Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Systemic analysis of stress transcriptomics of Synechocystis reveals common stress genes and their universal triggers†
M. A.
Sinetova
and
D. A.
Los
 *
*
Institute of Plant Physiology, Russian Academy of Sciences, Botanicheskaya street 35, Moscow, 127276, Russia. E-mail: losda@ippras.ru
First published on 18th August 2016
Abstract
Systemic analysis of stress transcriptomes of the cyanobacterium Synechocystis revealed that all stress-induced genes can be separated into two groups: one is clustered around heat-shock- and another – around cold-shock inducible genes. Genes for so-called heat shock proteins (HSPs) are induced by various stressors, e.g. heat, salt, hyperosmotic environment, reactive oxygen species (ROS), changes in light intensity and quality, or in the redox potential of the photosynthetic electron transport chain components. The number of specifically heat-induced genes is rather limited and their functions are mostly unknown. Genes induced by cold overlap with other set of genes induced by all above mentioned stressors with the exception of heat shock. The analysis shows that ROS and redox changes may function as universal triggers for stress responses in cyanobacteria.
Cyanobacteria represent the most ancient groups of organisms that perform oxygenic photosynthesis. Earlier studies demonstrated that cyanobacteria respond to various abiotic stresses (heat shock, oxidative stress, salt stress, hyperosmotic stress, high-intensity illumination, UV irradiation) by induction of similar sets of genes. This makes it possible to assume that different stressors generate a universal intracellular signal of yet unknown nature that triggers adaptive cell responses. Here we systematically analyzed transcription of stress responsive genes of Synechocystis on the basis of DNA microarray data in order to generalize cell responses to stress factors. The transcriptomic data, criteria, and method of analysis are described in Table S1 (ESI†).
Heat shock and other abiotic stresses
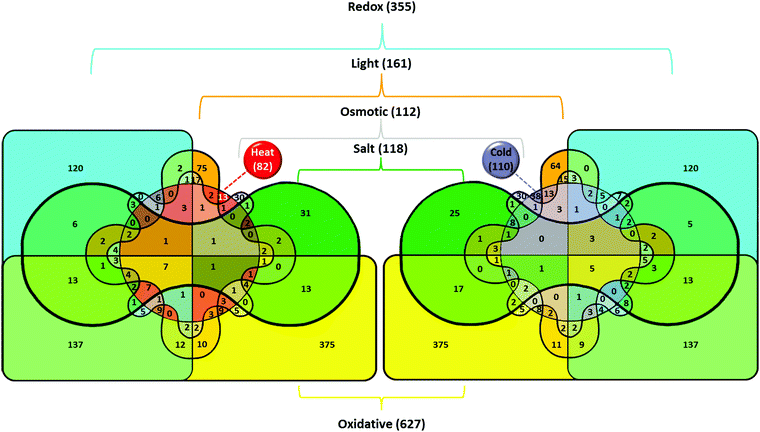
In Synechocystis sp. strain PCC 6803 (Synechocystis), short-time heat shock (15–20 min) induced ∼80 genes with the induction with factor (IF) of 2 and higher (Fig. 1).1–3 Most strongly induced genes belong to a group of so-called heat-shock protein (HSP) genes, e.g., genes for chaperones hspA, groES, groEL1, groEL2, dnaJ, htpG, dnaK, clpB1, protease htrA, sigma factors of RNA polymerase sigB and sigD, sensory histidine kinase hik34, superoxide dismutase sodB and some others (Fig. 2 and Table S2, ESI†).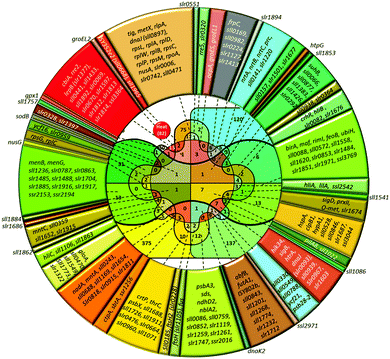 | ||
| Fig. 2 Venn diagram and lists of genes induced by heat and other abiotic stressors. Total number of genes upregulated in particular stress is shown in parentheses. The number of genes induced by the particular stress or by several stress types is shown without parentheses. Color definitions are the same as in Fig. 1. | ||
Besides heat shock, transcription of these genes is strongly induced by hyperosmotic and salt stress,4,5 high-light intensity,6 ultraviolet (UV-B),7 oxidative stress,8,9 and at low pH.10,11 About a half of stress-induced genes belongs to a category of genes with unknown function. Among those 80 heat-shock induced genes, only seven genes respond to heat specifically, i.e. they are not induced by other stressors (lower part of Table S2, ESI†).
Strong light caused a rapid (15 min) induction of ∼100 genes with induction factor of 2 or more.6 These are genes for subunits of NADPH+ dehydrogenase subunits (ndh), genes encoding the components of the CO2 concentrating mechanism (ccm) and RuBisCo. Probably, the expression of ndh and ccm genes is induced by certain signals that operate in similar manner under an increase of light intensity and a decrease in of CO2 concentration in the culture medium. Strong light also induced similar set of HSP genes: hspA, clpB1, dnaK2, htpG, groES, groEL1, and groEL2. The high-light-inducible (hli) genes were activated only during the first 15–20 min after exposure to strong light, whereas transcription of groES, groEL, and groEL2, remained at a high level even after several hours.6
The effect of UV-B on transcriptome of Synechocystis was estimated with custom-made DNA microarrays;7 therefore, the results cannot be quantitatively compared to those obtained with commercial arrays. Qualitative analysis shows that 20 min UV-B irradiation induced 55 genes with IF of 2 and more (Table S2, ESI†). UV-B enhanced transcription of two genes encoding the precursors of the main Photosystem II (PSII) protein, D1 (psbA2 and psbA3), genes for proteases (ctpA and ftsH) that are involved in the maturation and degradation of the D1 protein, and nblB gene, whose product is involved in the disassembly phycobilisomes.12 The list of induced genes includes HSP genes (groES, groEL1, cpn60, hspA, clpB, htpG, dnaK2, dnaJ, htpG) and as well as regulatory hik34, sigB, and sigD.
Salt stress (0.5 M NaCl for 15 min) upregulated ∼120 genes with the IF ≥ 4.5 Evidently, the list included all the representatives of HSP genes. A set of NaCl-specific genes limits to ABC-type transporters (cbiQ, norM, ycf85); synthesis of quinones (menB, menG, ubiH), biotin (birA), NAD (nadC), and riboflavin (ribF) metabolism, acetylation of proteins (rimI), protease hhoB, one translation initiation factor (infC), and several genes of unknown function.
Similarly, hyperosmotic stress (0.5 M sorbitol for 15 min) caused rapid and strong induction HSP genes (hspA, htpG, dnaK2, dnaJ, clpB1, and cpn60), as well as genes for a protease (htrA), the regulatory genes (sigB and sigD), and a gene for superoxide dismutase (sodB).4,13 Short-term (15–20 min) salt and hyperosmotic stresses induce essentially the same set of genes, probably, because of a rapid decrease in the cytoplasmic volume due to water loss at the initial stages of both stresses.4,5,13 However, there is a possibility of regulation that has emerged due to stress-induced ROS formation, or due to redox changes in the main pools of photosynthetic electron transport chain (ETC).
Genes specifically induced by sorbitol include those for phosphate (pstS), nitrate/nitrite (ntrABC) transporters, ribosomal proteins (rplI and rplY), periplasmic protease hhoA, genes involved in lipid metabolism (sll0208 and sll1377), and several genes of unknown function.4,13
Thus, in fact, so-called HSP genes are not strictly the genes for heat-shock-induced proteins. Since these genes respond non-specifically to the stressors of different nature at transcriptional1–13 and protein14,15 level, they should be rather referred to as general stress-induced genes, and the corresponding proteins – general stress proteins (GSPs).
Cold shock and other abiotic stresses
A limited number of genes are induced by short-term (15–30 min) heat and cold stresses: sigD for an alternative sigma factor of RNA polymerase, sbtA for a bicarbonate transporter, nblB for a phycobilisome degradation protein, fdx for ferredoxin, ISY352e2 for a transposase, and two genes of unknown function (Tables S2 and S3, ESI†). The groESL1 operon that codes for two GSPs, 60 kDa chaperonin GroEL1 and 10 kDa co-chaperonin GroES is also induced by cold.3 Such induction, however, is never detected under short-term cold shock. This allows a dissection of early stages of stress signal perception and transduction based on consideration that the lists of heat- and cold-stress induced genes do not overlap (Fig. 1).About 100 genes were induced by short-term (20–30 min) cold stress in Synechocystis with IF ≥ 2.1,3,16,17 These genes may be grouped into major functional categories, such as signal perception and transduction; transcription and translation; cell wall and membrane maintenance; photosynthesis and respiration; various cellular functions (cofactor biosynthesis, nucleotide metabolism); unknown functions (Table S3, ESI†).
Cold-induced regulatory genes are limited to two genes for the histidine kinases (hik3 and hik31), two genes for the DNA-binding transcriptional regulators (rre5 and sfsA), and a gene for the sigma factor of RNA polymerase (sigD). Hik3 is equipped with PAS/PAC and GAF sensory domains.18 Hik31 (CopS) is a transmembrane multisensory histidine kinase that regulates autotrophic growth, responses to light, and resistance to bivalent cations, Cd2+, Cu2+, and Zn2+.19–23 Hik31 controls genes that are involved in photosynthesis and genes for ribosomal proteins. Fluctuations in photosynthetic electron transport of different nature that oxidize plastocyanin promote the release of Cu2+, which binds to Hik31 and trigger the regulatory cascade.22,23 Rre5 belongs to a PatA subfamily of two component response regulators, which is involved in CO2 uptake and associated pH homeostasis. SfsA-like protein is involved in regulation of sugar catabolism, but its exact function in cyanobacteria is unknown.
SigD (RpoD) belongs to a Group 2 RNA polymerase σ factor, and it is the only σ factor abundant in the dark.24 Transcription of sigD is induced by heat, UV-B, and strong light,25 by the inhibitor of photosynthetic electron transfer chain, DCMU24 and by H2O2.26 These data suggest that SigD may be involved in light-dependent redox regulation of transcription.
The cofactor of Rho transcriptional terminator, NusG operates in combination with histone-like nucleoid-structuring protein H-NS and Rho-dependent transcriptional terminators to diminish genome-wide antisense transcription under cold stress.27
Low temperatures also activate genes for ribosomal machinery: 50S ribosomal proteins L20 and L11, 30S proteins S12, S21, and ribosome chaperone trigger factor (Tig). The latter supports early folding events and prevents misfolding and aggregation of proteins. The smpB gene encodes the protein that is required for rescue of ribosomes stalled on defective messages.28
The crhR gene for a DEAD (Asp-Glu-Ala-Asp) box family RNA helicase is rapidly and transiently induced by cold stress. Noteworthy, under cold stress, wild-type cells maintain the PQ pool in reduced state, while the crhR-deficient cells – in oxidized state.29 This implies the regulation via redox changes in PQ pool.
The cold acclimation of prokaryotes depends on the ability of cells to desaturate the fatty acids (FAs) in the membrane lipids and to adjust the membrane fluidity to changing ambient temperature. In cyanobacterial cells, the desaturation reactions occur in FAs that are bound to glycerolipids. Under cold stress, the de novo synthesis of FAs ceases for several hours. During this period, the existing FAs of lipids are desaturated to a certain level, after which cells begin to grow again. An increase in cold-induced FA desaturation is caused by induction of the expression of three known genes for the fatty acid desaturases, FADs (desA for Δ12-desaturase, desB for ω3-desaturase, and desD for Δ6-desaturase), accompanied by an increase in their mRNA stability.30 The stability of these three desaturase mRNAs (desA, desB, and desD) at low temperatures is ensured by RbpA3, a member of a family of RNA binding proteins.31
Some cold-induced genes are strictly light-dependent, e.g. two of four hli (high light induced) genes, hliA and hliB,32ycf39 for a PSII assembly factor,33lilA for a light-harvesting protein,34 or ndhD2 for a subunit of NADH dehydrogenase. The latter is controlled by PedR – a redox-sensitive transcription regulator.35,36 The gshB gene for glutathione synthase, which is involved in redox regulation and acclimation under changing light36 is also induced by cold.
Another group of cold-induced genes includes a family that encodes proteins for nucleotide binding and modification. Their functions in thermal acclimation are barely known. About a half of cold-induced genes is not functionally characterized and their role in cold stress responses and cold acclimation is absolutely unknown.
Among ∼100 cold-induced genes, only 38 genes are specifically induced by cold (marked by asterisks in Table S3, ESI†). This list is limited to genes of rbpA-family that encode RNA-binding proteins (RbpA2 and RbpA3); smpB, rplT, and pfbB involved in translation; era and mbpA for GTP-binding protein and DNA methylase; cbiF for the first step of cobalamin biosynthesis; ams1 for α-mannosidase; and several genes of unknown function.
Other cold-induced genes respond to two or more abiotic stresses, including salt, hyperosmotic, oxidative, acidic stresses, strong light and UV, chemical agents that alter the redox status of the PQ pool (Table S3, ESI†). The induction of the same genes by different stressors suggests the existence of some universal trigger(s) of stress responses. In the photosynthetic cells or organisms, the primary candidates for such universal triggers might be electric signaling via redox-responsive systems or chemical signaling with ROS or NO-derived reactive species.
Oxidative stress and redox regulation
Maintaining an optimal redox status within the cells is important for regulation of gene expression. Changes in redox status in the thylakoid membranes as a result of stress factors directly affect the functioning of the photosynthetic ETC. The crucial roles in the maintenance of the redox homeostasis play thiol molecules: glutathione, thioredoxin and glutaredoxin peroxiredoxins.37 Table S2 (ESI†) shows that, for the most part, the GSP genes are also regulated via changes in the redox state of plastoquinone (PQ) pool of PSII. Such changes can be simulated by the addition of the inhibitors that interrupt with a flow of electrons before the PQ pool and oxidize it (DCMU), or after the PQ pool and reduce it (DBMIB). The addition of DBMIB strongly induces transcription of the GSP genes.8,9 Changes in the redox state PQ pool may be also caused by an increase in light intensity (see above) or by changing the pH of the environment.The induction of GSP genes may be controlled by the reactive oxygen species (ROS) as well. The formation of ROS provoked by different stressors may influence the redox state of a cell. ROS are present in several forms: singlet oxygen (1O2), superoxide anion (O2˙−), hydrogen peroxide (H2O2), or hydroxyl radical (OH˙).38 Singlet oxygen and hydroxyl radical have an extremely short life-time and high degree of reactivity with surrounding proteins, lipids, and pigments. Superoxide radical cannot diffuse through the membrane due its negative charge. H2O2 is less reactive, but may recover to hydroxyl radical via the Fenton reaction and, thus, become destructive for the cells. The addition of 0.25 mM H2O2 to a culture medium of Synechocystis induces the transcription of 118 genes with the IF ≥ 3,39 among which there are GSP genes (hik34, sigB, hspA, dnaJ, clpB1, dnaK2, ctpA, hypA1, htpG, etc.) that, as indicated above, are induced by heat, strong light, ultraviolet light, salt, and sorbitol (Table S2, ESI†).
Similarly to heat shock and other stressors, ROS induce genes that are necessary for stress neutralization. These include genes for peroxiredoxin aphC (sll1621), catalase katG (sll1987), and thioredoxin trx (sll0755). ROS induce transcriptional regulator perR (slr1738),26 which is directly regulated by H2O2via oxidation of His-37 and His-91 residues, which leads to dissociation of PerR·DNA complex.40
The stress transcriptome analysis suggests that ROS (in particular, H2O2) and redox changes of the components of the photosynthetic electron chain may function as universal triggers for stress responses in cyanobacteria.
Redox regulation system consists of components of photosynthetic electron transport chain and the regulatory proteins that may perceive the difference of electric potentials, for example, transcription factor PedR (Ssl0564)35,36 or sensor kinase Hik31.19,22 Stress-induced ROS should have sufficient life-time to perform their regulatory functions.38 Electron transport inhibitors of photosynthetic electron transport (DCMU, DBMIB, or methylviologen) and the generator of ROS response (H2O2) evidence in favour of the suggestion about their universal triggers roles in the initiation of stress responses. Systematic analysis of transcriptome shows that redox- and ROS-regulatory pathways play important role in stress-induced gene expression. A redox potential of cells may change due to the emergence of ROS.41 In such case, about a quarter of genes induced by H2O2 and by a derivative of benzoquinone, DBMIB, overlaps. Some part of genes falls under the control of the ROS, and another part – under redox control (Fig. 1–3). In addition, there is a relatively minor set of stress-induced genes that does not depend on the ROS or redox regulatory systems. We have previously found that a part of the stress-dependent genes can be induced by changing the degree of supercoiling of the genomic DNA.3 This type of regulation does not require any regulators with SH-groups that can undergo oxidation or reduction cycles. Further experimental studies are necessary to directly confirm or refute the hypothesis about the universal triggers of stress responses in photosynthetic cells.
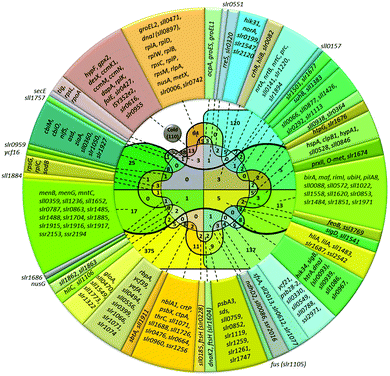 | ||
| Fig. 3 Venn diagram and lists of genes induced by cold and other abiotic stressors. Total number of genes upregulated in particular stress is shown in parentheses. The number of genes induced by the particular stress or by several stress types is shown without parentheses. Color definitions are the same as in Fig. 1. | ||
Conclusions
Transcriptomic analysis shows that some universal chemical or electric stimuli may exist that trigger the expression of stress-dependent genes regardless the physical nature of a stressor. The candidates for such triggers are reactive oxygen species and changes in redox status of plastoquinone pool. Application of the inhibitors (DBMIB, DCMU) or ROS (H2O2) evidence that redox status of PQ pool and ROS may be involved in the initiation of stress responses as universal triggers.Conflict of interest
Authors declare no conflict of interest.Acknowledgements
This work was supported by a grant from Russian Science Foundation to D. A. L. (no. 14-24-00020).Notes and references
- M. Inaba, I. Suzuki, B. Szalontai, Y. Kanesaki, D. A. Los, H. Hayashi and N. Murata, J. Biol. Chem., 2003, 278, 12191–12198 CrossRef CAS PubMed.
- I. Suzuki, Y. Kanesaki, H. Hayashi, J. J. Hall, W. J. Simon, A. R. Slabas and N. Murata, Plant Physiol., 2005, 138, 1409–1421 CrossRef CAS PubMed.
- J. S. Prakash, M. Sinetova, E. Kupriyanova, A. Zorina, I. Suzuki, N. Murata and D. A. Los, Mol. BioSyst., 2009, 5, 1904–1912 RSC.
- K. Paithoonrangsarid, M. A. Shumskaya, Y. Kanesaki, S. Satoh, S. Tabata, D. A. Los, V. V. Zinchenko, H. Hayashi, M. Tanticharoen, I. Suzuki and N. Murata, J. Biol. Chem., 2004, 279, 53078–53086 CrossRef CAS PubMed.
- M. A. Shumskaya, K. Paithoonrangsarid, Y. Kanesaki, D. A. Los, V. V. Zinchenko, M. Tanticharoen, I. Suzuki and N. Murata, J. Biol. Chem., 2005, 280, 21531–21538 CrossRef PubMed.
- Y. Hihara, A. Kamei, M. Kanehisa, A. Kaplan and M. Ikeuchi, Plant Cell, 2001, 13, 793–806 CrossRef CAS.
- L. Huang, M. P. McCluskey, H. Ni and R. A. LaRossa, J. Bacteriol., 2003, 184, 6845–6858 CrossRef.
- Y. Hihara, K. Sonoike, M. Kanehisa and M. Ikeuchi, J. Bacteriol., 2003, 185, 1719–1725 CrossRef CAS PubMed.
- A. K. Singh, H. Li and L. A. Sherman, Physiol. Plant., 2004, 120, 27–35 CrossRef CAS PubMed.
- H. Ohta, Y. Shibata, Y. Haseyama, Y. Yoshino, T. Suzuki, T. Kagasawa, A. Kamei, M. Ikeuchi and I. Enami, Photosynth. Res., 2005, 84, 225–230 CrossRef CAS PubMed.
- J. Uchiyama, R. Asakura, M. Kimura, A. Moriyama, H. Tahara, Y. Kobayashi, Y. Kubo, T. Yoshihara and H. Ohta, Biochim. Biophys. Acta, Bioenerg., 2012, 1817, 1270–1276 CrossRef CAS PubMed.
- H. Y. Hsiao, Q. He, L. G. Van Waasbergen and A. R. Grossman, J. Bacteriol., 2004, 186, 3882–3888 CrossRef CAS PubMed.
- A. Shapiguzov, A. A. Lyukevich, S. I. Allakhverdiev, T. V. Sergeyenko, I. Suzuki, N. Murata and D. A. Los, Microbiology, 2005, 151, 447–455 CrossRef CAS PubMed.
- J. Qiao, M. Shao, L. Chen, J. Wang, G. Wu, X. Tian, J. Liu, S. Huang and W. Zhang, Gene, 2013, 512, 6–15 CrossRef CAS PubMed.
- L. Gao, J. Wang, H. Ge, L. Fang, Y. Zhang, X. Huang and Y. Wang, Photosynth. Res., 2015, 126, 203–219 CrossRef CAS PubMed.
- I. Suzuki, Y. Kanesaki, K. Mikami, M. Kanehisa and N. Murata, Mol. Microbiol., 2001, 40, 235–244 CrossRef CAS PubMed.
- Z. Liu, H. Cui, Z. Liu, Y. Wang, Y. Cui, Z. Liu and S. Qin, Chin. J. Oceanol. Limnol., 2014, 32, 403–418 CrossRef CAS.
- H. Takala, A. Björling, O. Berntsson, H. Lehtivuori, S. Niebling, M. Hoernke, I. Kosheleva, R. Henning, A. Menzel, J. A. Ihalainen and S. Westenhoff, Nature, 2014, 509, 245–248 CrossRef CAS PubMed.
- S. Kahlon, K. Beeri, H. Ohkawa, Y. Hihara, O. Murik, I. Suzuki, T. Ogawa and A. Kaplan, Microbiology, 2006, 152, 647–655 CrossRef CAS PubMed.
- S. Nagarajan, D. M. Sherman, I. Shaw and L. A. Sherman, J. Bacteriol., 2012, 194, 448–459 CrossRef CAS PubMed.
- S. Nagarajan, S. Srivastava and L. A. Sherman, Mol. Microbiol., 2014, 91, 79–97 CrossRef CAS PubMed.
- J. Giner-Lamia, L. López-Maury, J. C. Reyes and F. J. Florencio, Plant Physiol., 2012, 159, 1806–1818 CrossRef CAS PubMed.
- J. Giner-Lamia, L. López-Maury and F. J. Florencio, PLoS One, 2014, 9, e108912 Search PubMed.
- I. Tuominen, E. Tyystjärvi and T. Tyystjärvi, J. Bacteriol., 2003, 185, 1116–1119 CrossRef CAS PubMed.
- S. Koskinen, K. Hakkila, L. Gunnelius, J. Kurkela, H. Wada and T. Tyystjärvi, Mol. Microbiol., 2016, 99, 43–54 CrossRef CAS PubMed.
- H. Li, A. K. Singh, L. M. McIntyre and L. A. Sherman, J. Bacteriol., 2004, 186, 3331–3345 CrossRef CAS PubMed.
- J. M. Peters, R. A. Mooney, J. A. Grass, E. D. Jessen, F. Tran and R. Landick, Genes Dev., 2012, 26, 2621–2633 CrossRef CAS PubMed.
- A. W. Karzai, M. M. Susskind and R. T. Sauer, EMBO J., 1999, 18, 3793–3799 CrossRef CAS PubMed.
- K. Sireesha, B. Radharani, P. S. Krishna, N. Sreedhar, R. Subramanyam, P. Mohanty and J. S. Prakash, Biochim. Biophys. Acta, Bioenerg., 2012, 1817, 1525–1536 CrossRef CAS PubMed.
- D. A. Los, M. K. Ray and N. Murata, Mol. Microbiol., 1997, 25, 1167–1175 CrossRef CAS PubMed.
- Q. Tang, X. Tan and X. Xu, Chin. Sci. Bull., 2010, 55, 2416–2421 CrossRef CAS.
- Q. He, N. Dolganov, O. Bjorkman and A. R. Grossman, J. Biol. Chem., 2001, 276, 306–314 CrossRef CAS PubMed.
- J. W. Chidgey, M. Linhartová, J. Komenda, P. J. Jackson, M. J. Dickman, D. P. Canniffe, P. Koník, J. Pilný, C. N. Hunter and R. Sobotka, Plant Cell, 2014, 26, 1267–1279 CrossRef CAS PubMed.
- G. Kufryk, M. A. Hernandez-Prieto, T. Kieselbach, H. Miranda, W. Vermaas and C. Funk, Photosynth. Res., 2008, 95, 135–145 CrossRef CAS PubMed.
- K. Nakamura and Y. Hihara, J. Biol. Chem., 2006, 281, 36758–36766 CrossRef CAS PubMed.
- M. Horiuchi, K. Nakamura, K. Kojima, Y. Nishiyama, W. Hatakeyama, T. Hisabori and Y. Hihara, Biochem. J., 2010, 431, 135–140 CrossRef CAS PubMed.
- J. C. Cameron and H. B. Pakrasi, Plant Physiol., 2010, 154, 1672–1685 CrossRef CAS PubMed.
- F. J. Schmitt, G. Renger, T. Friedrich, V. D. Kreslavski, S. K. Zharmukhamedov, D. A. Los, V. V. Kuznetsov and S. I. Allakhverdiev, Biochim. Biophys. Acta, Bioenerg., 2014, 1837, 835–848 CrossRef CAS PubMed.
- Y. Kanesaki, H. Yamamoto, K. Paithoonrangsarid, M. Shoumskaya, I. Suzuki, H. Hayashi and N. Murata, Plant J., 2007, 49, 313–324 CrossRef CAS PubMed.
- J. W. Lee and J. D. Helmann, Nature, 2006, 440, 363–367 CrossRef CAS PubMed.
- H. Liu, R. Colavitti, I. I. Rovira and T. Finkel, Redox-dependent transcriptional regulation, Circ. Res., 2005, 97, 967–974 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6mb00551a |
| This journal is © The Royal Society of Chemistry 2016 |