Open Access Article
Open Access ArticleUnprecedented staining of polar lipids by a luminescent rhenium complex revealed by FTIR microspectroscopy in adipocytes†
C. A.
Bader
a,
E. A.
Carter
b,
A.
Safitri
b,
P. V.
Simpson
c,
P.
Wright
c,
S.
Stagni
d,
M.
Massi
c,
P. A.
Lay
b,
D. A.
Brooks
a and
S. E.
Plush
*a
aMechanisms in Cell Biology and Disease Research Group, School of Pharmacy and Medical Sciences/Sansom Institute for Health Research, University of South Australia, Adelaide, Australia. E-mail: sally.plush@unisa.edu.au
bVibrational Spectroscopy Core Facility and School of Chemistry, The University of Sydney, Sydney, Australia
cSchool of Chemistry, Curtin University, Perth, Australia
dDepartment of Industrial Chemistry “Toso Montanari”, University of Bologna, Bologna, Italy
First published on 3rd May 2016
Abstract
Fourier transform infrared (FTIR) microspectroscopy and confocal imaging have been used to demonstrate that the neutral rhenium(I) tricarbonyl 1,10-phenanthroline complex bound to 4-cyanophenyltetrazolate as the ancillary ligand is able to localise in regions with high concentrations of polar lipids such as phosphatidylethanolamine (PE), sphingomyelin, sphingosphine and lysophosphatidic acid (LPA) in mammalian adipocytes.
Live cell fluorescent imaging provides a valuable tool for the visualisation of cellular structure and function. One key aspect of this research area is the design of luminescent molecules, either organic fluorophores or phosphorescent metal complexes (each of which possess unique advantages and disadvantages),1–7 that are able to target specific organelles in live cells or specific analytes of metabolic relevance. While significant advances have been reported in biological targeting, the specific staining of lipids (excluding those belonging to membrane layers) in live cells is an undoubtedly underdeveloped area. Lipids are critical for membrane structure, to compartmentalise cellular function, to orchestrate structural events at the membrane interface and to mediate signalling, but they also have a critical role in metabolic processes.8 Lipid distribution, degradation, homeostasis and metabolism have direct significance for major diseases, such as inflammation, diabetes, cancer, obesity, and heart disease.9 The availability of a diverse family of lipid markers, especially for visualising lipid orientated process in real time,10 therefore represents an unmet need in live cell imaging.
Advances in mass spectrometry have allowed high-sensitivity and high throughput lipid analysis for the study of lipid composition.11 However, the compositional analysis of lipids by electrospray mass spectrometry relies heavily on the ability to separate cellular components, which limits the biological relevance of the information obtained when using this technology. For example, the analysis of lipids found in lipid droplet organelles requires separation of the phospholipid membrane from the hydrophobic core and from other cellular components.12,13 Fluorescent dyes, such as Oil Red O, Nile Red, Fillipin III, BODIPY and LipidTox, are also commonly used for the imaging of lipid biology.14–16 However, the need for cell fixation for effective staining with Oil Red O, Nile Red, and Fillipin III results in lipid depletion.17 BODIPY has shown promise for live cell imaging of lipids. However, dual imaging can be difficult with emission potential in the red excitation channel as well as its expected green excitation.18 BODIPY and derivatives thereof also have a small Stokes-shift, which can lead to self-quenching and can be modestly cytotoxic.19 LipidTOX is amenable to live cell imaging, however, it is prone to photo-bleaching and suffers from low staining efficiency.20 Furthermore, the interaction of these dyes with lipids is non-specific by nature (e.g., Fillipin III has a higher specificity for gangliosides (GM1) in contrast to cholesterol;21 emission from Nile Red is dependent on hydrophobicity22), with the vast majority localising with neutral lipids or phospholipids. Consequently, there is a defined need for the development of new molecular probes that exclusively enable live cell imaging of polar lipids, particularly in conjunction with other biomarkers.23
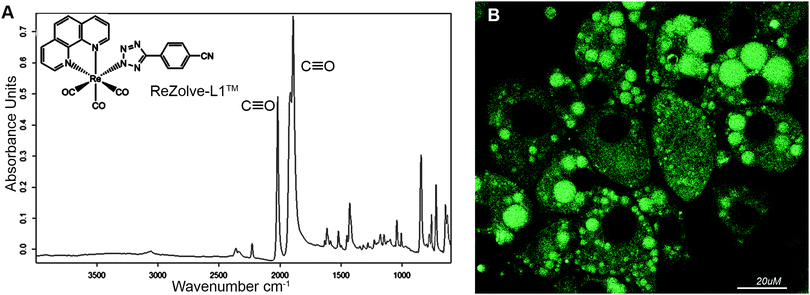
We have recently reported the first example of a luminescent metal complex capable of staining lipid droplets in live Drosophila tissue and live adipocytes.24 This species is composed of the neutral Re(I) tricarbonyl 4′-cyanophenyltetrazolato complex, herein designated as ReZolve-L1™ (Fig. 1A).‡ The staining of lipid droplets in live cells is significant as cellular dysfunction involving lipid droplets is increasingly being linked to major diseases.25,26 Lipid droplets are intracellular organelles that have important functions in membrane formation, cellular and lipid homeostasis (storage), metabolism (hydrolysis), and trafficking (signalling and transport).27,28
The observed staining of lipid droplets by ReZolve-L1™ (Fig. 1B) suggests that this metal complex has an affinity for one of the many lipid families found within lipid droplets. It is classically accepted that the lipid droplet cores are enriched in neutral lipids, like triacylglycerides and sterol esters for energy storage and the outer limiting membrane is a lipid monolayer mainly composed of polar lipids that can include unesterified cholesterol, sphingomyelin and phosphatidylethanolamine.29,30 The polar lipids in the outer membrane of the lipid droplet are integrally involved in lipid droplet dynamics, intracellular signalling, and lipid compartmentalisation, as well as lipid sequestration and trafficking.31,32 However, the localisation of lipids, especially polar lipids, in the lipid droplet structure has yet to be fully defined.33–35 Therefore, it is vital that the ‘selectivity’ of ReZolve-L1™ for these various lipid classes be defined.
Interestingly, ReZolve-L1™ was found to also be distributed to other regions of cells (Fig. 1B). A comparison of staining patterns of ReZolve-L1™ using Fillipin III as a co-stain or Oil Red O (reported in Bader et al.24 and reproduced as Fig. S1 in ESI†) suggests that ReZolve-L1™ may have an affinity for lipids distinct to that stained by either Fillipin III (cholesterol and GM1) or Oil Red O (neutral lipids). As there is a high demand for stains that are able to selectively discriminate polar from neutral lipids, the lipid ‘specificity’ of ReZolve-L1™ warranted further investigation.
Herein we demonstrate by means of IR microspectroscopy and confocal imaging36–38 the potential of this unique rhenium complex to localise with polar lipids in cells. Our results highlight the first example of a marker that is able to preferentially localise in areas with high polar lipid concentration, thus offering a novel biological tool for live cell imaging of lipids.
The FTIR spectrum of solid ReZolve-L1™ (Fig. 1A) is dominated by the strong C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O stretching bands centred at ∼2027 cm−1 and the doublet 1915/1893 cm−1, characteristic of facial tricarbonyl complexes.39 This is a spectral region where vibrational modes due to biochemical components are conveniently negligible.40 There is a range of additional bands for the complex in the region between 1800–600 cm−1 and, while these overlap with signals from cellular components, their relative intensities compared with those of the C
O stretching bands centred at ∼2027 cm−1 and the doublet 1915/1893 cm−1, characteristic of facial tricarbonyl complexes.39 This is a spectral region where vibrational modes due to biochemical components are conveniently negligible.40 There is a range of additional bands for the complex in the region between 1800–600 cm−1 and, while these overlap with signals from cellular components, their relative intensities compared with those of the C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O bands are such that they have not been used in the spatial analysis of the complex.
O bands are such that they have not been used in the spatial analysis of the complex.
To facilitate the imaging of lipid droplets (where ReZolve-L1™ has the highest intensity staining pattern), a mammalian adipocyte 3T3-L1 cell line was chosen, as these cells have large (up to 150 μm) lipid droplets when compared to other mammalian cells (0.1–2 μm).41 To prepare the sample for FTIR analysis, the 3T3-L1 cells were grown on a silicon nitride substrate,38 then rapidly fixed using cold methanol and air dried38,42 before being incubated with ReZolve-L1™ (10 μM). Importantly, direct growth of the cells on the silicon nitride substrate, followed by rapid cold MeOH fixation eliminates biochemical changes that can be induced by removing adherent cells from cell culture by trypsin, so cells grow with normal morphology.27 The cells prepared in this manner also retain the lipid droplets observed under optical microscopy and enable diverse biospectroscopic studies on adipocytes with minimal introduction of sampling artifacts.38,43
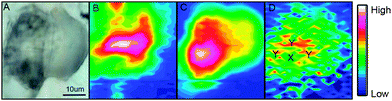
A bright field image of an adipocyte following incubation with ReZolve-L1™ is shown in Fig. 2A and depicts large characteristic lipid droplets. IR images generated by calculating the area under the protein amide I band (1690–1600 cm−1), the lipid ester C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O band (1712–1775 cm−1) and the ReZolve-L1™ C
O band (1712–1775 cm−1) and the ReZolve-L1™ C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O band (1942–1910 cm−1) from this cell are shown in Fig. 2B–D, respectively (see ESI,† Fig. S2 for a control cell). The FTIR image of the lipid ester, Fig. 2C, shows the highest integrated area coincides with the largest lipid droplet in the bright-field image Fig. 2A. In addition, a comparison of the highest integrated intensities from the amide I protein map against the ν(C
O band (1942–1910 cm−1) from this cell are shown in Fig. 2B–D, respectively (see ESI,† Fig. S2 for a control cell). The FTIR image of the lipid ester, Fig. 2C, shows the highest integrated area coincides with the largest lipid droplet in the bright-field image Fig. 2A. In addition, a comparison of the highest integrated intensities from the amide I protein map against the ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) lipid map showed that there was an area of high protein content adjacent to the lipid droplet location. This was assigned to the nucleus because of the high histone protein concentration in chromatin and is in close proximity to the major lipid droplet.
O) lipid map showed that there was an area of high protein content adjacent to the lipid droplet location. This was assigned to the nucleus because of the high histone protein concentration in chromatin and is in close proximity to the major lipid droplet.
Lipid droplets and nuclear proteins are often found in close proximity during the nascent formation of lipid droplets as the endoplasmic reticulum is an organelle for lipid synthesis.34,41 ReZolve-L1™ was detected throughout the cell, which was in agreement with the localisation of lipids and confocal microscopy (Fig. 1B). Increased intensities of the bands due to ReZolve-L1™ were observed in regions of high lipid content with some overlap in the area where lipids and high protein content were in close proximity, Fig. 2D. IR spectra were then extracted from two regions within the cell and averaged (Fig. 3; see ESI,† Fig. S3 for spectra from each region with correlating 2nd derivative analysis). These regions were selected based on the ReZolve-L1™ IR map shown in Fig. 2D in combination with confocal and epifluorescence images of both fixed and live adipocytes (Fig. 1B),24 which showed the highest accumulation of ReZolve-L1™ in these regions. In both extracted regions the relative absorbance from the ReZolve-L1™ ν(C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O) bands was relatively low in comparison with the other cellular functional groups. This is not unexpected as the cells were incubated with the same concentration of ReZolve-L1™ that is used in optical imaging experiments to ensure the same bio-distribution was obtained. Both extracted spectral averages show intense ν(CH) and ν(C
O) bands was relatively low in comparison with the other cellular functional groups. This is not unexpected as the cells were incubated with the same concentration of ReZolve-L1™ that is used in optical imaging experiments to ensure the same bio-distribution was obtained. Both extracted spectral averages show intense ν(CH) and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) lipid bands that can be attributed to high lipid concentration, in comparison to the smaller contribution from the ν(C
O) lipid bands that can be attributed to high lipid concentration, in comparison to the smaller contribution from the ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and ν(NH) protein bands. This relationship suggests that the spectra were extracted from the lipid droplet (hydrophobic core and membrane) and not the nuclei or cytoplasmic regions. As expected from the integrated intensities image shown in Fig. 2D, the spectra extracted from the area surrounding the lipid droplet marked Y in Fig. 2D (spectra shown in Fig. 3), including the lipid droplet membrane, had the highest relative ratio of absorbance attributed to ReZolve-L1™ when compared to known cellular functional groups. While the spectrum extracted from the region marked X in Fig. 2D (spectra shown in Fig. 3), inside the centre of the lipid droplet, showed only a relatively small concentration of ReZolve-L1™ in comparison to cellular functional groups. This suggested that ReZolve-L1™ may be localising in higher concentration with lipids found in the lipid droplet membrane rather than the neutral lipids (e.g., triglycerides).
O) and ν(NH) protein bands. This relationship suggests that the spectra were extracted from the lipid droplet (hydrophobic core and membrane) and not the nuclei or cytoplasmic regions. As expected from the integrated intensities image shown in Fig. 2D, the spectra extracted from the area surrounding the lipid droplet marked Y in Fig. 2D (spectra shown in Fig. 3), including the lipid droplet membrane, had the highest relative ratio of absorbance attributed to ReZolve-L1™ when compared to known cellular functional groups. While the spectrum extracted from the region marked X in Fig. 2D (spectra shown in Fig. 3), inside the centre of the lipid droplet, showed only a relatively small concentration of ReZolve-L1™ in comparison to cellular functional groups. This suggested that ReZolve-L1™ may be localising in higher concentration with lipids found in the lipid droplet membrane rather than the neutral lipids (e.g., triglycerides).
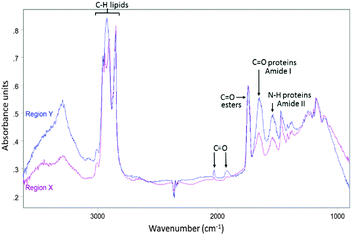 | ||
| Fig. 3 Average IR spectrum collected from region Y in Fig. 2D (blue) overlaid with average IR spectrum taken from region X in Fig. 2D (purple). | ||
To investigate if ReZolve-L1™ localises with specific lipids the spectra from the extracted regions in Fig. 3 were compared with spectra obtained from pure lipids. A large number of lipid structures are known to reside in cells, hence, a representative series of lipids from each of the main lipid families were chosen for analysis (see ESI,† Fig. S4 for reference IR spectra; see ESI,† Fig. S5 for peak assignments). Peaks were assigned for the extracted regions X and Y from second derivative spectra which allowed for the comparison of bands in the CH stretching region (3000–2700 cm−1) and the lipid references, see Fig. 4. Fig. 4 shows that the greatest correlation between the spectra extracted from high probe concentration (region Y) is with polar lipids. The bands at 2956, 2921, and 2848 cm−1 in the spectrum from region Y are coincident with the bands in the spectra collected from phosphatidylcholine, phosphatidylethanolamine, sphingomyelin, sphingosine and lysophosphatidic acid.
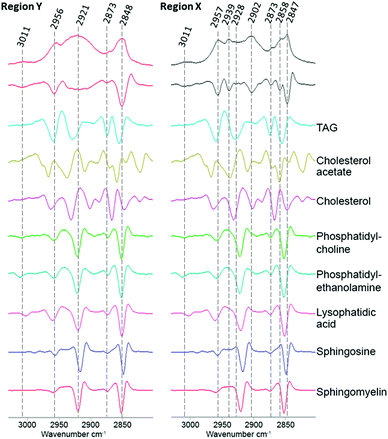 | ||
| Fig. 4 Comparison of the FTIR spectra from the CH stretching region (2750–3020 cm−1) extracted from region X and region Y (Fig. 2D) with second derivative spectra shown below followed by a range of pure lipid spectra as assigned. | ||
There was significantly less correlation with the non-polar lipids, such as cholesterol esters and triglycerides with the spectra from region Y. As expected the spectra extracted from region X showed high peak correlation with triglycerides and cholesterol acetate (2957, 2928 and 2858 cm−1). This suggests that ReZolve-L1™ mainly localised to areas directly adjacent to the hydrophobic core of the lipid droplet, corresponding to the limiting membrane. Moreover this indicates that ReZolve-L1™ is predominantly staining polar lipids and not neutral lipids in contrast to Oil Red O, Nile Red, BODIPY or LipidTOX (or derivatives thereof). Interestingly, the spectra extracted from region X did still show overlap with some of the polar lipids, albeit to a much smaller and less confident extent. It can be expected that the signal from the lipids will be more intense on the edges of the droplets as IR light is passed through many layers on the side (i.e. region Y) as opposed to only two layers at the top and bottom of the lipid droplet when the spectra are extracted from the middle (i.e. region X). Alternatively, there may be a more heterogeneous distribution of polar lipids in lipid droplets than is classically accepted. The latter correlates more with confocal microscopy Z-stack analysis which shows that while ReZolve-L1™ staining can be detected throughout the lipid droplet, it is more intense at the edges of the lipid droplet.
ReZolve-L1™, therefore, provides new and extremely important biochemical information that has not been obtained previously with staining and fluorescence microscopies. Future work will explore how the distribution of different lipids influences the localisation of ReZolve-L1™ and which other lipids might directly interact with this molecular probe during live cell imaging.
Current commercially available imaging reagents for the live cell imaging of polar lipids are mainly based on lipid analogues conjugated to fluorescent tags, such as BODIPY or pyrene.44–47 Although these have assisted the investigation of lipid trafficking in cells, it is unclear if these analogue species interact with other cellular structures in the same way as their endogenous counterparts.45 Issues with photobleaching and toxicity can also perturb time-resolved imaging using these reagents.19,45 Alternatively live-cell imaging of lipids can be achieved by fluorescently tagged proteins with lipid specific binding domains, which overcomes the issue of detecting endogenous lipids. However limited availability of specific proteins for this application combined with time consuming and costly transfection protocols reduces applicability.48 To the best of our knowledge, ReZolve-L1™ is the first example of a live cell compatible molecular probe that localises to areas of high endogenous polar lipid concentration in preference to non-polar lipids. Furthermore, ReZolve-L1™ has low toxicity,24 can be used in conjunction with other dyes, shows no to minimal photo-bleaching and has a high staining efficiency which are some of the drawbacks associated with other lipid dyes.19,20
Conclusions
We have provided FTIR imaging evidence that ReZolve-L1™ localises to the lipid droplet region of adipocytes, which confirmed conclusions drawn from fluorescence confocal microscopy imaging. We ascertained that this molecular probe does not predominantly localise with neutral lipids, such as triglycerides, but may be more closely aligned with polar lipids such as phosphatidylcholine, phosphatidylethanolamine, sphingomyelin, sphingosine and lysophosphatidic acid which are present in higher concentrations in lipid droplet membranes. This provided strong evidence that ReZolve-L1™ has an affinity for polar lipids and the unprecedented potential to image these lipids in live cells.Acknowledgements
This work was supported by funding from the University of South Australia, an ITEK catalyst grant and a BioSA Innovation grant awarded to DAB, SP and MM. MM also wishes to thank the ARC for funding (FT130100033) and an ARC LIEF Grant to PAL and EAC for the FTIR instrumentation (LE0883036) and ARC Discovery grants to PAL (DP130103566 and DP140100176). AS is supported by an Australia Awards Scholarship.Notes and references
- K. K.-W. Lo, Acc. Chem. Res., 2015, 48, 2985–2995 CrossRef CAS PubMed.
- M. P. Coogan and V. Fernandez-Moreira, Chem. Commun., 2014, 50, 384–399 RSC.
- H. Kobayashi, M. R. Longmire, M. Ogawa and P. L. Choyke, Chem. Soc. Rev., 2011, 40, 4626–4648 RSC.
- E. J. New, D. Parker, D. G. Smith and J. W. Walton, Curr. Opin. Chem. Biol., 2010, 14, 238–246 CrossRef CAS PubMed.
- M. R. Gill and J. A. Thomas, Chem. Soc. Rev., 2012, 41, 3179–3192 RSC.
- J. A. Thomas, Chem. Soc. Rev., 2015, 44, 4494–4500 RSC.
- M. Mauro, A. Aliprandi, D. Septiadi, N. S. Kehr and L. De Cola, Chem. Soc. Rev., 2014, 43, 4144–4166 RSC.
- S. Takatori, R. Mesman and T. Fujimoto, Biochemistry, 2014, 53, 639–653 CrossRef CAS PubMed.
- K. Reue, J. Lipid Res., 2011, 52, 1865–1868 CrossRef CAS PubMed.
- G. van Meer, EMBO J., 2005, 24, 3159–3165 CrossRef CAS PubMed.
- L. Li, J. Han, Z. Wang, J. A. Liu, J. Wei, S. Xiong and Z. Zhao, Int. J. Mol. Sci., 2014, 15, 10492 CrossRef PubMed.
- A. Reis, A. Rudnitskaya, G. J. Blackburn, N. M. Fauzi, A. R. Pitt and C. M. Spickett, J. Lipid Res., 2013, 54, 1812–1824 CrossRef CAS PubMed.
- A. R. Thiam, R. V. Farese, Jr. and T. C. Walther, Nat. Rev. Mol. Cell Biol., 2013, 14, 775–786 CrossRef CAS PubMed.
- R. N. Melo, H. D'Ávila, P. Bozza and P. Weller, in Light Microscopy, ed. H. Chiarini-Garcia and R. C. N. Melo, Humana Press, 2011, vol. 689, ch. 9, pp. 149–161 Search PubMed.
- P. Greenspan, E. P. Mayer and S. D. Fowler, J. Cell Biol., 1985, 100, 965–973 CrossRef CAS PubMed.
- R. Koopman, G. Schaart and M. K. Hesselink, Histochem. Cell Biol., 2001, 116, 63–68 CAS.
- S. Fukumoto and T. Fujimoto, Histochem. Cell Biol., 2002, 118, 423–428 CrossRef CAS PubMed.
- Y. Ohsaki, Y. Shinohara, M. Suzuki and T. Fujimoto, Histochem. Cell Biol., 2010, 133, 477–480 CrossRef CAS PubMed.
- R. D. Moriarty, A. Martin, K. Adamson, E. O'Reilly, P. Mollard, R. J. Forster and T. E. Keyes, J. Microsc., 2014, 253, 204–218 CrossRef CAS PubMed.
- E. J. O'Rourke, A. A. Soukas, C. E. Carr and G. Ruvkun, Cell Metab., 2009, 10, 430–435 CrossRef PubMed.
- J. R. Arthur, K. A. Heinecke and T. N. Seyfried, J. Lipid Res., 2011, 52, 1345–1351 CrossRef CAS PubMed.
- D. L. Sackett and J. Wolff, Anal. Biochem., 1987, 167, 228–234 CrossRef CAS PubMed.
- J. Spandl, D. J. White, J. Peychl and C. Thiele, Traffic, 2009, 10, 1579–1584 CrossRef CAS PubMed.
- C. A. Bader, R. D. Brooks, Y. S. Ng, A. Sorvina, M. V. Werrett, P. J. Wright, A. G. Anwer, D. A. Brooks, S. Stagni, S. Muzzioli, M. Silberstein, B. W. Skelton, E. M. Goldys, S. E. Plush, T. Shandala and M. Massi, RSC Adv., 2014, 4, 16345–16351 RSC.
- N. Krahmer, R. V. Farese, Jr. and T. C. Walther, EMBO Mol. Med., 2013, 5, 905–915 CrossRef CAS PubMed.
- D. A. Gross and D. L. Silver, Crit. Rev. Biochem. Mol. Biol., 2014, 49, 304–326 CrossRef CAS PubMed.
- S. D. Kohlwein, M. Veenhuis and I. J. van der Klei, Genetics, 2013, 193, 1–50 CrossRef CAS PubMed.
- M. Beller, K. Thiel, P. J. Thul and H. Jäckle, FEBS Lett., 2010, 584, 2176–2182 CrossRef CAS PubMed.
- H. Abramczyk, J. Surmacki, M. Kopec, A. K. Olejnik, K. Lubecka-Pietruszewska and K. Fabianowska-Majewska, Analyst, 2015, 140, 2224–2235 RSC.
- D. J. Murphy, Prog. Lipid Res., 2001, 40, 325–438 CrossRef CAS PubMed.
- I. Dugail and E. Hajduch, Cell. Mol. Life Sci., 2007, 64, 2452–2458 CrossRef CAS PubMed.
- Z. Sun, J. Gong, L. Wu and P. Li, in Methods in Cell Biology, ed. Y. Hongyuan and L. Peng, Academic Press, 2013, vol. 116, pp. 253–268 Search PubMed.
- E. Muro, G. E. Atilla-Gokcumen and U. S. Eggert, Mol. Biol. Cell, 2014, 25, 1819–1823 CrossRef PubMed.
- A. Pol, S. P. Gross and R. G. Parton, J. Cell Biol., 2014, 204, 635–646 CrossRef CAS PubMed.
- F. Wilfling, J. T. Haas, T. C. Walther and R. V. F. Jr, Curr. Opin. Cell Biol., 2014, 29, 39–45 CrossRef CAS PubMed.
- E. A. Carter, K. K. Tam, R. S. Armstrong and P. A. Lay, Biophys. Rev., 2009, 1, 95–103 CrossRef.
- M. Diem, M. Romeo, S. Boydston-White, M. Miljkovic and C. Matthaus, Analyst, 2004, 129, 880–885 RSC.
- E. A. Carter, B. S. Rayner, A. I. McLeod, L. E. Wu, C. P. Marshall, A. Levina, J. B. Aitken, P. K. Witting, B. Lai, Z. Cai, S. Vogt, Y.-C. Lee, C.-I. Chen, M. J. Tobin, H. H. Harris and P. A. Lay, Mol. BioSyst., 2010, 6, 1316–1322 RSC.
- M. V. Werrett, D. Chartrand, J. D. Gale, G. S. Hanan, J. G. MacLellan, M. Massi, S. Muzzioli, P. Raiteri, B. W. Skelton, M. Silberstein and S. Stagni, Inorg. Chem., 2011, 50, 1229–1241 CrossRef CAS PubMed.
- K. Meister, J. Niesel, U. Schatzschneider, N. Metzler-Nolte, D. A. Schmidt and M. Havenith, Angew. Chem., Int. Ed., 2010, 49, 3310–3312 CrossRef CAS PubMed.
- M. Digel, R. Ehehalt and J. Füllekrug, FEBS Lett., 2010, 584, 2168–2175 CrossRef CAS PubMed.
- V. Zohdi, D. R. Whelan, B. R. Wood, J. T. Pearson, K. R. Bambery and M. J. Black, PLoS One, 2015, 10, e0116491 Search PubMed.
- L. E. Wu, A. Levina, H. H. Harris, Z. Cai, B. Lai, S. Vogt, D. E. James and P. A. Lay, Angew. Chem., Int. Ed., 2016, 55, 1742–1745 CrossRef CAS PubMed.
- K. Tanhuanpää, J. Virtanen and P. Somerharju, Biochim. Biophys. Acta, Mol. Cell Res., 2000, 1497, 308–320 CrossRef.
- P. Somerharju, Chem. Phys. Lipids, 2002, 116, 57–74 CrossRef CAS PubMed.
- M. Holtta-Vuori, R. L. Uronen, J. Repakova, E. Salonen, I. Vattulainen, P. Panula, Z. Li, R. Bittman and E. Ikonen, Traffic, 2008, 9, 1839–1849 CrossRef CAS PubMed.
- D. L. Marks, R. Bittman and R. E. Pagano, Histochem. Cell Biol., 2008, 130, 819–832 CrossRef CAS PubMed.
- Y. Mély and G. Duportail, Fluorescent Methods to Study Biological Membranes, Springer, Berlin, Heidelberg, 2012 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6mb00242k |
| ‡ The authors declare that Rezolve-L1™ is the subject of a patent (PCT/AU2015/000159), which has commercial potential and has been licensed to ReZolve Scientific for commercial sales as a molecular probe. DAB, SP, MM, CAB and SS may be future shareholders in ReZolve Scientific. |
| This journal is © The Royal Society of Chemistry 2016 |