Open Access Article
Open Access ArticleCyclic dinucleotide detection with riboswitch–G-quadruplex hybrid†
Genichiro
Tsuji‡
a and
Herman O.
Sintim‡
*ab
aDepartment of Chemistry and Biochemistry, University of Maryland, College Park, MD 20742, USA
bCenter for Drug Discovery, Purdue University, West Lafayette, IN 47907, USA. E-mail: hsintim@purdue.edu
First published on 21st December 2015
Abstract
A cyclic dinucleotide riboswitch has been fused with a G-quadruplex motif to produce a conditional riboswitch-peroxidase-mimicking sensor that oxidizes both colorimetric and fluorogenic substrates in the presence of c-di-GMP. We find that signal-to-noise ratio could be improved by using a two-, not three-, floor split G-quadruplex for this conditional peroxidase-mimicking riboswitch.
Nucleotides play important roles in both prokaryotes and eukaryotes and act as precursors for the synthesis of RNA and DNA, phosphoryl transfer reagents to modulate protein and small molecule function and also act as important second messengers.1 Cyclic mononucleotides were discovered in the late 1950s2,3 and are now known to regulate many key processes in both eukaryotes4 and prokaryotes.5 Ligands that target phosphodiesterases,6 such as sildenafil and vardenafil, are now used to treat erectile dysfunction.7 Recently new classes of cyclic nucleotides, which contain dinucleotides,8 have emerged as interesting second messengers in both bacterial and eukaryotic cells.9,10 In most Gram negative bacteria, high intracellular concentrations of c-di-GMP lead to enhanced ability to form biofilms whereas reduced c-di-GMP concentration results in increased production in virulence factors in some bacteria.11 Other bacterial phenotypes, such as motility, phage resistance, cell morphology and heavy metal resistance are also controlled by c-di-GMP.10,12 Due to the importance of this signaling molecule, interest in understanding how nutrient availability and environmental factors affect c-di-GMP signaling has increased. Also medicinal chemists desire to develop small molecules that can be used to perturb the intracellular concentration of c-di-GMP; such molecules could have antibacterial properties. We have been interested in developing different detection schemes to detect various cyclic dinucleotides to facilitate dinucleotide research and inhibitor discovery.13–16 Recently we reported that a cyclic dinucleotide riboswitch that was appended to RNA aptamers that bind to fluorogenic substrates (such as spinach) via a communication module, could detect nanomolar concentrations of c-di-GMP.17,18 A limitation of our approach however is that the excitation and emission profile of the spinach fluorophore (DFHBI)19–21 overlapped with several fluorophores in biological matrices as well as the fluorescence of several small molecules in our small molecule libraries, which we use for identifying cyclic dinucleotide synthesis or degradation inhibitors.22 We therefore initiated a program to expand the detection modality of cyclic dinucleotide riboswitches by appending a conditional G-quadruplex peroxidase23–26 unit to the riboswitch, i.e. a split-G-quadruplex unit that could form a proficient peroxidase-mimicking enzyme only when c-di-GMP was present, see Fig. 1 for design. Since G-quadruplex peroxidases are known to oxidize a variety of chromophores27,28 and fluorogenic compounds,29,30 we anticipated that this new riboswitch sensor could give different outputs, including colorimetric and fluorescence with a wide range of emission wavelengths and could complement other detection modes, such as electrochemical detection.31
 | ||
| Fig. 1 A cyclic dinucleotide riboswitch, Vc2 RNA,32–34 fused to a G-quadruplex. [S] = reduced substrate, [S*] = oxidized substrate. | ||
Oligonucleotides containing at least four tracts of guanine (such as (GyGN)x; where y ≥ 1 and x ≥ 4 and N could be any nucleobase) have a high propensity to form intramolecular G-quadruplexes in the presence of monovalent as well as a few divalent cations.35,36 The propensity to form G-quadruplexes however decrease as the length of the nucleotides between the guanine tracts (N) increases.37–39 With this knowledge, we hypothesized that a c-di-GMP riboswitch that contains guanine tracts (less than four) at both 3′- and 5′-ends would be unable to form G-quadruplex but upon binding to c-di-GMP, the separate guanine tracts would be close to each other to fold into a G-quadruplex, which could potentially catalyze the oxidation of substrates.40 The stability of such G-quadruplex will depend on the “communication module”41 in the riboswitch–G-quadruplex fusion, a duplex region that connects the c-di-GMP binding site to the G-quadruplex moiety. We therefore investigated several types of communication modules (see Fig. 2), which we had earlier observed to be appropriate for transducing c-di-GMP binding to a spinach binding domain.17 Two types of c-di-GMP riboswitch–G-quadruplex fusions, symmetric and asymmetric, were investigated. In the symmetric version two tracts of guanine (2 × GGG or GG) were placed at both the 3′- and 5′-ends, whereas in the asymmetric version the 3′-end contained only one tract of guanine (GGG) whereas the 5′-end contained three tracts of guanine (Fig. 1).
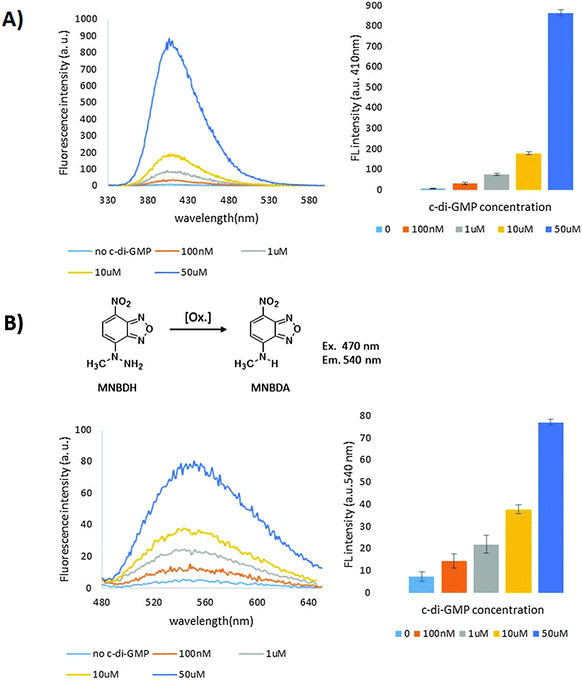
To evaluate the various c-di-GMP riboswitch–G-quadruplex fusions, we used tyramine.42 Tyramine has low fluorescence and can be oxidized into highly fluorescent dimers and polymers with hydrogen peroxide and peroxidase (Fig. 3). The asymmetric constructs were poor peroxidases in the absence or presence of c-di-GMP, whereas the symmetric versions (with the exception of the ST1 construct) were good peroxidases (Fig. 4). The ST2 and ST5 symmetric constructs (denoted as F3-ST2Sym and F3-ST5Sym, respectively) could discriminate between the presence and absence of c-di-GMP (see Fig. 4 and 5), whereas ST3 and ST4 symmetric constructs were equally active in the presence and absence of c-di-GMP. The ST7 construct was not active, even in the presence of c-di-GMP. Although F3-ST2Sym and F3-ST5Sym could detect c-di-GMP, the signal-to-noise (S/N) ratio was deemed low. We rationalized that perhaps a riboswitch–G-quadruplex fusion, which had only two G-tetrad floors and hence less likely to fold into a G-quadruplex than the three G-tetrad floor, would have a lower background noise and hence higher S/N. Peroxidation reaction using the RNA with two G-tetrad floors (denoted as F2-ST2Sym) displayed lower background fluorescence compared to RNA with three G-tetrad floors (F3-ST2Sym), Fig. 5 and 6. The background fluorescence intensities of F2-ST5Sym (two G-tetrad floors) and F3-ST5Sym (three G-tetrad floors) were similar; it appears that the factors that control background noise are complex and involve both the stem sequence and/or number of G-tetrad floors. Surprisingly, in the presence of c-di-GMP the RNA–G-quadruplexes with two floors were catalytically more proficient than those with three G-tetrad floors (compare F2-ST2Sym with F3-ST2Sym and F2-ST5Sym with F3-ST5Sym, Fig. 4). Therefore the S/N ratios of the two G-tetrad floors constructs were better than the three G-tetrad floors constructs. We currently do not have a hypothesis to explain the enhanced catalytic proficiency of the two G-tetrad floors RNA construct. Future work, beyond the scope of this work, which would investigate more of the two G-tetrad floors RNA constructs will reveal if this trend is indeed general or specific to this case. Having identified F2-ST2Sym as a c-di-GMP sensor (Fig. 4 and Fig. S1, ESI†), we proceeded to investigate if it could detect the dinucleotide in a concentration-dependent manner. Pleasingly the intensity of the emission at 410 nm increased as the concentration of c-di-GMP was increased and the signal from 100 nM c-di-GMP was nearly five times higher than when no c-di-GMP was added (Fig. 6A).
 | ||
| Fig. 5 Peroxidase reaction of tyramine using ST2Sym riboswitch–G4 fused RNA [3 floors G-tetrads (left) and 2 floors G-tetrads (right)] in the presence or absence of c-di-GMP. Conditions: same as Fig. 4. Fluorescence intensities at 410 nm in the absence and presence of c-di-GMP are 13.1 and 70.9 respectively for 3 floors G-tetrads, 6.8 and 177.4 respectively for 2 floors G-tetrads. | ||
A motivation for our program is to develop a platform that could be potentially used in challenging environments, where biological fluorophores (such as NADH43) could cause interference. Thus, having established that ST2Sym could be used to detect c-di-GMP, using tyramine, we next evaluated other peroxidation substrates (N-methyl-4-hydrazino-7-nitrobenzofurazan (MNBDH),44 10-acetyl-3,7-dihydroxyphenoxazine (Amplex Red)29 and 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS)). Both oxidized products from MNBDH and Amplex red oxidations (MNBDA and resorufin, see Fig. 6B and Fig. S1 ESI,† for structures) have higher emission wavelengths than both spinach 1 and 2 fluorophores (compare emission maxima of 540 and 585 nm for MNBDA and resorufin respectively with 496 and 498 for spinach 1 and 2 fluorophores, 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI)),45 which had been used for earlier riboswitch-based detection of c-di-GMP.17 Therefore the ability to detect c-di-GMP with these two “more red” fluorophores could extend the utility of riboswitch-based cyclic dinucleotide sensing and allow for detections in environments where either biological components or other small molecules also have fluorescence. Pleasingly, MNBDH and Amplex red could be oxidized by ST2Sym and the fluorescence intensities of the products were higher in the presence of c-di-GMP than when no c-di-GMP was present (see Fig. 6B and Fig. S2, S3, ESI†). The c-di-GMP riboswitch–G-quadruplex hybrid could also oxidize ABTS into the radical cation, which can be detected via UV absorption (see Fig. S4, ESI†) and hence adds colorimetric detection mode to the riboswitch sensing platform.
In conclusion, we have extended the detection modality of cyclic dinucleotide-based riboswitch sensors to include colorimetric detection and also extend the emission fluorescence range. The detection of metabolites using RNA riboswitches and aptamers is gaining increased attention but the method is limited by lack of appropriate fluorophores and aptamer units that bind to fluorophore units to increase fluorescence. The method described herein is general and could be applied to any fluorogenic substrate that could be oxidized by a peroxidase-mimicking G-quadruplex so the method has the potential to increase the emission wavelength range that could be used in RNA/DNA based sensors. Current efforts are focused on screening other fluorogenic compounds and/or developing compounds that could be oxidized into products with emission fluorescence in the red range and will be reported in due course.
Acknowledgements
We are grateful to NSF (CHEM1307218) and Camille Dreyfus Foundation (Teacher-Scholar fellowship to HOS) for funding.References
- D. Kalia, S. Nakayama, M. Guo, J. Zhou, Y. Zheng, Y. Luo, B. Roembke, G. Merey and H. O. Sintim, Chem. Soc. Rev., 2013, 42, 305–341 RSC.
- K. D. Bhoola and M. J. Lemin, J. Biol. Chem., 1958, 232, 1077–1091 Search PubMed.
- T. W. Rall and E. W. Sutherland, J. Biol. Chem., 1958, 232, 1065–1076 CAS.
- A. Y. Kots, E. Martin, I. G. Sharina and F. Murad, Handb. Exp. Pharmacol., 2009, 191, 1–14 CAS.
- M. Gomelsky, Mol. Microbiol., 2011, 79, 562–565 CrossRef CAS PubMed.
- D. H. Maurice, H. Ke, F. Ahmad, Y. Wang, J. Chung and V. C. Manganiello, Nat. Rev. Drug Discovery, 2014, 13, 290–314 CrossRef CAS PubMed.
- R. Bruzziches, D. Francomano, P. Gareri, A. Lenzi and A. Aversa, Expert Opin. Pharmacother., 2013, 14, 1333–1344 CrossRef CAS PubMed.
- P. Ross, H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinberger-Ohana, D. Marel, J. H. Boom and M. Benziman, Nature, 1987, 325, 279–281 CrossRef CAS PubMed.
- U. Romling, Sci. Signaling, 2008, 1, pe39 CrossRef PubMed.
- U. Romling, M. Y. Galperin and M. Gomelsky, Microbiol. Mol. Biol. Rev., 2013, 77, 1–52 CrossRef PubMed.
- A. D. Tischler and A. Camilli, Mol. Microbiol., 2004, 53, 857–869 CrossRef CAS PubMed.
- U. Romling, Sci. Signaling, 2008, 1, pe39 CrossRef PubMed.
- S. Nakayama, I. Kelsey, J. Wang, K. Roelofs, B. Stefane, Y. Luo, V. T. Lee and H. O. Sintim, J. Am. Chem. Soc., 2011, 133, 4856–4864 CrossRef CAS PubMed.
- S. Nakayama, K. G. Roelofs, V. T. Lee and H. O. Sintim, Mol. BioSyst., 2012, 8, 726–729 RSC.
- B. Roembke, J. Wang, S. Nakayama, J. Zhou and H. O. Sintim, RSC Adv., 2013, 3, 6305–6310 RSC.
- B. Roembke, J. Zhou, Y. Zheng, D. Sayre, A. Lizardo, L. Bernard and H. O. Sintim, Mol. BioSyst., 2014, 10, 1568–1575 RSC.
- S. Nakayama, Y. Luo, J. Zhou, T. K. Dayie and H. O. Sintim, Chem. Commun., 2012, 48, 9059–9061 RSC.
- Y. Luo, B. Chen, J. Zhou, H. O. Sintim and T. K. Dayie, Mol. BioSyst., 2014, 10, 384–390 RSC.
- J. S. Paige, K. Y. Wu and S. R. Jaffrey, Science, 2011, 333, 642–646 CrossRef CAS PubMed.
- K. Y. Han, B. J. Leslie, J. Fei, J. Zhang and T. Ha, J. Am. Chem. Soc., 2013, 135, 19033–19038 CrossRef CAS PubMed.
- W. Song, R. L. Strack, N. Svensen and S. R. Jaffrey, J. Am. Chem. Soc., 2014, 136, 1198–1201 CrossRef CAS PubMed.
- Y. Zheng, J. Zhou, D. A. Sayre and H. O. Sintim, Chem. Commun., 2014, 50, 11234–11237 RSC.
- P. Travascio, Y. Li and D. Sen, Chem. Biol., 1998, 5, 505–517 CrossRef CAS PubMed.
- E. Golub, C.-H. Lu and I. Willner, J. Porphyrins Phthalocyanines, 2015, 19, 65–91 CrossRef CAS.
- J. Kosman and B. Juskowiak, Anal. Chim. Acta, 2011, 707, 7–17 CrossRef CAS PubMed.
- L. Stefan, T. Lavergne, N. Spinelli, E. Defrancq and D. Monchaud, Nanoscale, 2014, 6, 2693–2701 RSC.
- D. Sen and L. C. H. Poon, Crit. Rev. Biochem. Mol. Biol., 2011, 46, 478–492 CAS.
- S. Nakayama and H. O. Sintim, Anal. Chim. Acta, 2012, 747, 1–6 CrossRef CAS PubMed.
- S. Nakayama and H. O. Sintim, Mol. BioSyst., 2010, 6, 89–91 RSC.
- (a) B. T. Roembke, S. Nakayama and H. O. Sintim, Methods, 2013, 64, 185–198 CrossRef CAS PubMed; (b) S. Nakayama and H. O. Sintim, Methods Mol. Biol., 2013, 1039, 153–159 CrossRef.
- Q. Xie, F. Zhao, H. Liu, Y. Shan and F. Liu, Anal. Chim. Acta, 2015, 882, 22–26 CrossRef CAS PubMed.
- N. Sudarsan, E. R. Lee, Z. Weinberg, R. H. Moy, J. N. Kim, K. H. Link and R. R. Breaker, Science, 2008, 321, 411–413 CrossRef CAS PubMed.
- N. Kulshina, N. J. Baird and A. R. Ferré-D’Amaré, Nat. Struct. Mol. Biol., 2009, 16, 1212–1217 CAS.
- K. D. Smith, S. V. Lipchock, T. D. Ames, J. Wang, R. R. Breaker and S. A. Strobel, Nat. Struct. Mol. Biol., 2009, 16, 1218–1223 CAS.
- M. L. Bochman, K. Paeschke and V. A. Zakian, Nat. Rev. Genet., 2012, 13, 770–780 CrossRef CAS PubMed.
- D. Yang and K. Okamoto, Future Med. Chem., 2010, 2, 619–646 CrossRef CAS PubMed.
- A. N. Lane, J. B. Chaires, R. D. Gray and J. O. Trent, Nucleic Acids Res., 2008, 36, 5482–5515 CrossRef CAS PubMed.
- E. Hatzakis, K. Okamoto and D. Yang, Biochemistry, 2010, 49, 9152–9160 CrossRef CAS PubMed.
- A. Y. Q. Zhang and S. Balasubramanian, J. Am. Chem. Soc., 2012, 134, 19297–19308 CrossRef CAS PubMed.
- S. Nakayama and H. O. Sintim, J. Am. Chem. Soc., 2009, 131, 10320–10333 CrossRef CAS PubMed.
- M. N. Stojanovic and D. M. Kolpashchikov, J. Am. Chem. Soc., 2004, 126, 9266–9270 CrossRef CAS PubMed.
- S. Nakayama, J. Wang and H. O. Sintim, Chem. – Eur. J., 2011, 17, 5691–5698 CrossRef CAS PubMed.
- J. R. Lakowicz, Principles Fluorescence Spectroscopy, Springer, New York, NY, USA, 3rd edn, 2006 Search PubMed.
- J. Kosman, Y.-T. Wu, A. Gluszynska and B. Juskowiak, Anal. Bioanal. Chem., 2014, 406, 7049–7057 CrossRef CAS PubMed.
- R. L. Strack, M. D. Disney and S. R. Jaffrey, Nat. Methods, 2013, 10, 1219–1224 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental section including sequences and preparation of RNA used in this study, general preparation method of samples for oxidase reaction. See DOI: 10.1039/c5mb00751h |
| ‡ Present address: Department of Chemistry, Purdue University, 560 Oval Drive, West Lafayette, IN 47907, USA. |
| This journal is © The Royal Society of Chemistry 2016 |