Open Access Article
Open Access ArticleComparative lipidomic study of urothelial cancer models: association with urothelial cancer cell invasiveness†
Yang
Yu
a,
Matej
Skočaj
bc,
Mateja Erdani
Kreft
b,
Nataša
Resnik
b,
Peter
Veranič
b,
Pietro
Franceschi
d,
Kristina
Sepčić
c and
Graziano
Guella
*ae
aBioorganic Chemistry Laboratory, Department of Physics, University of Trento, Trento, Italy. E-mail: graziano.guella@unitn.it; Fax: +39 461281696; Tel: +39 461 281536
bInstitute of Cell Biology, Faculty of Medicine, University of Ljubljana, Ljubljana, Slovenia
cDepartment of Biology, Biotechnical Faculty, University of Ljubljana, Ljubljana, Slovenia
dBiostatistics and Data Management, Research and Innovation Centre-Fondazione Edmund Mach, S. Michele all'Adige, TN, Italy
eCNR, Institute of Biophysics, Povo, Trento, Italy
First published on 16th August 2016
Abstract
Comparative lipidomic studies were performed across the RT4 versus T24 urothelial cancer cell lines, as models for noninvasive urothelial papilloma cells (with a relatively high level of differentiation) and invasive urothelial carcinoma cells (with low level of differentiation), respectively. The aim was to investigate the differences in lipid profile associated with different levels of urothelial cancer cell invasiveness. The cellular lipidomes were characterized using our previously developed joint methodology of liquid chromatography-mass spectrometry and high-resolution nuclear magnetic resonance, which included analysis of the phospholipids and ceramide-based glycosphingolipids. This study shows that the invasive T24 cells have 3-fold lower levels of 1-alkyl (ether)-2-acyl phosphocholine species, which are accompanied by greater length and higher unsaturation of acyl chains of several lipid classes. Moreover, d18:1-based glycosphingolipids show different profiles; in particular, α-hydroxylated glucosylceramides have lower levels in the T24 cells, along with increased lactosyl ceramides. These differences between RT4 and T24 cells suggest significantly different organization of the cellular membranes, which can affect the membrane fluidity and membrane-dependent functions, and contribute to the lower stiffness of plasma membrane and reduced cell–cell adhesion required for movement and invasiveness of these T24 urothelial carcinoma cells with a high metastatic potential.
Introduction
Urothelial cells cover the mammalian urinary tract from the renal pelvis to the proximal urethra and their specific structural characteristics provide the tight organization of the urothelium, which forms the blood–urine barrier, i.e. the tightest and most impermeable barrier in the body.1,2 As a special type of epithelium, the structure and function of the proteins in urothelial cells have been broadly studied also with respect to cancer transformation.3–7 However, the general characterization of the lipidic component of urothelium still remains far behind, with no studies to date on the lipid hallmarks of its relevant pathological states. Our recent study revealed that the amount of cholesterol, as well as the amount of sphingomyelin/cholesterol membrane domains is highly increased in urothelial cancer cells in comparison to normal urothelial cells.8 To associate the different cell properties of noninvasive and invasive urothelial cancer cells (reference) with their lipid composition a more precise study of the lipid composition of these two types of cells was required.Urinary bladder cancer is the eighth cause of deaths among different types of cancer and for men it is the fourth common cancer causing 8% of malignancies with 3% of deaths as well as causing 3% of malignancies with 1.5% of deaths in women.9 The urothelial carcinoma can be classified according to several grades, from more differentiated and poorly aggressive (grade I) up to less differentiated and more invasive or even metastatic urothelial carcinoma cells (grade III). These grades are associated to the broad spectrum of invasiveness of urothelial carcinoma cells according to the development and progression of the carcinoma. Urothelial carcinoma cell invasiveness and metastasis formation are proposed to connect with aberrant lipid biosynthesis and metabolism involved in carcinoma development.10 In addition to enwrapping the cytoplasmic components, the plasma membrane is involved in communication of the cell with the environment, as well as having important roles in cell–cell and/or cell–matrix adhesion and has increased mechanical stability during cell division. However, cancer cells usually show reduced cell adhesion, and there is a growing evidence that correlates this reduction with the invasive properties and metastasis formation of a carcinoma.11 Along with increased invasiveness and enhanced motility, another important signature for epithelial carcinoma is the separation of individual cells from the epithelial colonies. Thus, most critically, the flexible features of the membrane lipids can promote decreased shear forces in this separation and penetration through the tissue.12,13 Also, the plasma membrane provides specific domains for protein–lipid interactions, some of which are involved in structural and signaling alterations during cell proliferation, motility, and death.13,14 For example, proteins that are members of the receptor tyrosine kinases family and other signaling proteins like Ras, caveolins and CD44, have been demonstrated to mediate cell motility and migration in association with lipid rafts.15–17 However, despite these important roles of lipids, systematic studies on the lipid characteristics and variations have rarely been investigated for urothelial carcinoma. Furthermore, with the close relationship between the changes in lipid profiles and urothelial carcinoma cell invasiveness, there is now great interest in lipidomic studies in urinary bladder cancers.
With respect to urothelial cancer cell lines, we used two types of in vitro model systems: (1) human cancer urothelial cell line RT4 as a model of noninvasive papilloma cells, and (2) human cancer urothelial cell line T24 as invasive urothelial carcinoma cells.8,18 RT4 cells are less aggressive displaying similar growth and motility characteristics to normal epithelial cells.19–21 T24 cells derive from poorly differentiated (grade III) urinary bladder carcinoma,22,23 in which cells display higher metastatic potential characteristics such as separating during cell motility and non-self-limited growth with extensive invasiveness and pervasion. The urothelial papilloma cancer cells that RT4 cells represent are the most relevant for the treatment of urinary bladder cancer because they represent 90% of bladder tumors, and are the only bladder tumors that are locally treated. On the other hand T24 cells represent a model for less frequent but life threatening metastatic urothelial carcinoma. Thus, the two cancer urothelial cell lines, RT4 and T24 that we used are relevant model systems for any diagnosis and treatment in urological oncology. To date, these two representative models of cancer urothelial cells have been broadly used in urinary bladder cancer research, such as mutation-related studies and for morphological comparisons;20,21,24 however, to the best of our knowledge, no systematic lipid studies have been performed yet with these RT4 and T24.
Interestingly, several studies have linked glycosphingolipids (GSLs) to cancer pathogenesis and progression.25,26 Furthermore, epithelial carcinoma motility, invasiveness, and morphology changes have been proposed to be associated with abnormal glycosylation and the subsequent GSL variations.26,27 Moreover, biophysical characterization of RT4 and T24 cancer urothelial cells demonstrated that T24 cells have lower stiffness due to reorganization of their cell cytoskeleton.22 However, no further studies have looked at the global lipid variations that might contribute to either the urothelial cancer cell invasiveness or the biophysical alterations in the perspective of urinary bladder cancer. Thus, more information needs to be acquired from global lipid profile analysis, including GSLs characterization.
To address the changes in the lipid profiles of cancer urothelial cells related to cell invasiveness, we performed a comparative lipidomic study using RT4 and T24 cells as the model systems. The further aims were to determine any patterns in the associated lipid variations, and to open investigation on the relevant cellular functions and on the development of their therapeutic potential in the treatment of patients with urinary bladder cancer.
First, we investigated RT4 and T24 cells to characterize their main phospholipids and GSLs (with a focus on the d18:1-based GSLs) classes, the individual molecular species, their triacylglycerols (TAGs) and cholesterol. To tackle this analytical problem, we exploited our previously developed multi-technique methodology, wherein high resolution nuclear magnetic resonance (NMR) is combined with classical high-performance liquid chromatography (HPLC)-electrospray ionization (ESI) tandem mass spectrometry (MS/MS).28 Furthermore, multivariate data analysis (MVDA)29 was applied to explore our extensive lipidomic dataset, highlighting the most prominent patterns. Finally univariate statistical testing was used to identify potential lipid markers which differentiate RT4 and T24 cells and which could be potentially associated to urothelial cancer cell invasiveness. These analyses have demonstrated powerful applicability to lipidomics data interpretation, and can highlight important patterns hidden in such measurements.
To sum up, our data show for the first time the differences of lipid profiles for RT4 cells representing noninvasive urothelial papilloma cells and T24 cells representing invasive urothelial carcinoma cells, which are relevant model systems for any diagnosis and treatment in urinary bladder cancer, and they provide a preliminary insight into the role played by lipids in the mechanisms of cancer invasiveness.
Material and methods
Chemicals
Methanol was liquid chromatography-mass spectrometry (LC-MS) hypergrade (99.9% purity; Merck). Chloroform and water were HPLC grade (99.8% purity; VWR Chemicals). Ultrapure water was produced using a Milli-Q system (Millipore). Perdeuterated methanol (CD3OD; ≥ 99.8% deuteration; Merck) was used for NMR spectroscopy. The lipid standard of 1,2-dilauroyl-sn-glycero-3-phosphocholine [i.e., PC(12:0/12:0)] and the GSL standard of C16 galactosyl (β) ceramide (d18:1/16:0) (D-galactosyl-β-1,1′ N-palmitoyl-D-erythro-sphingosine) were used (>99% purity; Avanti Polar Lipids; Alabaster, USA). All standard and extracted cellular lipids were prepared and dissolved in glass vials.RT4 and T24 urothelial cancer cell culture
The RT4 and T24 urothelial cancer cell cultures were maintained as in Imani et al. (2012).24 Briefly, both of these human urothelial cancer cell lines were cultured in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of Advanced-Dulbecco's Modified Essential Medium (ADMEM, Invitrogen, Gibco, Paisley, UK) and Ham's F-12 medium (Sigma-Aldrich, St. Louis, MO, USA), supplemented with 5% fetal bovine serum (Gibco, Invitrogen, Carlsbad, CA, USA), 100 μg ml−1 streptomycin and 100 units per ml penicillin. The RT4 and T24 urothelial cancer cell cultures were plated simultaneously with a seeding density of 5 × 104 cells per cm2. To achieve sufficient biomass for NMR analysis, two flasks (2 × 75 cm2) of RT4 cultures and three flasks (3 × 75 cm2) of T24 cultures were necessary (approximately 3 × 107 cells in each case). The cells were incubated at 37 °C in a humidified atmosphere of 5% CO2 for 1 week to achieve confluence. The cells were then detached with TrypLE Select (Gibco, Invitrogen), resuspended in the cell growth medium, and centrifuged at 200 × g for 5 min. The cell pellets were collected for lipid extraction and analysis. RT4 and T24 cells were both grown repeatedly in 3 different months.
1 mixture of Advanced-Dulbecco's Modified Essential Medium (ADMEM, Invitrogen, Gibco, Paisley, UK) and Ham's F-12 medium (Sigma-Aldrich, St. Louis, MO, USA), supplemented with 5% fetal bovine serum (Gibco, Invitrogen, Carlsbad, CA, USA), 100 μg ml−1 streptomycin and 100 units per ml penicillin. The RT4 and T24 urothelial cancer cell cultures were plated simultaneously with a seeding density of 5 × 104 cells per cm2. To achieve sufficient biomass for NMR analysis, two flasks (2 × 75 cm2) of RT4 cultures and three flasks (3 × 75 cm2) of T24 cultures were necessary (approximately 3 × 107 cells in each case). The cells were incubated at 37 °C in a humidified atmosphere of 5% CO2 for 1 week to achieve confluence. The cells were then detached with TrypLE Select (Gibco, Invitrogen), resuspended in the cell growth medium, and centrifuged at 200 × g for 5 min. The cell pellets were collected for lipid extraction and analysis. RT4 and T24 cells were both grown repeatedly in 3 different months.
Fluorescent labeling and microscopy
RT4 and T24 cells were grown on cover glasses in conditions as described above, fixed with 2% paraformaldehyde for 30 minutes then labeled with ostreolysinA-mCherry (1 μM),30 for 10 minutes and fixed again for 10 minutes with 2% paraformaldehyde. After that cells were labeled with phalloidin–FITC (16 μg ml−1) for 30 minutes. The coverslips were mounted in Vectashield with 4,6-diamidino-2-phenylindole (DAPI) for nuclear staining, and analysed using an oil-immersion objective (63× oil/NA 1.40) under an AxioImager Z1 fluorescent microscope (Carl Zeiss, Germany) with an ApoTome device (Carl Zeiss, Germany) for generation of optical sections. Images were acquired using the Axio-Vision programme (Carl Zeiss, Germany).Lipid extraction and sample preparation
The extraction procedure for the total lipids given in our previous study was modified as a 2-step protocol with the lipid recovery expanded to also include more highly polar lipids from the biological matrix (e.g., GSLs).31,32 The collected cell pellets were resuspended in 1 ml Milli-Q water by gently pipetting in and out, and moved to glass bottles. Before lipid extraction, 60 μl of 100 ng μl−1 lipid standard PC (12:0/12:0) was added to the samples, which is a lipid that is different from all previously described natural cellular lipids. Five milliliters of chloroform/methanol (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) was firstly added to the resuspended cell pellets, followed by sonication and shaking for 2 h. The samples were then centrifuged at 2500 × g for 10 min. The organic (lower) phase was carefully collected into a glass vial, and the upper (aqueous) phase was extracted again with the addition of 3 ml chloroform/methanol (2
1, v/v) was firstly added to the resuspended cell pellets, followed by sonication and shaking for 2 h. The samples were then centrifuged at 2500 × g for 10 min. The organic (lower) phase was carefully collected into a glass vial, and the upper (aqueous) phase was extracted again with the addition of 3 ml chloroform/methanol (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), with shaking for 1 h, and repeated centrifugation and lower phase collection. Finally, the combined organic extracts were evaporated in a rotary evaporator (Rotavapor R-134, V700, Büchi, Switzerland), dried under N2 and stored at −20 °C.
1, v/v), with shaking for 1 h, and repeated centrifugation and lower phase collection. Finally, the combined organic extracts were evaporated in a rotary evaporator (Rotavapor R-134, V700, Büchi, Switzerland), dried under N2 and stored at −20 °C.
High resolution NMR measurements
NMR setup was similar to that reported in detail in our previous study.28 Briefly, 1H-NMR (400 MHz) and 31P-NMR (162 MHz) spectra of the lipid standard and the cell–lipid extracts (3 replicates which were grown, extracted and analyzed independently in different months) dissolved in CD3OD were recorded at 300 K on a Bruker-Avance 400 MHz NMR spectrometer, with a 5 mm BBI probe outfit with pulsed-gradient field utility. The 1H-90° proton pulse length was 9.3 μs with a transmission power of 0 db, and the 31P-90° proton pulse length was set up as 17 μs with a transmission power of −3 db. Calibration of the chemical shift scale (δ) was carried out on the residual proton signal of the CD3OD at δH 3.310 ppm and δC 49.00 ppm, and the phosphocholine (PC) signal at δP −0.550 ppm was used for the calibration of the 31P-NMR δ scale. The following measurements were performed (with the acquired information): 1H-NMR (proton chemical shifts and scalar couplings J); 31P-NMR; 1H–1H TOCSY (proton–proton scalar correlation); 1H–13C HSQC (proton–carbon one-bond correlation); 1H–13C HMBC (proton–carbon multiple-bond correlation). NMR spectra (1H and 31P) of the cell–lipid samples were processed using the MestreNova 10.1 software (Mestrelab Research S.L., Escondido, CA). The identification of lipid classes from NMR data was achieved by comparisons with our previous NMR measurements carried out on commercially available lipid standards.Optimization of GSL standard for ESI MS/MS analysis
The characteristic fragmentation of the GSL standard, C16 β-galactosyl-ceramide (d18:1/16:0), was studied and optimized using ESI triple-quadrupole MS/MS (API 3000; Applied Biosystems, Italy). Full scans and MS/MS scans were performed in both positive and negative ionization modes. Class-specific precursor ion scanning (PIS) of the sphingosine-based (d18:1 backbone) ions at m/z 264 and of the dihydrosphingosine-based (d18:0 backbone) ions at m/z 266 in ESI-positive ion mode33 were optimized to characterize the d18:1-based and d18:0-based GSLs, as indicated by the precursor ion form of [M + H]+. Both daughter ions are attributable to the sphingosine (264 Da) or sphinganine (266 Da) backbones that were derived from the loss of the N-acyl chain and two water molecules from the corresponding [M + H]+ parent ion.HPLC-ESI MS/MS measurements
The previously developed methods for LC-MS/MS analysis28 were used with modifications, which here also included GSL analysis. The optimized class-specific MS/MS head group survey scan with PIS and neutral loss scanning (NLS) combined with High Performance LCTM (CBM-20 A; Shimadzu, Italy), equipped with a binary pump (LC-20AB; Shimadzu, Italy) were used to separate and analyze the phospholipid and GSL extracts from RT4 and T24 cells. In short, cellular lipid extracts were separated using C18 columns (Kinetex; pore size, 100 Å; inner diameter, 4.6 mm; particle size, 2.6 μm; column length, 10 cm; Phenomenex, Italy) with mobile phase A as methanol/H2O (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, v/v) + 10 mM ammonium acetate and mobile phase B as methanol + 10 mM ammonium acetate. The gradient started from 70% B to reach 100% B in 45 min, and was then maintained at 100% B for 20 min, which was then followed by re-equilibration to the starting conditions for 15 min. The samples (10 μl) were injected with a flow rate of 1 ml min−1. The MS/MS analyses were performed in positive ionization mode with optimized analytical parameters as follows: NEB (Nebulizer Gas) 9, CUR (Curtain Gas) 10, TEM (Temperature) 300 °C, IS (IonSpray Voltage) 5 kV, CAD (collision gas) 4, DP (Declustering Potential) 65, FP (Focusing Potential) 250, EP (Entrance Potential) 5, CXP (Collision Cell Exit Potential) 18 and CE (Collision Energy) 50 for GSL/CE 40 for phospholipid analysis. Q1 and Q3 were both set with unit resolution and step size 0.1 amu. PC, lyso PC (LPC), ether PC (e-PC) and sphingomyellins (SMs) were characterized by PIS on m/z 184, which indicated the precursor ion form of [M + H]+. To evaluate the reproducibility of the LC-MS/MS analysis, the lipid extracts of RT4 and T24 cells which were grown, extracted and analyzed repeatedly in 3 different months were injected in duplicate. Phosphoserine (PS) with precursor ion [M + H]+, phosphatidic acid (PA) with precursor ion [M + NH4+]+, phosphoglycerol (PG) with the form of [M + NH4+]+, and all phosphoethanolamine (PE) with ([M + H]+) and phosphoinositol (PI) with ([M + NH4+]+) as the precursor ion forms, were detected and analyzed by NLS of 185, 115, 189, 141 and 277 Da, respectively.
3, v/v) + 10 mM ammonium acetate and mobile phase B as methanol + 10 mM ammonium acetate. The gradient started from 70% B to reach 100% B in 45 min, and was then maintained at 100% B for 20 min, which was then followed by re-equilibration to the starting conditions for 15 min. The samples (10 μl) were injected with a flow rate of 1 ml min−1. The MS/MS analyses were performed in positive ionization mode with optimized analytical parameters as follows: NEB (Nebulizer Gas) 9, CUR (Curtain Gas) 10, TEM (Temperature) 300 °C, IS (IonSpray Voltage) 5 kV, CAD (collision gas) 4, DP (Declustering Potential) 65, FP (Focusing Potential) 250, EP (Entrance Potential) 5, CXP (Collision Cell Exit Potential) 18 and CE (Collision Energy) 50 for GSL/CE 40 for phospholipid analysis. Q1 and Q3 were both set with unit resolution and step size 0.1 amu. PC, lyso PC (LPC), ether PC (e-PC) and sphingomyellins (SMs) were characterized by PIS on m/z 184, which indicated the precursor ion form of [M + H]+. To evaluate the reproducibility of the LC-MS/MS analysis, the lipid extracts of RT4 and T24 cells which were grown, extracted and analyzed repeatedly in 3 different months were injected in duplicate. Phosphoserine (PS) with precursor ion [M + H]+, phosphatidic acid (PA) with precursor ion [M + NH4+]+, phosphoglycerol (PG) with the form of [M + NH4+]+, and all phosphoethanolamine (PE) with ([M + H]+) and phosphoinositol (PI) with ([M + NH4+]+) as the precursor ion forms, were detected and analyzed by NLS of 185, 115, 189, 141 and 277 Da, respectively.
LC-MS data processing
The acquired raw MS/MS spectra were processed using Analyst (Version 1.42), with extraction of the reference ions from the ‘total ion chromatogram’ (TIC), and integration of the peak areas from the ‘extracting ion chromatogram’ upon aligning the mass tolerance and peak smoothing. To reduce the impact of the analytical variability, the relative percent amount of every species belonging to a given phospholipid class was normalized to the total TIC signal of this lipid class in each sample; for example, the relative molar fraction of each PC species in a given sample was obtained by the ratio of its area to the total area of PC lipids. The same procedure was followed for all the lipid classes that were investigated. It is important to point out that this type of normalization allows evaluating the intra-class relative abundance of each lipid species, but is not suitable to compare the relative abundances of lipids belonging to different classes. The lipid molecular species, identified by scanning (i.e., PIS, NLS) data from LC-MS/MS measurements, were also annotated on the basis of their retention times and through inquiries of ‘Lipid Mass Spec. Prediction’ (LIPID MAPS) by surveying the parameters of ‘Mass’ (the m/z value of a lipid species), ‘Mass tolerance’ (±0.5 m/z), ‘Head group’ (PC or PE, PI, PS etc.), and ‘Precursor ion’ ([M + H]+ or [M + NH4+]+). Specifically, identification of various GSLs (including glucosylceramide [GlcCer], α-hydroxylated glucosylceramide [GlcCer(h)] and lactosyl ceramide [LacCer]) was achieved through their retention times and the LIPID MAPS Structure Database.Each lipid species was considered as a variable, and hence a data matrix comprised of rows that represented the different samples and columns of variables as the relative ratios of individual lipids was subjected to MVDA, using SIMCA-P 13.0 (Umetrics, Italy). Principal component analysis (PCA), an unsupervised multivariate method, was first used to visualize the preprocessed MS/MS data, reduce the dimensionality of the datasets and to highlight systematic patterns, sample groupings, trends of variation, and differences between the RT4 and T24 cells. PCA was performed on the Pareto-scaled data matrix. In order to identify the most important lipid species which differentiate the RT4 and T24 cells, univariate unpaired t-test with Bonferroni's multiple testing correction at the 0.05 level was carried out with the GraphPad Prism 5 software. Testing was performed after averaging the lipid concentration of each analytical replicate. Finally, the distribution/variation of fatty acyl carbon chain length and double bonds (db) were evaluated. The Unsaturation Index (UI) analysis was performed, wherein the relative abundance of individual lipid species was multiplied by their corresponding unsaturation number.
Results and discussion
Morphological comparison between RT4 and T24 cells
Initially, the morphological features of the noninvasive urothelial papilloma RT4 cells and invasive urothelial carcinoma T24 cells were compared (Fig. 1). The RT4 cells firmly bound to each other, showing the typical polygonal epithelial shape. In contrast, the invasive urothelial carcinoma cells, the T24 cells, are well spread, with wider intercellular spaces and only occasional contacts mainly by multiple interconnecting nanotubes (Fig. 1, right panel). Fluorescent labelling of actin filaments with phalloidin showed clear difference in distribution of actin filaments forming short, condensed stress fibers at the basal part of RT4 cells and thin, dense actin cortex close to the plasma membrane in the middle region of the cells. On the other hand the T24 cells have thicker stress fibers and clearly distinguished network close to the plasma membrane seen in optical sections within the middle region of the cells (Fig. 1). Labeling of liquid ordered raft domains with ostreolysine mCherry showed that T24 cells are anchored to the growing substrate with much longer and more frequent tubular protrusions in T24 cells in comparison to RT4 cells (Fig. 1). The morphological comparisons between the RT4 and T24 cells showed characteristics, which are likely to be associated with different abilities in forming intercellular connections between more and between less invasive tumor cells and with differences in the set of lipids that make up these membrane structures.341H-NMR and 31P-NMR analysis of RT4 and T24 lipid extracts
The lipids extracted from the cultured RT4 and T24 cells were dissolved in CD3OD for 1D and 2D-NMR determinations at 300 K. The 1H-NMR spectrum (128 transients, 5 s relaxation delay) provided information to study the overall lipid profiles of the cell membrane, including triacylglycerols (TAGs) and cholesterol (Chol). A careful inspection of 1H-NMR spectra of these lipid extracts showed a very significant difference in the overall distribution of unsaturations in the acyl chains. Indeed, whilst in the T24 cell extract (blue line) the peak area of the signals at δH ∼ 2.84, attributable to bis-allylic protons (present only in PUFA chains), was much higher than the corresponding peak area in the RT4 cell extract (red line), the opposite change was observed for the allylic protons, present in all the unsaturated chains, at δH ∼ 2.04. Concerning the positions of the double bonds within the PUFA chains, 1H-NMR spectra clearly indicated that they must be ω-6 type, since the downfield triplet expected at δH 0.97 for the terminal methyl in ω-3 type was undetectable (both for RT4 and T24) and no long-range coupling was found in this region neither with olefinic protons (δH = 5.34) nor with bis-allylic protons (δH = 2.84) in the corresponding TOCSY-2D spectrum. Thus, strong evidences indicated that, during the development of urothelial carcinoma cell invasiveness, the FA chains are significantly rewired at first by the action of Δ12-desaturase enzyme. Another striking difference emerging from the 1H-spectra analysis was the relative amount of TAGs (Fig. 2B) as estimated by the relative peak area of the signals at δH 4.36 (dd), attributable to the isochronous protons at the C-1 and C3 positions of the TAG glycerol backbone. The TAG relative amount was found much greater in RT4 than in T24 cell extracts; in the latter, TAGs were almost totally absent if we consider that residual signals present in this region (δH ∼ 4.36) in 1H-NMR spectrum of T24 cells extract is mainly due (TOCSY-2D analysis) to the H–C(1) olefinic proton of 1Z-alkenyl chains of plasmenyl-phospholipids (p-PC + p-PE). Other minor differences were also detected, such as a small increase in SM, but the main structural information was essentially that in T24 cells, the ω-6 PUFA lipids were significantly increased, while the TAG neutral lipids were significantly decreased. Worth of note, the molar ratio of [Chol]/[all PC + SM] as obtained by peak-normalized-area ratio of the corresponding methyls signals was slightly lower for T24 cells (0.50 ± 0.01) than RT4 cells (0.54 ± 0.01) (Fig. 2A). Since the cholesterol content (expressed as μg cholesterol per mg of cell protein) was found slightly higher in T24 (28.0 ± 0.51) in comparison to RT4 (22.9 ± 1.3),8 our NMR analysis suggests an increased relative amount of PC and SM lipids in invasive cells, thus further reinforcing (in the overlay of Fig. 2A and B, the blue line should be increased of about 15%) the conclusion that higher invasiveness (T24) is related to an higher relative amount of unsaturated FA.The last outcome finds support in the corresponding 31P-NMR spectra (Fig. 2C) that shows its complementarity to 1H-NMR spectra by (i) allowing an inter-classes PL relative distribution and (ii) adding another important piece of information concerning the relative amount of 1-alkyl (ether)-2-acyl PC lipids (e-PC). In brief, for RT4 cells, the e-PC lipids represented a significant part (∼17%) of the overall PLs, while they were almost depleted for T24 cells. Moreover, this analysis confirmed the information, only guessed by 1H-NMR, that not only the relative molar fraction of SM was higher in T24 (∼6.5%) than RT4 cells (∼3.5%) but also that the area of the signal attributable to unresolved PC + plasmenyl-PC at δP = −0.55 was higher for T24 cells (∼47%) than for RT4 cells (∼40%) thus accounting for the overall strong reduction in e-PC lipids observed for T24 cells. Although further biosynthetic investigations are required to understand the mechanisms of lipid trafficking, our data suggest that at least partially, the progression in urothelial carcinoma invasiveness leads also to a rewiring of 1-alkyl, 2-acyl PC into 1,2-diacyl PC.
As the first snapshot of the cell–lipid profile, NMR analysis provided useful information in several aspects including structural alterations in the fatty acyl chains, estimations of the phospholipid and TAG ratios, and quantitative variations in the phospholipid classes. To summarize, compared with the noninvasive urothelial papilloma cells (RT4), the invasive urothelial carcinoma cells (T24) have molar ratios (i) much lower for TAGs, as defined by the [TAG]/[all PC + SM] ratio, (ii) much lower for 1-alkyl (ether)-2-acyl PC, (iii) similar for 1-alkenyl-2-acyl (plasmenyl) phospholipids, (iv) higher for ω-6 PUFA phospholipid acyl chains, (v) slightly higher for SM (18:1 based-ceramides), as defined by the SM/[all PC + SM] ratio, (vi) slightly lower for cholesterol, as defined by the [Chol]/[all PC + SM] ratio; and finally (vii) slightly higher for the total (PC + plasmenyl PC), as defined by the [PC + plasmenyl PC]/[all PC + SM] ratio.
As the changes in the TAGs and ω-6 PUFAs were inversely related, the increased ω-6 PUFAs in urothelial cancer cells with enhanced cell invasiveness and metastatic potential (i.e., T24 cells) cannot be attributed to the TAG acyl chains, but instead to an ‘unsaturation rewiring’ of the acyl chains of the phospholipids and/or SMs. Such interesting earmarks might be indications of the progression of the urothelial carcinoma invasive cell phenotype. Similar investigations were performed by Stenman K. et al. (2009) to characterize the ω-6 PUFAs in human malignant prostate tissue, where they had proposed the relevant factors of malignancy gain to be dietary effects and variations in mitochondrial membrane breakdown during cell death and/or fast turnover of lipid regions.35 As we discussed in our previous study,28 the increase in the overall lipid unsaturation (i.e., increased unsaturation index) might determine the altered membrane fluidity and the enhanced endoplasmic reticulum activity (which contains primarily unsaturated phospholipid species), in comparison to plasma membrane activity, where sphingolipids, sterols and saturated phospholipid species are more abundant. In order to confirm this, however, lipidomic analyses of subcellular fractions should be performed, in the future. In a word, the urothelial carcinoma cells with higher metastatic potential (T24 cells) would have higher levels of ω-6 PUFA acyl chains together with reduced TAGs content, contributing to the increased membrane fluidity that facilitates their invasiveness and motility.
Despite the important structural information obtained by the NMR measurements here, more comprehensive information on the lipid molecular species (e.g., lipid identity, chain length and number of unsaturations) was obtained using HPLC-ESI MS/MS.
Cellular lipids comparison between RT4 and T24 cells by HPLC-ESI MS/MS measurements
As shown for the TIC in the top panel of Fig. 3A, there were large changes in the relative peak intensities of the choline-containing phospholipid species for the RT4 cells (Fig. 3A, red line) compared to the T24 cells (Fig. 3A, blue line). First of all, great variations occurred in PC species with short chain lengths, among which the MS spectrum of PC 28:0 is shown in the middle panel of Fig. 3A: in the T24 cells in particular, these species were particularly low. Another significant difference is shown in the bottom panel of Fig. 3A (retention time window, 20.2–20.4 min) for the identified lipid species e-PC 34:1 belonging to the ether-PC class (alkyl 18:0/acyl 16:1). With the class-specific survey scanning, quantitative data were generated from peak area of the detected precursor ions on the assumption that all the species belonging to the same class had nearly identical ESI-MS response factors.36 As shown in Fig. 3B, the amount of e-PC decreased significantly in T24 cells, an outcome in fair agreement with the above-discussed 31P-NMR data.It is known that specific genetic alterations that accompany the progress of malignant transformation can modify the synthesis and breakdown of choline-containing phospholipids, with higher levels of choline metabolites used as a diagnostic marker of malignancy for breast cancer.37 With the development of metastatic potential of cancer cells, choline metabolism and choline-derived metabolites can go through alterations via various metabolic pathways. The study of Katz-Brull et al. (2002)37 demonstrated that the reduced levels of a choline-ether phospholipid might serve as a metabolic marker for breast cancer. Similarly, our HPLC-MS/MS results showed a decrease (∼3-fold) in 1-alkyl, 2-acyl PC species in the invasive urothelial carcinoma cells (i.e., T24 cells) compared with their noninvasive counterpart (i.e., RT4 cells). The role of e-PCs in different levels of urothelial cancer cell invasiveness is not clear, but their strong reduction with enhanced urothelial cell motility and invasiveness might find a reasonable explanation within the biochemical scenario highlighted in recent reports in the literature.38,39 Indeed, according to these reports, the more rapid choline uptake and kinase activity in cells with higher metastatic potential is related to the overproduction of the biochemical precursor free phosphocholine (−O3POCH2CH2N+(CH3)3), which leads to strong perturbation of the biosynthesis of all the PC species, including their ether analogues (i.e., e-PCs). Thus, the substantially decreased e-PC in the invasive T24 urothelial carcinoma cells can not only be used as a biochemical feature of urothelial cancer cell invasiveness, but can also indicate that in the progression of the invasive phenotype, the biosynthesis of 1-alkyl, 2-acyl PC is strongly suppressed. On the other hand, the biosynthesis of the ether phospholipids starts in peroxisomes, with the acylation of dihydroxyacetonephosphate (DHAP) by the enzyme DHAP acyltransferase, followed by the formation of the ether linkage by the enzyme alkyl-DHAP synthase, which catalyzes the exchange of the acyl chain in acyl-DHAP for a long-chain fatty alcohol.40 Furthermore, e-PC lipids have been demonstrated to be the biogenetic precursors of the strong biologically active 1-alkyl, 2-acetyl PC (i.e., platelet-activating factor), a family of naturally occurring acetylated phospholipid mediators that are involved in inflammation processes.41
As previously mentioned, ceramides and GSLs are important lipid classes especially in epithelia and the abnormal synthesis/expression of these altered lipid variants has been extensively shown to be involved in cancer pathogenesis; thus, an RPLC-ESI (+) MS/MS method was developed also for their characterization. Direct comparisons were carried out for the precursor ion chromatograms that indicated d18:1-based ceramide (Cer), glucosylceramide (GlcCer), their α-hydroxylated analogues (GlcCer(h)), and lactosyl ceramide (LacCer) from both RT4 (Fig. 4A) and T24 (Fig. 4B) cells. Various ceramide species had higher abundance in both cell lines, such as Cer(d18:1/16:0) at the nominal mass 539 Da shown in the middle panel of Fig. 4A and B. Despite common Cer and GlcCer lipids, RT4 and T24 cells have distinct profiles for GlcCer(h) and LacCer. In fact, several highly abundant GlcCer(h) species for RT4 cells such as hydroxylated-GlcCer(d18:1/16:0) at the nominal mass 717 Da shown in the bottom panel of Fig. 4A were completely absent for T24 cells; on the other hand, most of the LacCer species for T24 cells (such as LacCer(d18:1/24:1) in the bottom panel of Fig. 4B,) were below the limits of detection for RT4 cells. Quantitative data of these changes, shown in Fig. 4C, suggest a significant reduction of GlcCer(h) along with increased LacCer levels in T24 cells.
GSLs have been reported as tumor-associated cell–surface antigens correlated with cell adhesion and modulators of signal transduction42,43 as well as involved in cell differentiation, carcinogenesis and cancer pathogenesis such as epithelial ovarian cancer.44–47 The variations in LacCer might be relevant for altered (down-regulated or reduced) cell–cell adhesion, which is an important feature for carcinoma cell invasiveness. GlcCer(h) is an important GSL, although despite its brief description for the nervous system, epidermis and kidney, its physiological and pathological roles have been overlooked and are mostly unknown. As described by Coderch L. et al.,48 major epidermal hydroxylated ceramide and GlcCer can be classified as α-hydroxylated or ω-hydroxylated. Also GlcCer(h) with fatty acyl chain lengths of <26 carbons usually have α-hydroxy structures, while the ω-hydroxy fatty acyl structure is seen for GlcCer(h) with fatty acyl chain lengths of 30–40 carbons. However, the profiles and the functions of GlcCer(h) in biological tissue other than skin and the nervous system have rarely been studied. Here, we thus propose that depletion of GlcCer(h) for the urothelial carcinoma T24 cells with invasive properties is highly correlated with the abnormal permeability barrier structure,49 and thus non-functional blood–urine barrier. Moreover, the unique enzyme fatty acid 2-hydroxylase for GlcCer(h) biosynthesis and the only known peroxisomal α-oxidation pathway for GlcCer(h) degradation have been described.50 Therefore, alterations in the enzyme or genes related to these pathways might contribute to the mechanisms behind the GlcCer(h) differences between RT4 and T24 cells.
Multivariate data analysis and analysis for potential individual markers
According to our results shown in Fig. 3, there were relevant changes in choline-containing lipid species with shorter chain lengths and in 1-alkyl, 2-acyl PC. In order to reveal such variation patterns and to further analyze differences in lipids between RT4 and T24 cells, MVDA was performed by systematically looking at the individual lipid species. Fig. 5 illustrates the MVDA that was conducted on the 42 PC species characterized for both types of cells. The PCA scores plot (Fig. 5A) shows a clear-cut separation between RT4 and T24 cells in the principal component (PC1) dimension, which suggests that PC lipid profiles varied significantly between the noninvasive and invasive cells. The first two components of the PCA model cumulatively accounted for 96% of the total variability. The first principal component (% of variance) clearly separated the two types of cells, while the second (% of variance) was associated with the biological variability of the samples and the technical reproducibility of the data. | ||
| Fig. 5 Multivariate data analysis of 42 cellular phosphatidylcholine (PC) species in RT4 and T24 cells. (A) PCA scores plot. (B) PCA biplot. | ||
The contribution of each lipid species to the separation of RT4 and T24 cells in the PCA scores plot was further investigated in the corresponding PCA biplot (Fig. 5B), which shows the RT4 and T24 replicates and 42 PC species as variables in the same graph. The variables, which were displayed along the first component in correspondence of RT4 cells, represented the PC species that were more abundant in the noninvasive RT4 cells, while on the opposite side were reported the featured PC species in the invasive T24 cells. The PC species further away from the central part of Fig. 5B represented as more featured variables for the corresponding separated cell types and showed the most substantial difference between two cell types. As seen in Fig. 5B, PC species with shorter chain length and low unsaturation index (0 or just one double bond) were much more representative for RT4 cells, while PC species with relatively longer chain length and higher unsaturation index were enriched in T24 cells.
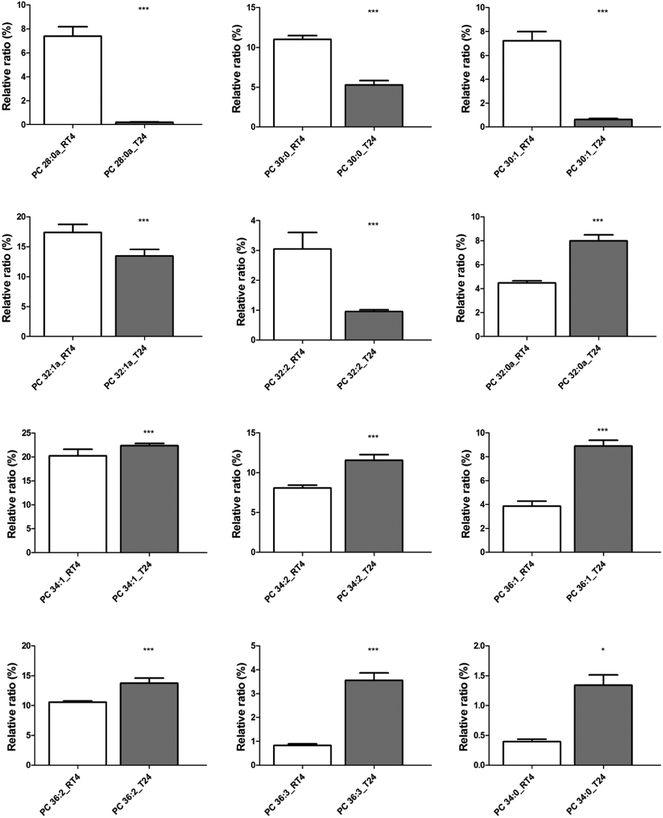
In order to verify the presence of statistically significant differences in the concentration of the main PC lipid markers revealed from the PCA biplot (Fig. 5B), t-test with Bonferroni's multiple testing correction at the 0.05 level was performed.51 As presented in the analysis of individual PC species shown in Fig. 6, the increased urothelial carcinoma invasiveness was accompanied by particularly lower levels of PC species bearing fatty acids with shorter chain (<32 carbons) and lower unsaturation index (PC 28:0a, PC 30:0, PC 30:1, PC 32:1). An exception was seen for PC 32:2, which has two double bonds in its fatty acyl chain. At odd, PC 32:0a (i.e. an isomer of PC 32:0) was highly enriched in T24 cells. However, except for this shorter saturated species, in comparison with RT4 cells, the T24 cells show a higher concentration of fatty acyl chain length as well as an increased unsaturation level. MVDA and univariate data analysis of the phospholipid molecular species from other classes were carried out in a similar workflow. The identified lipid species were listed in Table S1 (ESI†). And similar to PC lipids, the T24 cells show a higher concentration of PE and ether PE (e-PE) lipids which have longer fatty acyl chain length and increased unsaturation level.
Fatty acyl (FA) carbon chain length, double bonds (db) distribution/variation and unsaturation index (UI) analysis
According to these analyses, in order to highlight general trends in the two model systems, we have compared the distribution and the variation of fatty acyl carbon chain length (Fig. S1, ESI†), unsaturations (double bonds, Fig. S2, ESI†), and Unsaturation Index (UI, Fig. S3, ESI†). As indicated in Fig. S1A (ESI†), the PC lipids had fatty acyl chains with carbon numbers ranging from 28 to 40, among which the dominant ones were those with 34 carbons (C). It is clearly shown in these analyses that the proportion of these PC lipids with less than 32 C dramatically decreased in the invasive T24 cells compared with RT4 cells, whereas the PC lipids with 34–40 C significantly increased in T24 cells. A very similar trend was also seen for ether PC (e-PC), except that e-PC lipids with 42 C and 44 C decreased again in T24 cells (Fig. S1B, ESI†). As far as SMs are concerned, there were three trends in different carbon number distribution blocks (Fig. S1C, ESI†). The proportion of SMs with 32–34 C decreased in T24 cells, whereas SMs with 36–42 C were significantly enriched upon increased invasiveness of T24 cells; SMs with the longest chains (43, 44 C) decreased again in the invasive T24 cells compared with the noninvasive counterpart RT4 cells.Apart from the carbon number distributions and differences, another important feature of the lipids, the unsaturation distribution (i.e., double bonds, db) was also worth investigation (Fig. S2, ESI†). A clearer pattern and trend in unsaturation (db) differences due to increased urothelial cancer cell invasiveness was established: the invasive T24 cells with metastatic potential had lower fractions of saturated or mono-unsaturated (0, 1 db) PC species (Fig. S2A, ESI†) as well as the ether forms (e-PC, Fig. S2B, ESI†), while di-unsaturated and polyunsaturated PC and e-PC species were much more enriched in T24 cells (2–7 db), compared with RT4 cells. A further UI analysis was also performed to determine the total unsaturation variation in T24 cells with respect to RT4 cells (Fig. S3, ESI†) and we found overall higher UI in T24 cells. Since, as above reported, the PUFA chains responsible for this UI increase in the T24 cell extracts are of ω-6 type, our findings indicate that the over-production of ω-6 PUFAs represents not only a marker of urinary bladder carcinoma but also of its increased invasiveness.
The differences between the two cell types with their corresponding responsible PC and PE lipid species have been revealed by MVDA and the further univariate analysis of the main PC and PE lipid markers. And to sum up, the general trends from the above analyses of fatty acyl (FA) carbon chain length indicate that T24 cells have reduced levels of shorter lipids, and enriched longer lipids, but reduced proportions of the longest lipids. A possible explanation is that to facilitate their invasiveness to the surrounding cells or environment, the invasive urothelial carcinoma cells (e.g., T24 cells) keep an optimal plasma membrane and other cellular membrane fitness stiffness. The difference between the optimal carbon numbers of SM and PC (and e-PC) might be due to ceramide backbone specificity. As illustrated in Fig. S2 and S3 (ESI†), T24 cells had higher unsaturation (db) in their lipid fatty acyl chains, thus suggesting that the invasive urothelial carcinoma cells might require less stiffness for their movement and invasiveness.22 Such findings are in agreement with previous NMR analyses that T24 cells have higher levels of phospholipids with ω-6 PUFA acyl chains. Although the correlation between the higher levels of ω-6 PUFA phospholipids and urothelial cancer cell invasiveness is not clear, some studies have indicated that dietary fish-oil-derived ω-3 PUFAs (e.g., docosahexaenoic acid in particular) can reduce the invasive phenotype of bladder and pancreatic carcinoma cells.52 In agreement with above-reported analysis, shorter and saturated or mono-unsaturated lipids are dominant in the noninvasive urothelial papilloma cell type (RT4), while the urothelial carcinoma cells with invasive and metastatic potential (T24) have lipids with longer (but not the longest FA) and more unsaturated fatty acyl chains.
Conclusions and perspectives
The previously developed NMR and RPLC-MS/MS-based methodology was used here to characterize cellular lipids. The in vitro models of two human urothelial cancer cell lines were used to study the differences in the lipid profiles associated with different levels of urothelial cancer cell invasiveness. In this joint NMR and LC-MS/MS analysis, these data show that the increased urothelial cancer cell invasiveness for T24 cells is associated with: (a) a strong (3 fold at least) decrease of the relative levels of e-PC lipids, together with the increased ω-6 PUFA acyl chains of the phospholipids; (b) selectively decreased lipid species with shorter and saturated/mono-unsaturated acyl chains; (c) significantly increased lipid species with longer chain lengths, and with di-unsaturated and polyunsaturated fatty acyl chains; and (d) a depletion of GlcCer(h) and increased levels of LacCer. To summarize, our results demonstrate that the two types of cells, papilloma and carcinoma urothelial cells, express some significant differences of cellular membranes including changes in their compositional lipid structures. The molecular pathways whereby cells undergo these large changes remain to be determined, with extensive biosynthetic and biochemical investigations required to explain these findings further. Seventy percent of urothelial cancers at diagnosis are noninvasive papilloma types with low frequency for progression to highly invasive or metastatic carcinomas.52 Thus our data as discussed above may help to explore extensive therapeutic targets and agents for new therapies.Acknowledgements
This research was supported by University of Trento, Department of Physics. The authors would like to thank Adriano Sterni (Univ. of Trento) for special LC-MS measurements and helpful technical assistance. We also thank CIBIO-UNITN for financing the Doctoral Scholarship for Yang Yu and acknowledge Dr Christopher Berrie for critical reading of the manuscript. The Slovenian authors gratefully acknowledge the Slovenian Research Agency for financial support (grants J4-7162, J3-7494 and P1-0207, P3-0108).References
- R. Romih, P. Korošec, W. de Mello, Jr. and K. Jezernik, Cell Tissue Res., 2005, 320, 259–268 CrossRef PubMed.
- M. E. Kreft, S. Hudoklin, K. Jezernik and R. Romih, Protoplasma, 2010, 246, 3–14 CrossRef PubMed.
- I. Sterle, D. Zupančič and R. Romih, BioMed Res. Int., 2014, 2014, 805236 Search PubMed.
- K. Klopfer, B. Delahunt, M. Adamson and H. Samaratunga, Anticancer Res., 2014, 34, 6779–6784 Search PubMed.
- W. Li, Y. Liang, M. T. Deavers, A. M. Kamat, S. F. Matin, C. P. Dinney, B. Czerniak and C. C. Guo, Am. J. Clin. Pathol., 2014, 142, 864–871 CrossRef PubMed.
- W. Tian, G. Guner, H. Miyamoto, A. Cimino-Mathews, N. Gonzalez-Roibon, P. Argani, X. Li, R. Sharma, A. P. Subhawong, K. Rezaei, T. J. Bivalacqua, J. I. Epstein, J. A. Bishop and G. J. Netto, Hum. Pathol., 2015, 46, 58–64 CrossRef CAS PubMed.
- H. Tsumura, K. Matsumoto, M. Ikeda, K. Yanagita, S. Hirano, M. Hagiwara, R. Nagashio, T. Fujita, Y. Sato and M. Iwamura, Asian Pac. J. Cancer Prev., 2015, 16, 1539–1543 CrossRef PubMed.
- N. Resnik, U. Repnik, M. E. Kreft, K. Sepčić, P. Maček, B. Turk and P. Veranič, PLoS One, 2015, 10, e0137878 Search PubMed.
- A. Rezaianzadeh, A. Mohammadbeigi, J. Mobaleghi and N. Mohammadsalehi, J. Mid-life Health, 2012, 3, 88–92 CrossRef PubMed.
- F. Baenke, B. Peck, H. Miess and A. Schulze, Dis. Models & Mech., 2013, 6, 1353–1363 CAS.
- U. Cavallaro and G. Christofori, Biochim. Biophys. Acta, Rev. Cancer, 2001, 1552, 39–45 CrossRef CAS.
- L. Hu, W. Hittelman, T. Lu, P. Ji, R. Arlinghaus, I. Shmulevich, S. R. Hamilton and W. Zhang, Lab. Invest., 2009, 89, 531–548 CrossRef CAS PubMed.
- S. K. Patra, in Signal Transduction in Cancer Metastasis, ed. W.-S. Wu and C.-T. Hu, Springer Netherlands, Dordrecht, 2010, ch 11, pp. 209–224 Search PubMed.
- S. K. Patra, Biochim. Biophys. Acta, Rev. Cancer, 2008, 1785, 182–206 CrossRef CAS PubMed.
- S. K. Patra, Exp. Cell Res., 2008, 314, 1193–1201 CrossRef CAS PubMed.
- T. Murai, Int. J. Cell Biol., 2012, 2012, 763283 Search PubMed.
- I. S. Babina, E. A. McSherry, S. Donatello, A. D. Hill and A. M. Hopkins, Breast Cancer Res., 2014, 16, 1–14 CrossRef PubMed.
- R. Imani, P. Veranič, A. Iglič, M. E. Kreft, M. Pazoki and S. Hudoklin, Photochem. Photobiol. Sci., 2015, 14, 583–590 CAS.
- D. Kabaso, M. Lokar, V. Kralj-Iglič, P. Veranič and A. Iglič, Int. J. Nanomed., 2011, 6, 495–509 CrossRef CAS PubMed.
- C. Booth, P. Harnden, L. K. Trejdosiewicz, S. Scriven, P. J. Selby and J. Southgate, Lab. Invest., 1997, 76, 843–857 CAS.
- E. D. Lobban, B. A. Smith, G. D. Hall, P. Harnden, P. Roberts, P. J. Selby, L. K. Trejdosiewicz and J. Southgate, Am. J. Pathol., 1998, 153, 1957–1967 CrossRef CAS PubMed.
- H. Liu, Q. Tan, W. R. Geddie, M. A. Jewett, N. Phillips, D. Ke, C. A. Simmons and Y. Sun, Cell Biochem. Biophys., 2014, 68, 241–246 CrossRef CAS PubMed.
- J. Bubenik, M. Baresova, V. Viklicky, J. Jakoubkova, H. Sainerova and J. Donner, Int. J. Cancer, 1973, 11, 765–773 CrossRef CAS PubMed.
- R. Imani, D. Kabaso, M. Erdani Kreft, E. Gongadze, S. Penič, K. Eleršič, A. Kos, P. Veranič, R. Zorec and A. Iglič, Croat. Med. J., 2012, 53, 577–585 CrossRef CAS PubMed.
- S. Lahiri and A. H. Futerman, Cell. Mol. Life Sci., 2007, 64, 2270–2284 CrossRef CAS PubMed.
- Y. Igarashi and R. Kannagi, J. Biochem., 2010, 147, 3–8 CrossRef CAS PubMed.
- R. Kannagi, M. Izawa, T. Koike, K. Miyazaki and N. Kimura, Cancer Sci., 2004, 95, 377–384 CrossRef CAS PubMed.
- Y. Yu, L. Vidalino, A. Anesi, P. Macchi and G. Guella, Mol. BioSyst., 2014, 10, 878–890 RSC.
- M. Orešič, Eur. J. Lipid Sci. Technol., 2009, 111, 99–106 CrossRef.
- M. Skočaj, N. Resnik, M. Grundner, K. Ota, N. Rojko, V. Hodnik, G. Anderluh, A. Sobota, P. Maček, P. Veranič and K. Sepčić, PLoS One, 2014, 9, e92783 Search PubMed.
- C. S. Ejsing, J. L. Sampaio, V. Surendranath, E. Duchoslav, K. Ekroos, R. W. Klemm, K. Simons and A. Shevchenko, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 2136–2141 CrossRef CAS PubMed.
- J. L. Sampaio, M. J. Gerl, C. Klose, C. S. Ejsing, H. Beug, K. Simons and A. Shevchenko, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 1903–1907 CrossRef CAS PubMed.
- R. L. Shaner, J. C. Allegood, H. Park, E. Wang, S. Kelly, C. A. Haynes, M. C. Sullards and A. H. Merrill, J. Lipid Res., 2009, 50, 1692–1707 CrossRef CAS PubMed.
- N. Resnik, K. Sepčić, A. Plemenitaš, R. Windoffer, R. Leube and P. Veranič, J. Biol. Chem., 2011, 286, 1499–1507 CrossRef CAS PubMed.
- K. Stenman, J. Hauksson, G. Gröbner, P. Stattin, A. Bergh and K. Riklund, Magn. Reson. Mater. Phys., Biol. Med., 2009, 22, 327–331 CrossRef CAS PubMed.
- R. Taguchi, T. Houjou, H. Nakanishi, T. Yamazaki, M. Ishida, M. Imagawa and T. Shimizu, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2005, 823, 26–36 CrossRef CAS PubMed.
- R. Katz-Brull, D. Seger, D. Rivenson-Segal, E. Rushkin and H. Degani, Cancer Res., 2002, 62, 1966–1970 CAS.
- G. A. Ruiter, M. Verheij, S. F. Zerp and W. J. van Blitterswijk, Int. J. Radiat. Oncol., Biol., Phys., 2001, 49, 415–419 CrossRef CAS.
- T. Shah, F. Wildes, M. F. Penet, P. T. Winnard, Jr., K. Glunde, D. Artemov, E. Ackerstaff, B. Gimi, S. Kakkad, V. Raman and Z. M. Bhujwalla, NMR Biomed., 2010, 23, 633–642 CrossRef CAS PubMed.
- A. K. Hajra, Prog. Lipid Res., 1995, 34, 343–364 CrossRef CAS PubMed.
- F. Maria and H. Ralf, Lipid Oxidation in Health and Disease, CRC Press, 2015, ch. 8, pp. 151–172 Search PubMed.
- S. Hakomori, Biochem. Soc. Trans., 1993, 21, 583–595 CrossRef CAS PubMed.
- S. Hakomori, K. Handa, K. Iwabuchi, S. Yamamura and A. Prinetti, Glycobiology, 1998, 8, xi–xix CrossRef CAS PubMed.
- S. Hakomori, Acta Anat., 1998, 161, 79–90 CrossRef CAS PubMed.
- T. Pochechueva, F. Jacob, A. Fedier and V. Heinzelmann-Schwarz, Metabolites, 2012, 2, 913–939 CrossRef CAS PubMed.
- S. Lahiri and A. H. Futerman, Cell. Mol. Life Sci., 2007, 64, 2270–2284 CrossRef CAS PubMed.
- S. Basu, R. Ma, B. Mikulla, M. Bradley, C. Moulton, M. Basu, S. Banerjee and J. Inokuchi, Glycoconjugate J., 2004, 20, 157–168 CrossRef CAS PubMed.
- L. Coderch, O. López, A. de la Maza and J. Parra, American Journal of Clinical Dermatology, 2003, 4, 107–129 CrossRef PubMed.
- N. Amen, Doctor of Natural Sciences, University of Heidelberg, 2013.
- V. Foulon, M. Sniekers, E. Huysmans, S. Asselberghs, V. Mahieu, G. P. Mannaerts, P. P. Van Veldhoven and M. Casteels, J. Biol. Chem., 2005, 280, 9802–9812 CrossRef CAS PubMed.
- P. Franceschi, M. Giordan and R. Wehrens, TrAC, Trends Anal. Chem., 2013, 50, 11–21 CrossRef CAS.
- D. D'Eliseo, L. Manzi, N. Merendino and F. Velotti, J. Nutr. Biochem., 2012, 23, 452–457 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6mb00477f |
| This journal is © The Royal Society of Chemistry 2016 |