of serum haptoglobin in gastric cancer.
Conclusions
This report is the first study to elucidate specific glycan structures as well as glycan variations of serum haptoglobin associated with gastric cancer using an LC/MS-based in-depth glycomic approach. We built up an analytical platform to profile haptoglobin glycans using PGC chip-based nano LC/MS following antibody-assisted targeted purification of haptoglobin, for the purposes of a biomarker discovery of gastric cancer. Glycans were successfully analyzed and the major components of serum haptoglobin are bi- and tri-antennary sialylation, an established hallmark of haptoglobin glycosylation regardless of age, sex, and gastric cancer. However, significant differences between patients with gastric cancer and the healthy controls were easily observable. Interestingly, we found that haptoglobin glycans highly branched and decorated with fucosylation and sialylation were correlated with gastric cancer. Moreover, especially antennae fucosylation in tri- and tetra-antennary sialylated complex type N-glycan, which can be categorized as SLea or SLex, was revealed as the leading glycan signature associated with gastric cancer.The current study demonstrates that shifting from the current paradigm of serum glycan profiling to targeted glycoprotein profiling dramatically enhances the specificity of potential glyco-biomarkers for disease. Further analysis will be performed to obtain site-specific profiling which provides more biological information and thus serves as a significantly larger source of potential glyco-markers. This was a small study with only 60 samples tested. Further testing of these methods will need to be performed before more rigorous training and blinded studies can be performed, but these early results using serum glycomic for gastric cancer show great promise for an eventual use in the clinic.
Author contributions
Conceived and designed the experiments: Hyun Joo An, Jung Hoe Kim, Sung Hyeon Lee. Performed the experiments: Sung Hyeon Lee, In Seok Yeo, Seunghyup Jeong, Jua Lee. Analyzed the data: Jae-Han Kim, Seunghyup Jeong, Jua Lee. Contributed reagents/materials/analysis tools: Myung Jin Oh, Unyong Kim, Seung-Yeol Park. Contributed human serum samples/IRB approval: Se Hoon Park. Wrote the manuscript: Sung Hyeon Lee, Seunghyup Jeong, Jua Lee, Hyun Joo An. Helped to prepare figure and table: Sumin Kim, Su Hee Kim.Abbreviations
| AAL | Aleuria aurantia lectin |
| PHA-E | Phaseolus vulgaris-E lectin |
| WGA | Wheat germ agglutinin |
| Con A | Concanavalin A |
| Hex | Hexose |
| HexNAc | N-Acetylhexosamine |
| Fuc | Fucose |
| NeuAc | N-Acetylneuraminic acid (sialic acid) |
| Man | Mannose |
| PGC | Porous graphitized carbon |
| CID | Collision-induced dissociation |
| m/z | Mass to charge ratio |
| MS | Mass spectrometry |
| RT | Room temperature |
Acknowledgements
This project was sponsored by Glycan Co., Ltd in Republic of Korea.References
- L. A. Torre, F. Bray, R. L. Siegel, J. Ferlay, J. Lortet-Tieulent and A. Jemal, Ca-Cancer J. Clin., 2015, 65, 87–108 CrossRef PubMed.
- D. Compare, A. Rocco and G. Nardone, Eur. Rev. Med. Pharmacol. Sci., 2010, 14, 302–308 CAS.
- N. Duraker, A. Naci Celik and N. Gencler, European journal of surgical oncology, 2002, 28, 844–849 CrossRef CAS PubMed.
- K. Miki, M. Ichinose, N. Kawamura, M. Matsushima, H. B. Ahmad, M. Kimura, J. Sano, T. Tashiro, N. Kakei and H. Oka, et al. , Jpn. J. Cancer Res., 1989, 80, 111–114 CrossRef CAS PubMed.
- J. M. Kang, N. Kim, J. Y. Yoo, Y. S. Park, D. H. Lee, H. Y. Kim, H. S. Lee, G. Choe, J. S. Kim, H. C. Jung and I. S. Song, Helicobacter, 2008, 13, 146–156 CrossRef PubMed.
- M. Z. Qiu, J. Z. Lin, Z. Q. Wang, F. H. Wang, Z. Z. Pan, H. Y. Luo, Y. H. Li, Z. W. Zhou, Y. J. He and R. H. Xu, Int. J. Biol. Markers, 2009, 24, 258–264 CAS.
- Y. Hao, Y. Yu, L. Wang, M. Yan, J. Ji, Y. Qu, J. Zhang, B. Liu and Z. Zhu, J. Proteome Res., 2008, 7, 3668–3677 CrossRef CAS PubMed.
- D. Santini, B. Vincenzi, M. E. Fratto, G. Perrone, R. Lai, V. Catalano, C. Cass, P. A. Ruffini, C. Spoto, P. Muretto, S. Rizzo, A. O. Muda, J. R. Mackey, A. Russo, G. Tonini and F. Graziano, J. Cell. Physiol., 2010, 223, 384–388 CAS.
- N. Oue, K. Sentani, T. Noguchi, S. Ohara, N. Sakamoto, T. Hayashi, K. Anami, J. Motoshita, M. Ito, S. Tanaka, K. Yoshida and W. Yasui, Int. J. Cancer, 2009, 125, 2383–2392 CrossRef CAS PubMed.
- W. Liu, B. Liu, L. Xin, Y. Zhang, X. Chen, Z. Zhu and Y. Lin, Clin. Chim. Acta, 2007, 377, 119–126 CrossRef CAS PubMed.
- G. W. Hart and R. J. Copeland, Cell, 2010, 143, 672–676 CrossRef CAS PubMed.
- R. Apweiler, H. Hermjakob and N. Sharon, Biochim. Biophys. Acta, 1999, 1473, 4–8 CrossRef CAS.
- K. W. Moremen, M. Tiemeyer and A. V. Nairn, Nat. Rev. Mol. Cell Biol., 2012, 13, 448–462 CrossRef CAS PubMed.
- S. R. Stowell, T. Ju and R. D. Cummings, Annu. Rev. Pathol.: Mech. Dis., 2015, 10, 473–510 CrossRef CAS PubMed.
- S. S. Pinho and C. A. Reis, Nat. Rev. Cancer, 2015, 15, 540–555 CrossRef CAS PubMed.
- D. Kolarich, B. Lepenies and P. H. Seeberger, Curr. Opin. Chem. Biol., 2012, 16, 214–220 CrossRef CAS PubMed.
- Z. Kyselova, Y. Mechref, P. Kang, J. A. Goetz, L. E. Dobrolecki, G. W. Sledge, L. Schnaper, R. J. Hickey, L. H. Malkas and M. V. Novotny, Clin. Chem., 2008, 54, 1166–1175 CAS.
- S. Ozcan, D. A. Barkauskas, L. R. Ruhaak, J. Torres, C. L. Cooke, H. J. An, S. Hua, C. C. Williams, L. M. Dimapasoc and J. H. Kim, Cancer Prev. Res., 2014, 7, 226–235 CrossRef CAS PubMed.
- H. J. An, S. R. Kronewitter, M. L. de Leoz and C. B. Lebrilla, Curr. Opin. Chem. Biol., 2009, 13, 601–607 CrossRef CAS PubMed.
- R. Plomp, A. Bondt, N. de Haan, Y. Rombouts and M. Wuhrer, Mol. Cell. Proteomics, 2016, 15, 2217–2228 CAS.
- P. Shah, X. Wang, W. Yang, S. Toghi Eshghi, S. Sun, N. Hoti, L. Chen, S. Yang, J. Pasay, A. Rubin and H. Zhang, Mol. Cell. Proteomics, 2015, 14, 2753–2763 CAS.
- H. Kaji, M. Ocho, A. Togayachi, A. Kuno, M. Sogabe, T. Ohkura, H. Nozaki, T. Angata, Y. Chiba, H. Ozaki, J. Hirabayashi, Y. Tanaka, M. Mizokami, Y. Ikehara and H. Narimatsu, J. Proteome Res., 2013, 12, 2630–2640 CrossRef CAS PubMed.
- A. Matsuda, A. Kuno, H. Matsuzaki, T. Kawamoto, T. Shikanai, Y. Nakanuma, M. Yamamoto, N. Ohkohchi, Y. Ikehara, J. Shoda, J. Hirabayashi and H. Narimatsu, J. Proteomics, 2013, 85, 1–11 CrossRef CAS PubMed.
- S. Hua and H. J. An, BMB Rep., 2012, 45, 323–330 CrossRef CAS PubMed.
- T. Kumada, H. Toyoda, T. Tada, S. Kiriyama, M. Tanikawa, Y. Hisanaga, A. Kanamori, J. Tanaka, C. Kagebayashi and S. Satomura, J. Gastroenterol., 2014, 49, 555–563 CrossRef CAS PubMed.
- H. Oka, A. Saito, K. Ito, T. Kumada, S. Satomura, H. Kasugai, Y. Osaki, T. Seki, M. Kudo and M. Tanaka, J. Gastroenterol. Hepatol., 2001, 16, 1378–1383 CrossRef CAS PubMed.
- K. Oda, A. Ido, T. Tamai, M. Matsushita, K. Kumagai, S. Mawatari, A. Saishoji, T. Kure, K. Ohno, E. Toyokura, D. Imanaka, A. Moriuchi, H. Uto, M. Oketani, T. Hashiguchi and H. Tsubouchi, Oncol. Rep., 2011, 26, 1227–1233 CAS.
- M. R. Langlois and J. R. Delanghe, Clin. Chem., 1996, 42, 1589–1600 CAS.
- D. J. Schaer, F. Vinchi, G. Ingoglia, E. Tolosano and P. W. Buehler, Frontiers in physiology, 2014, 5, 415 CrossRef PubMed.
- K. Ratanasopa, S. Chakane, M. Ilyas, C. Nantasenamat and L. Bulow, Antioxid. Redox Signaling, 2013, 18, 2364–2374 CrossRef CAS PubMed.
- W. Dobryszycka, Eur. J. Clin. Chem. Clin. Biochem., 1997, 35, 647–654 CAS.
- K. B. Chandler, P. Pompach, R. Goldman and N. Edwards, J. Proteome Res., 2013, 12, 3652–3666 CrossRef CAS PubMed.
- T. Fujimura, Y. Shinohara, B. Tissot, P. C. Pang, M. Kurogochi, S. Saito, Y. Arai, M. Sadilek, K. Murayama, A. Dell, S. Nishimura and S. I. Hakomori, Int. J. Cancer, 2008, 122, 39–49 CrossRef CAS PubMed.
- S. Zhang, S. Shang, W. Li, X. Qin and Y. Liu, Glycobiology, 2016, 26, 684–692 CrossRef CAS PubMed.
- M. Nakano, T. Nakagawa, T. Ito, T. Kitada, T. Hijioka, A. Kasahara, M. Tajiri, Y. Wada, N. Taniguchi and E. Miyoshi, Int. J. Cancer, 2008, 122, 2301–2309 CrossRef CAS PubMed.
- J. N. Arnold, R. Saldova, M. C. Galligan, T. B. Murphy, Y. Mimura-Kimura, J. E. Telford, A. K. Godwin and P. M. Rudd, J. Proteome Res., 2011, 10, 1755–1764 CrossRef CAS PubMed.
- D. Wang, M. Hincapie, T. Rejtar and B. L. Karger, Anal. Chem., 2011, 83, 2029–2037 CrossRef CAS PubMed.
- P. Pompach, Z. Brnakova, M. Sanda, J. Wu, N. Edwards and R. Goldman, Mol. Cell. Proteomics, 2013, 12, 1281–1293 CAS.
- A. Sarrats, R. Saldova, E. Pla, E. Fort, D. J. Harvey, W. B. Struwe, R. de Llorens, P. M. Rudd and R. Peracaula, Proteomics: Clin. Appl., 2010, 4, 432–448 CrossRef CAS PubMed.
- S. J. Yoon, S. Y. Park, P. C. Pang, J. Gallagher, J. E. Gottesman, A. Dell, J. H. Kim and S. I. Hakomori, Int. J. Oncol., 2010, 36, 193–203 CrossRef CAS PubMed.
- S. Y. Park, S. J. Yoon, S. I. Hakomori, J. M. Kim, J. Y. Kim, B. Bernert, T. Ullman, S. H. Itzkowitz and J. H. Kim, Int. J. Oncol., 2010, 36, 1291–1297 CAS.
- S. Y. Park, S. J. Yoon, Y. T. Jeong, J. M. Kim, J. Y. Kim, B. Bernert, T. Ullman, S. H. Itzkowitz, J. H. Kim and S. I. Hakomori, Int. J. Cancer, 2010, 126, 142–155 CrossRef CAS PubMed.
- S. Y. Park, S. H. Lee, N. Kawasaki, S. Itoh, K. Kang, S. Hee Ryu, N. Hashii, J. M. Kim, J. Y. Kim and J. Hoe Kim, Int. J. Cancer, 2012, 130, 2366–2376 CrossRef CAS PubMed.
- Z. Lin, D. M. Simeone, M. A. Anderson, R. E. Brand, X. Xie, K. A. Shedden, M. T. Ruffin and D. M. Lubman, J. Proteome Res., 2011, 10, 2602–2611 CrossRef CAS PubMed.
- E. Miyoshi, S. Shinzaki, K. Moriwaki and H. Matsumoto, Methods Enzymol., 2010, 478, 153–164 CAS.
- Y. Y. Bahk, S. A. Kim, J. S. Kim, H. J. Euh, G. H. Bai, S. N. Cho and Y. S. Kim, Proteomics, 2004, 4, 3299–3307 CrossRef CAS PubMed.
- N. Okuyama, Y. Ide, M. Nakano, T. Nakagawa, K. Yamanaka, K. Moriwaki, K. Murata, H. Ohigashi, S. Yokoyama, H. Eguchi, O. Ishikawa, T. Ito, M. Kato, A. Kasahara, S. Kawano, J. Gu, N. Taniguchi and E. Miyoshi, Int. J. Cancer, 2006, 118, 2803–2808 CrossRef CAS PubMed.
- S. Zhang, H. Shu, K. Luo, X. Kang, Y. Zhang, H. Lu and Y. Liu, Mol. BioSyst., 2011, 7, 1621–1628 RSC.
- C. I. Balog, K. Stavenhagen, W. L. Fung, C. A. Koeleman, L. A. McDonnell, A. Verhoeven, W. E. Mesker, R. A. Tollenaar, A. M. Deelder and M. Wuhrer, Mol. Cell. Proteomics, 2012, 11, 571–585 CAS.
- H. Nie, X. Liu, Y. Zhang, T. Li, C. Zhan, W. Huo, A. He, Y. Yao, Y. Jin, Y. Qu, X. L. Sun and Y. Li, Sci. Rep., 2015, 5, 16007 CrossRef PubMed.
- K. Moriwaki and E. Miyoshi, World journal of hepatology, 2010, 2, 151–161 CrossRef PubMed.
- H. J. An, P. Gip, J. Kim, S. Wu, K. W. Park, C. T. McVaugh, D. V. Schaffer, C. R. Bertozzi and C. B. Lebrilla, Mol. Cell. Proteomics, 2012, 11, M111 Search PubMed.
- S. R. Kronewitter, H. J. An, M. L. de Leoz, C. B. Lebrilla, S. Miyamoto and G. S. Leiserowitz, Proteomics, 2009, 9, 2986–2994 CrossRef CAS PubMed.
- C. A. Buck, M. C. Glick and L. Warren, Science, 1971, 172, 169–171 CAS.
- E. Miyoshi, K. Moriwaki, N. Terao, C. C. Tan, M. Terao, T. Nakagawa, H. Matsumoto, S. Shinzaki and Y. Kamada, Biomolecules, 2012, 2, 34–45 CrossRef CAS PubMed.
- E. Miyoshi, K. Moriwaki and T. Nakagawa, J. Biochem., 2008, 143, 725–729 CrossRef CAS PubMed.
- J. Bones, J. C. Byrne, N. O'Donoghue, C. McManus, C. Scaife, H. Boissin, A. Nastase and P. M. Rudd, J. Proteome Res., 2011, 10, 1246–1265 CrossRef CAS PubMed.
- N. Yoshihama, K. Yamaguchi, S. Chigita, M. Mine, M. Abe, K. Ishii, Y. Kobayashi, N. Akimoto, Y. Mori and T. Sugiura, PLoS One, 2015, 10, e0124743 Search PubMed.
- P. Stanley, H. Schachter and N. Taniguchi, in Essentials of Glycobiology, ed. A. Varki, R. D. Cummings, J. D. Esko, H. H. Freeze, P. Stanley, C. R. Bertozzi, G. W. Hart and M. E. Etzler, Cold Spring Harbor Laboratory Press, 2009 Search PubMed.
- S. D. Rosen and C. R. Bertozzi, Curr. Opin. Cell Biol., 1994, 6, 663–673 CrossRef CAS PubMed.
- E. L. Berg, M. K. Robinson, O. Mansson, E. C. Butcher and J. L. Magnani, J. Biol. Chem., 1991, 266, 14869–14872 CAS.
- S. Nakamori, M. Kameyama, S. Imaoka, H. Furukawa, O. Ishikawa, Y. Sasaki, T. Kabuto, T. Iwanaga, Y. Matsushita and T. Irimura, Cancer Res., 1993, 53, 3632–3637 CAS.
- T. Narita, H. Funahashi, Y. Satoh, T. Watanabe, J. Sakamoto and H. Takagi, Cancer, 1993, 71, 3044–3053 CrossRef CAS PubMed.
- M. Amado, F. Carneiro, M. Seixas, H. Clausen and M. Sobrinho-Simoes, Gastroenterology, 1998, 114, 462–470 CrossRef CAS.
- S. E. Baldus, T. K. Zirbes, S. P. Monig, S. Engel, E. Monaca, K. Rafiqpoor, F. G. Hanisch, C. Hanski, J. Thiele, H. Pichlmaier and H. P. Dienes, Tumor Biol., 1998, 19, 445–453 CrossRef CAS.
- A. Togayachi, T. Kudo, Y. Ikehara, H. Iwasaki, S. Nishihara, T. Andoh, M. Higashiyama, K. Kodama, S. Nakamori and H. Narimatsu, Int. J. Cancer, 1999, 83, 70–79 CrossRef CAS PubMed.
- T. Koike, N. Kimura, K. Miyazaki, T. Yabuta, K. Kumamoto, S. Takenoshita, J. Chen, M. Kobayashi, M. Hosokawa, A. Taniguchi, T. Kojima, N. Ishida, M. Kawakita, H. Yamamoto, H. Takematsu, A. Suzuki, Y. Kozutsumi and R. Kannagi, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 8132–8137 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6mb00559d |
| ‡ Jung Hoe Kim and Hyun Joo An contributed equally to this paper as co-corresponding authors. |
| This journal is © The Royal Society of Chemistry 2016 |