Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
One-step fabrication of an organ-on-a-chip with spatial heterogeneity using a 3D bioprinting technology
Hyungseok
Lee
and
Dong-Woo
Cho
*
Department of Mechanical Engineering, Pohang University of Science and Technology (POSTECH), San 31, Hyoja-dong, Nam-gu, Pohang, Gyungbuk 790-784, South Korea. E-mail: dwcho@postech.ac.kr; Fax: +82 54 279 5419; Tel: +82 54 279 2171
First published on 15th June 2016
Abstract
Although various types of organs-on-chips have been introduced recently as tools for drug discovery, the current studies are limited in terms of fabrication methods. The fabrication methods currently available not only need a secondary cell-seeding process and result in severe protein absorption due to the material used, but also have difficulties in providing various cell types and extracellular matrix (ECM) environments for spatial heterogeneity in the organs-on-chips. Therefore, in this research, we introduce a novel 3D bioprinting method for organ-on-a-chip applications. With our novel 3D bioprinting method, it was possible to prepare an organ-on-a-chip in a simple one-step fabrication process. Furthermore, protein absorption on the printed platform was very low, which will lead to accurate measurement of metabolism and drug sensitivity. Moreover, heterotypic cell types and biomaterials were successfully used and positioned at the desired position for various organ-on-a-chip applications, which will promote full mimicry of the natural conditions of the organs. The liver organ was selected for the evaluation of the developed method, and liver function was shown to be significantly enhanced on the liver-on-a-chip, which was prepared by 3D bioprinting. Consequently, the results demonstrate that the suggested 3D bioprinting method is easier and more versatile for production of organs-on-chips.
Introduction
In recent decades, the microfluidics-based cell culture platform has been revealed as an effective experimental tool for a wide range of biological applications, such as metabolomics,1 cell analysis,2,3 and organs-on-chips.4 In particular, organs-on-chips, which mimic the biological environment of the human body, are rising as innovative tools in the field of drug discovery.5 The current drug discovery process requires multiple screening steps, which include two-dimensional (2D) in vitro cell culture and animal model tests.6 However, it is generally known that the current 2D in vitro cell culture model, with cells only, cannot fully mimic the natural environment of human organs; the use of animal models also leads to several problems, such as ethical concerns, time consumption, and inefficient test results because of the huge differences compared with the human body.7 For these reasons, various types of organs-on-chips have been introduced for simple and precise drug screening in the drug discovery process.8Organs-on-chips have been mainly prepared using several microengineering methods, such as soft lithography, replica molding, and the microcontact printing technique.7,9 However, these methods not only require microengineering expertise in the fabrication process and exhibit severe protein absorption due to the material used, but also have several drawbacks in the aspect of biological structure preparation.9 For example, poor selectivity of various cell types for spatial heterogeneity and the difficulty of providing multiple types of extracellular matrix (ECM) environments for cell–ECM interactions are the main drawbacks regarding the biological structure preparation. Furthermore, living organisms have complex and organized 2D-to-3D microscale structures composed of multi-layers, cell types, ECMs, and many other elements,10 which could make the fabrication of organs-on-chips difficult using the current methods. Thus, establishing a novel microengineering method that can overcome the aforementioned drawbacks is very important.
These days, 3D printing is used to produce functional devices in diverse fields, such as tissue scaffolding,11–13 prototyping,14 electronics,15 sensors,16 and microfluidic17 research, because of its capabilities in producing designed, complex micro-architectures. Thus, a large number of studies have reported the fabrication of microfluidic devices with 3D printing technologies for chemical mixing, gradient generation, and sensing applications; however, most of them were based on a stereolithography-based 3D printing technology.17–19 Until now, no study has used cells and biomaterials directly during the 3D printing process to establish the organ-on-a-chip platforms. 3D bioprinting is an advanced 3D printing technology that uses cells and biomaterials as printing materials. The most promising advancement of 3D bioprinting is that biocompatible polymers, ECM-based hydrogels, and multiple cell types can be delivered simultaneously and positioned as intended to fabricate complex 3D biological constructs.20,21 There have been interesting studies that applied 3D bioprinting of a micro-organ on pre-prepared microfluidic platforms for organ-on-a-chip applications;22–24 however, there have been no reports describing the use of this technology for whole organ-on-a-chip fabrication including a microfluidic system with multiple cell types and biomaterials for optimal biomimicry. Until recently, we have actively developed 3D bioprinting systems and methods using multiple cell types and biomaterials for complex-shaped heterogeneous tissue models.25 Based on our previous experience and current work, we suggest that the 3D bioprinting method can be a good candidate to overcome the problems of current organs-on-chips.
Here, we verify the 3D bioprinting method for organ-on-a-chip platforms. Protein absorption on the organ-on-a-chip platform was evaluated by comparing it with a PDMS platform, and the developed method was confirmed with heterotypic cell types and biomaterials for application to various organ-on-a-chip platforms, which were prepared simply in a single printing process without a secondary cell-seeding process. Furthermore, our method was successfully applied to a liver-on-a-chip, showing that it is easier and more versatile in producing organs-on-chips.
Materials and methods
Materials preparation
As a platform material for an organ-on-a-chip, poly(ε-caprolactone) (PCL, MW = 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–50
000–50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, Polysciences Inc., Warrington, PA, USA) was used. PCL was printed with a printing speed of 200 mm min−1 and a pneumatic pressure of 500 kPa using a 200 μm nozzle. PCL was printed with a minimum line width of 175 μm. For the preparation of the hydrogels, 3% w/v gelatin (from porcine skin G6144-500G, Sigma-Aldrich, St. Louis, MO, USA) hydrogels were prepared by dissolving gelatin in serum-free M199 medium (Gibco Invitrogen, Grand Island, NY), and 2% w/v collagen type 1 hydrogels were prepared by dissolving a lyophilized collagen sponge (Dalim Corporation, Seoul, Korea) into 0.5 M of acetic acid and neutralized by 10 N sodium hydroxide. Lastly, 10-times concentrated Dulbecco's modified Eagle's culture medium (DMEM, Gibco BRL, NY, USA) was added to the pH-adjusted collagen hydrogel (1/10th of the volume) to supply the medium to the cells during the printing process. The printing conditions were adjusted depending on each hydrogel's property, and the collagen and gelatin hydrogels were printed with minimum line widths of 300 μm and 230 μm, respectively.
000, Polysciences Inc., Warrington, PA, USA) was used. PCL was printed with a printing speed of 200 mm min−1 and a pneumatic pressure of 500 kPa using a 200 μm nozzle. PCL was printed with a minimum line width of 175 μm. For the preparation of the hydrogels, 3% w/v gelatin (from porcine skin G6144-500G, Sigma-Aldrich, St. Louis, MO, USA) hydrogels were prepared by dissolving gelatin in serum-free M199 medium (Gibco Invitrogen, Grand Island, NY), and 2% w/v collagen type 1 hydrogels were prepared by dissolving a lyophilized collagen sponge (Dalim Corporation, Seoul, Korea) into 0.5 M of acetic acid and neutralized by 10 N sodium hydroxide. Lastly, 10-times concentrated Dulbecco's modified Eagle's culture medium (DMEM, Gibco BRL, NY, USA) was added to the pH-adjusted collagen hydrogel (1/10th of the volume) to supply the medium to the cells during the printing process. The printing conditions were adjusted depending on each hydrogel's property, and the collagen and gelatin hydrogels were printed with minimum line widths of 300 μm and 230 μm, respectively.
Design of an organ-on-a-chip platform
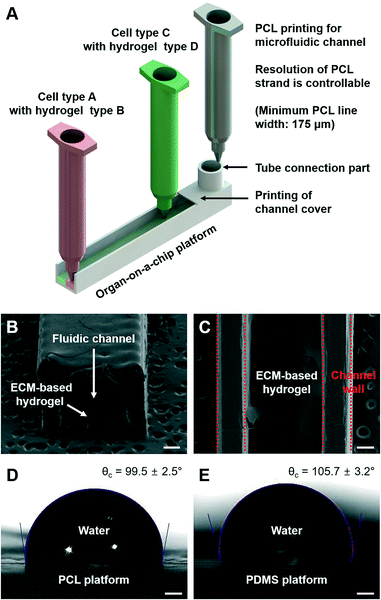
A 3D bioprinting code was generated for the printing system, and the organ-on-a-chip platforms were printed, which is an easy approach to fabrication of an organ-on-a-chip platform with a complex design via 3D bioprinting. A fluidic channel with internal dimensions of 1.5 mm × 1.5 mm × 15 mm was prepared for multiple applications: for various organ-on-a-chip platforms, protein absorption testing, and liver-on-a-chip application. ECM-based hydrogels for the 3D micro-environment were printed with a 400 μm thickness for various organ-on-a-chip platforms and the liver-on-a-chip. For the dye absorption test, a channel with the same internal size as the one above was prepared by the 3D printing method and polydimethylsiloxane (PDMS, Sylgard 184, Dow Corning Corporation, USA) replica molding.Measurement of the contact angle and protein absorption
Distilled water (5 μL) was dropped onto each of the PCL and PDMS platforms, and each contact angle was measured by a droplet analysis device (SmartDrop, Femtofab, Korea). To compare the dye absorption of the PCL-printed and PDMS replica-molded devices, a solution of Rhodamine B (1 μM in phosphate-buffered saline) was pumped through the devices. To perfuse through the devices, the dye was ejected at a flow rate of 5 μL min−1 from the perfusion pump (Ismatec UK Co. Weston-super-Mare, England). After perfusion for 12 hours, each channel was cut through the vertical section, and the absorption depth was visualized by using a confocal microscope (Olympus FluoView FV1000, Tokyo, Japan).Scanning electron microscopy
All printed constructs were dried under vacuum at room temperature, then coated with platinum in a sputter coater (Ion Sputter E-1045, Hitachi, Tokyo, Japan). An SEM system (SU-6600, Hitachi, Tokyo, Japan), operating at 15 kV, was used to examine the hydrogel position within the printed platform and to analyze the printed microfluidic channels of the organ-on-a-chip platform.Cell preparation, labeling, and encapsulation into hydrogels
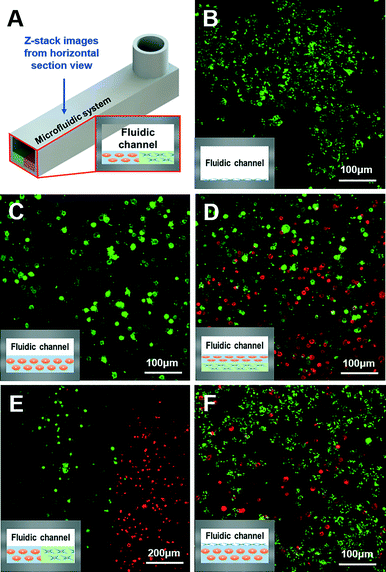
Human hepatocellular carcinoma (HepG2) cell lines and human umbilical vein endothelial cells (HUVEC) were purchased from ATCC (USA) and Lonza (Basel, Switzerland), respectively. HepG2 was cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco BRL, NY, USA) with 10% (v/v) fetal bovine serum (FBS, Gibco BRL, NY, USA) and 1% (v/v) penicillin/streptomycin (P/S, Sigma-Aldrich, St. Louis, MO, USA) at 37 °C in a humidified 5% CO2 atmosphere. HUVEC was cultured in a complete endothelium medium (EGM-2 BulletKit, Lonza) under the same conditions as the HepG2 cell culture. To label the cells, the HepG2 cells were incubated at 37 °C in a 5% CO2 incubator for 15 min with DiI (red) or DiO (green) fluorescent dyes. These HepG2-DiI-labeled cells and HepG2-DiO-labeled cells were then used as two different representative cell types. These green and red-labeled cells were encapsulated in collagen type 1 or gelatin hydrogels and used in preparing various organ-on-a-chip platforms. The concentration of cells used was from 1 to 5 × 106 cells ml−1. A confocal microscope (Olympus FluoView FV1000, Tokyo, Japan) was used for visualization.Printing of various organ-on-a-chip platforms
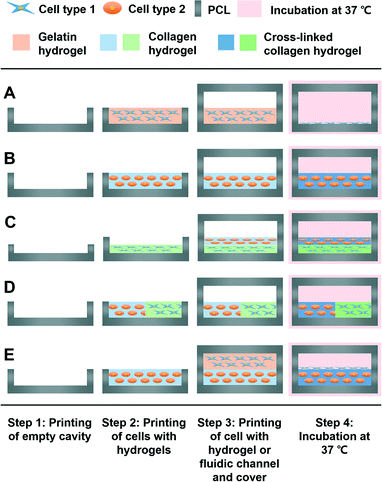
An empty cavity was first prepared, and cell types and ECM-based hydrogels for the micro-environment were printed in the prepared empty cavity. Secondly, with several layer-by-layer processes, the side walls of the fluidic channel were printed. The fluidic channel was also covered and sealed by printing the housing material in a manner wherein it is crossing above the pre-printed side walls of the fluidic channel to avoid fluid leakage. Lastly, the tube connection part for dynamic stimulation was printed. In the process of printing the tube connection, an optimal design was selected to avoid leakage. In the described process, 2D, 3D, 3D/3D vertical, 3D/3D horizontal, and 3D/2D models for various organ-on-a-chip platforms were prepared using the 3D bioprinting technology (Fig. 1).Application to a liver-on-a-chip
To establish the 3D bioprinted liver-on-a-chip, HepG2 and HUVEC cells were encapsulated in collagen type 1 and gelatin hydrogels, respectively. Cell–hydrogel mixtures were applied to each 2D and 3D part in the 3D/2D model. The concentration of HepG2 cells used was 2 × 107 cells ml−1, and the concentration of HUVEC cells used was 5 × 104 cells ml−1. Furthermore, a 20 μL min−1 continuous flow of the DMEM/EGM-2 medium was finally perfused to establish the liver-on-a-chip system.Liver function test
For functional assessment, each construct with the same number of hepatocytes was cultured for 6 days. During this period, the same amount of the sample medium was collected every 2 days in all experimental groups to compare the analysis results, and the medium was refreshed. The urea and albumin values were quantified from the collected medium. We used ELISA for the quantitative analysis of albumin (DuoSet ELISA development system, Genzyme) and a urea assay kit (BioVision Research Products, Mountain View, CA, USA) for the urea assay. For the hepatocyte viability test, samples were stained with Live/Dead kits (Life Technologies, Germany). The live and dead hepatocytes were imaged using a Zeiss LSM 510 Meta confocal microscope (Zeiss, Jena, Germany).Statistical analysis
All variables for urea synthesis and albumin secretion were expressed as means ± standard deviation (SD). Evaluation of the difference between the mean values of each group was performed by Student's t-test, in which a p value of <0.05 was considered significant.Results and discussion
3D bioprinting of an organ-on-a-chip platform
Fig. 2A shows a schematic illustration of the 3D bioprinting technology for an organ-on-a-chip. Among multiple candidates of biocompatible polymers for 3D bioprinting, PCL was chosen as a housing material for the organ-on-a-chip platforms. PCL is not only non-toxic, but also has a relatively low melting point of 60 °C compared with other biocompatible thermoplastics, resulting in high cell viability during the printing process.Fig. 2B shows the vertical section of a prepared channel with a width of 1.5 mm. The width of the channel can be controlled in the range from 200 μm to 2 mm. When the channel width was wider than 2 mm, it was hard to maintain the shape of the channel. When the PCL material is deposited during the 3D bioprinting process, the organ-on-a-chip platform was printed as a one-body structure. This is because the melted PCL material was printed above the solidified layer, and they attached and combined without an additional sealing process. In addition, multiple cellular components were printed and ECM-based hydrogels were used to provide the micro-environment to the cells. Fig. 2C shows the horizontal section view of an organ-on-a-chip fabricated with ECM-based hydrogels, where the hydrogels were located in the fluidic channel exactly as intended. Lastly, printing of the channel cover and the tube connection part was conducted to apply continuous dynamic stimulation to the cells, which could lead to better system functionality. Furthermore, the contact angle of the organ-on-a-chip platform printed by PCL was measured compared to that of the PDMS platform (Fig. 2D and E). The result shows that the printed organ-on-a-chip platform with PCL possessed hydrophobicity and had a slightly lower water contact angle value than the PDMS platform, which clearly indicate that confinement of the cell culture medium in an organ-on-a-chip can be done well.26
Evaluation of protein absorption
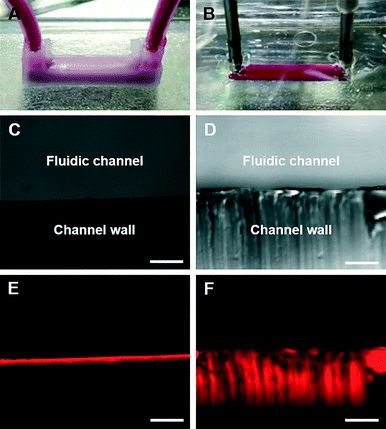
Protein absorption in a cell culture device is very important for the accurate measurement of cell metabolism and drug sensitivity. As mentioned before, most organs-on-chips have been fabricated by soft lithography and a PDMS replica molding process.7,9 However, it is generally known that for a non-specific/hydrophobic protein, less than approximately 500 Da of its molecular mass can be absorbed into PDMS.27 This protein absorption can cause major concern regarding the cell culture in the organ-on-a-chip platform, since the culture medium contains numerous proteins and growth factors. For example, non-specific protein absorption onto PDMS can change the protein level within the medium, which may severely affect the cell culture conditions and functions.28 Nevertheless, this issue has been ignored in many studies because of the lack of an alternative microengineering technique.29 To compare the protein absorption into the channel wall in our 3D bioprinted organ-on-a-chip platform with that in the PDMS platform, two microfluidic channels with the same internal dimensions of 1.5 mm × 1.5 mm × 15 mm were prepared with PCL and PDMS (Fig. 3A and B).In order for the components of the medium and drugs to permeate the cell membrane and reach the interior of the cells, the molecular mass of the molecules must be smaller than 500 Da.30 Therefore, rhodamine B dye with a molecular mass of 479 Da was used as a representative protein molecule with a molecular mass of 500 Da (the maximum molecular mass value that can permeate the cell membrane). In the PDMS channel platform, the absorption depth was about 400 μm; however, it was only about 50 μm in our printed channel platform (Fig. 3C–F). Furthermore, we have calculated the absorption ratio of the dye by collecting samples from the inlet and outlet of each platform. Compared with the inlet dye concentration, 3.4% of the dye was absorbed in the channel with PCL. In the channel with PDMS, 10.5% of the dye was absorbed, which had about a threefold higher value than that of PCL. The results indicate that the microfluidic channel printed using PCL can be a suitable candidate for maintaining the medium composition in an organ-on-a-chip, and it will allow the drug discovery process to be more accurate with 3D-bioprinted organs-on-chips. However, the PCL material shows low optical transparency compared to PDMS (Fig. 3C and D). The optical transparency of organs-on-chips with a 3D bioprinting technology is one other problem that has to be overcome in the near future. A printable biomaterial that possesses good optical transparency and less protein absorption has to be developed.
Printing of various organ-on-a-chip platforms
Living organisms have complex and organized 2D-to-3D microscale systems composed of multilayers and many other elements, such as cells and ECM components.10 Therefore, establishing an effective fabrication method to fully mimic the natural conditions of an organism is important in organ-on-a-chip preparation. Here, we have verified the possibilities of various organ-on-a-chip platforms using the green- and red-stained cells with ECM-based hydrogels.Gelatin hydrogels have unique thermo-sensitive properties. These materials are in the gel state at low temperature; however, they turn into liquid form at 37 °C. Using these gelatin hydrogels, gelatin hydrogels with cells were first printed. After the incubation process, only gelatin material in the liquid state was removed, and only the cell components remained. Thus, the use of gelatin hydrogels as a printing material followed by an incubation process at 37 °C made it possible to deliver the cells only in the 2D state in the printed microfluidic system (Fig. 4B). Collagen hydrogels are one of the most commonly used biomaterials as a representative ECM component in the bioengineering field, and have opposite thermo-sensitive properties compared with gelatin hydrogels. At a low temperature, the collagen material is in liquid form; however, at 37 °C, it turns into the gel state. Thus, it was possible to make the 3D micro-environment for the cells with encapsulation in the collagen hydrogels (Fig. 4C). By comparing the 2D and 3D models, the morphological cell differences were observed. The cells were observed with stretched and scattered shapes in the 2D model; however, the morphology of the cells in the 3D model was rounded in shape due to the 3D micro-environment caused by the collagen hydrogel the day after the 3D bioprinting process. In addition to the 2D or 3D model with a single cell type, the 3D/3D vertical and 3D/3D horizontal models with heterotypic cell types were fabricated, as shown in Fig. 4D and E, respectively. These heterotypic models could be engineered due to the advancement of the 3D bioprinting technology, which can deliver ECM-based hydrogels and multiple cell types in an intended space to fabricate complex 3D biological constructs. Lastly, by applying the gelatin and collagen hydrogels to the 2D and 3D models, respectively, the 3D/2D model was successfully prepared (Fig. 4F). The cells in the 2D and 3D parts of the 3D/2D model showed similar cell morphology to the individual 2D and 3D models. Thus, the results indicate that appropriate delivery of cells and ECM-based hydrogels to the desired positions in channels and effective preparation of heterotypic dimensional models are possible with a 3D bioprinting technology, which will lead to optimal biomimicry of native organisms with organs-on-chips. We have shown that printing of 2D cell patterns, 3D cell patterns, dividing 3D cell patterns, and stacking cell patterns in the microfluidic channel are possible. Therefore, not only the various organ-on-a-chip platforms suggested, but also much more complex platforms can be designed and printed through a 3D bioprinting technology. For example, the 3D/3D horizontal model and 3D/3D vertical model can be combined for a complex organ-on-a-chip platform. Additionally, any complex patterns can be designed and printed. Furthermore, all the cellular components were successfully delivered by simple one-step fabrication using 3D bioprinting with high efficiency. Thus, there was no need for a low-efficiency secondary cell-seeding process, which was required by most of the current organ-on-a-chip platforms prepared by the stereolithography-based fabrication methods.
Application to a liver-on-a-chip
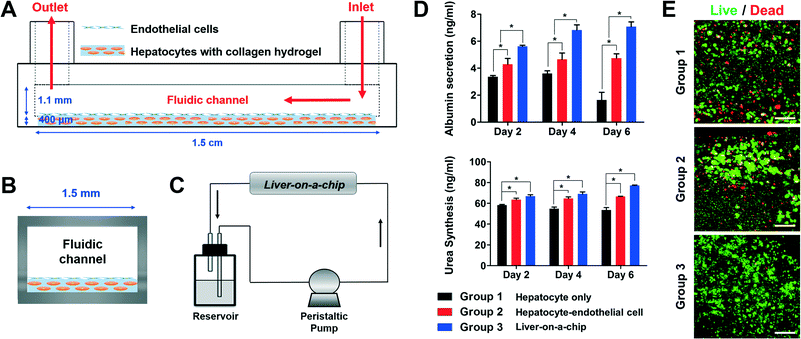
The liver is one of the largest internal organs in the human body and conducts numerous major activities, such as blood purification, detoxification, and protein synthesis.31 According to reports, many pharmaceutical products have been banned, withdrawn, or not approved because of drug-induced liver injury.32 Thus, establishing an in vivo-like liver-on-a-chip model could be a significant development in the drug discovery process. In practice, multiple studies have introduced the liver-on-a-chip;24,33–35 however, there still exist many limitations, such as difficulties in providing various cell types and ECM environments for spatial heterogeneity in the liver-on-a-chip. For this reason, we have selected the liver organ to confirm our 3D bioprinting technology for liver-on-a-chip applications using two different cell types and ECM-based hydrogels for spatial heterogeneity. Understanding the natural environment of the liver tissue is essential to prepare the liver-on-a-chip. Multiple cell types, such as hepatocytes, endothelial cells, and stellate cells, compose the liver in a complex order.36 In particular, the liver sinusoids in the liver micro-architecture, which are mainly composed of patterned/lined endothelial cells and hepatocytes, play a major role in hepatocyte metabolism and detoxification, for example.23,37 Therefore, the endothelial cells and hepatocytes were applied to each 2D and 3D part in the 3D/2D model (Fig. 5A and B) for high-fidelity biomimicry of the liver sinusoid architecture. Furthermore, a continuous flow of the medium was finally perfused to establish the liver-on-a-chip system (Fig. 5C). For the comparison groups, the following were used: 3D bioprinted groups of hepatocytes only in the 3D model with static culture (group 1) and hepatocytes–endothelial cells in the 3D/2D model with static culture (group 2). From the evaluation of the albumin and urea secretion with time, basic liver functions in the three experimental groups were observed (Fig. 5D). Basically, with the culture of hepatocytes only (group 1), the liver function gradually decreased with time. However, co-culturing the hepatocytes with endothelial cells (group 2) significantly enhanced the liver functions. In addition, the liver-on-a-chip, prepared by 3D bioprinting (group 3), showed higher absolute values of urea and albumin secretion than those of the co-cultured groups (group 2), which were fundamentally caused by constant perfusion of the medium through the printed microfluidic pathways in the liver-on-a-chip, mimicking the natural flow environment. With the perfusion of the medium, the liver-on-a-chip can create a more suitable culture environment by continuously refreshing the medium and removing waste products. Hepatocytes are the main cells that affect the secreted albumin and urea in the liver. Therefore, in addition to albumin/urea analysis, the hepatocyte viability of each group was also evaluated. Compared with the secreted albumin/urea values (Fig. 5D), we observed a similar tendency of hepatocyte viability in each experimental group on day 6 (Fig. 5E). Furthermore, there were reports that vascular formation is different from the shear stress induced by microfluidics.38,39 Compared with the static culture, it is reported that endothelial cells showed a stretched formation and direction related to the microfluidic pathway. Therefore, the endothelial cells in the liver-on-a-chip will appear more stretched and have specific direction compared with those in group 2, which will promote mimicking of the natural liver sinusoid structure. Consequently, the results clearly show that our 3D bioprinted liver-on-a-chip has better liver function than the static culture models, suggesting that a 3D bioprinting technology can be applied to various organ-on-a-chip platforms. However, in future development of organs-on-chips, the most important thing that organs-on-chips must possess is the much higher functionality that can actually resemble human organ functions. Therefore, developing and studying conditions that will show higher functionality have to be pursued in 3D-bioprinting-based organ-on-a-chip research.A 3D bioprinting technology also has the potential to be expanded to body-on-a-chip development. Until now, connecting complex organ-on-a-chip systems in a stable condition with mimicry of the human circulatory system has been one of the challenges yet to be conquered for body-on-a-chip research.40 Several attempts have been made, such as connecting each organ-on-a-chip platform with tubing and combining all cell-culture chambers on a single chip.41,42 However, problems of bubbles and leaks into the body-on-a-chip platforms and locating different cell types in each cell culture chamber remain as obstacles. However, the body-on-a-chip can be prepared simply in a single-chip platform, and the printing of different cell types in each cell culture chamber resembling human physiological conditions can be performed through one-step fabrication with a 3D bioprinting technology.
Conclusion
Establishing new micro-engineering methods for an organ-on-a-chip is important to overcome drawbacks that current organs-on-chips have. Here, we applied a 3D bioprinting technology for organ-on-a-chip fabrication. Our platform showed less protein absorption than the PDMS platform, such that it enables accurate measurement of metabolism and drug screening. Due to the advancement of the 3D bioprinting technology, heterotypic cell types and biomaterials were successfully used and positioned at the desired position, and one-step fabrication of an organ-on-a-chip was possible without a secondary cell-seeding process. Our suggested 3D bioprinting method was applied for various organ-on-a-chip platforms to show spatial heterogeneity, and the liver organ was selected for the technical evaluation. Liver function was shown to be significantly enhanced on the 3D bioprinted liver-on-a-chip. Thus, it is easier and more versatile in producing organs-on-chips with the suggested 3D bioprinting method. Our novel 3D bioprinting method will provide a new research paradigm in the field of organs-on-chips development and possibly lead to future applications for a body-on-a-chip.Acknowledgements
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (No. 2010-0018294). This work was supported by the Industrial Technology Innovation Program (No. 10048358) funded by the Ministry of Trade, Industry & Energy (MI, Korea).References
- P. N. Nge, C. I. Rogers and A. T. Woolley, Chem. Rev., 2013, 113, 2550–2583 CrossRef CAS PubMed.
- A. M. Taylor, M. Blurton-Jones, S. W. Rhee, D. H. Cribbs, C. W. Cotman and N. L. Jeon, Nat. Methods, 2005, 2, 599–605 CrossRef CAS PubMed.
- S. Kim, H. Lee, M. Chung and N. L. Jeon, Lab Chip, 2013, 13, 1489–1500 RSC.
- P. Neuži, S. Giselbrecht, K. Länge, T. J. Huang and A. Manz, Nat. Rev. Drug Discovery, 2012, 11, 620–632 CrossRef PubMed.
- D. Huh, B. D. Matthews, A. Mammoto, M. Montoya-Zavala, H. Y. Hsin and D. E. Ingber, Science, 2010, 328, 1662–1668 CrossRef CAS PubMed.
- K. K. Van Rompay, Antiviral Res., 2010, 85, 159–175 CrossRef CAS PubMed.
- D. Huh, G. A. Hamilton and D. E. Ingber, Trends Cell Biol., 2011, 21, 745–754 CrossRef CAS PubMed.
- Š. Selimović, M. R. Dokmeci and A. Khademhosseini, Curr. Opin. Pharmacol., 2013, 13, 829–833 CrossRef PubMed.
- S. N. Bhatia and D. E. Ingber, Nat. Biotechnol., 2014, 32, 760–772 CrossRef CAS PubMed.
- J. H. Yeon and J.-K. Park, BioChip J., 2007, 1, 17–27 Search PubMed.
- W.-Y. Yeong, C.-K. Chua, K.-F. Leong and M. Chandrasekaran, Trends Biotechnol., 2004, 22, 643–652 CrossRef CAS PubMed.
- Y.-J. Seol, T.-Y. Kang and D.-W. Cho, Soft Matter, 2012, 8, 1730–1735 RSC.
- B. R. Song, S. S. Yang, H. Jin, S. H. Lee, J. H. Lee, S. R. Park, S.-H. Park and B.-H. Min, Tissue Eng. Regener. Med., 2015, 12, 172–180 CrossRef CAS.
- N. N. Zein, I. A. Hanouneh, P. D. Bishop, M. Samaan, B. Eghtesad, C. Quintini, C. Miller, L. Yerian and R. Klatte, Liver Transpl., 2013, 19, 1304–1310 CrossRef PubMed.
- K. Sun, T. S. Wei, B. Y. Ahn, J. Y. Seo, S. J. Dillon and J. A. Lewis, Adv. Mater., 2013, 25, 4539–4543 CrossRef CAS PubMed.
- S. J. Leigh, R. J. Bradley, C. P. Purssell, D. R. Billson and D. A. Hutchins, PLoS One, 2012, 7, e49365 CAS.
- A. K. Au, W. Huynh, L. F. Horowitz and A. Folch, Angew. Chem., Int. Ed., 2016, 55, 3862–3881 CrossRef CAS PubMed.
- K. B. Anderson, S. Y. Lockwood, R. S. Martin and D. M. Spence, Anal. Chem., 2013, 85, 5622–5626 CrossRef CAS PubMed.
- A. I. Shallan, P. Smejkal, M. Corban, R. M. Guijt and M. C. Breadmore, Anal. Chem., 2014, 86, 3124–3130 CrossRef CAS PubMed.
- S. V. Murphy and A. Atala, Nat. Biotechnol., 2014, 32, 773–785 CrossRef CAS PubMed.
- D. B. Kolesky, R. L. Truby, A. Gladman, T. A. Busbee, K. A. Homan and J. A. Lewis, Adv. Mater., 2014, 26, 3124–3130 CrossRef CAS PubMed.
- R. Chang, J. Nam and W. Sun, Tissue Eng., Part C, 2008, 14, 157–166 CrossRef CAS PubMed.
- R. Chang, K. Emami, H. Wu and W. Sun, Biofabrication, 2010, 2, 045004 CrossRef PubMed.
- N. S. Bhise, V. Manoharan, S. Massa, A. Tamayol, M. Ghaderi, M. Miscuglio, Q. Lang, Y. S. Zhang, S. R. Shin and G. Calzone, Biofabrication, 2016, 8, 014101 CrossRef PubMed.
- F. Pati, J.-H. Shim, J.-S. Lee and D.-W. Cho, Manuf. Lett., 2013, 1, 49–53 CrossRef CAS.
- G. Comina, A. Suska and D. Filippini, Lab Chip, 2014, 14, 2978–2982 RSC.
- S. Halldorsson, E. Lucumi, R. Gómez-Sjöberg and R. M. Fleming, Biosens. Bioelectron., 2015, 63, 218–231 CrossRef CAS PubMed.
- J. M. Anderson, N. P. Ziats, A. Azeez, M. R. Brunstedt, S. Stack and T. L. Bonfield, J. Biomater. Sci., Polym. Ed., 1996, 7, 159–169 CrossRef.
- M.-H. Wu, S.-B. Huang and G.-B. Lee, Lab Chip, 2010, 10, 939–956 RSC.
- R. Gomez-Sjoberg, A. A. Leyrat, B. T. Houseman, K. Shokat and S. R. Quake, Anal. Chem., 2010, 82, 8954–8960 CrossRef CAS PubMed.
- T. Tao and J. Peng, J. Genet. Genomics, 2009, 36, 325–334 CrossRef CAS PubMed.
- E.-M. Materne, A. G. Tonevitsky and U. Marx, Lab Chip, 2013, 13, 3481–3495 RSC.
- P. J. Lee, P. J. Hung and L. P. Lee, Biotechnol. Bioeng., 2007, 97, 1340–1346 CrossRef CAS PubMed.
- B. J. Kane, M. J. Zinner, M. L. Yarmush and M. Toner, Anal. Chem., 2006, 78, 4291–4298 CrossRef CAS PubMed.
- C.-T. Ho, R.-Z. Lin, W.-Y. Chang, H.-Y. Chang and C.-H. Liu, Lab Chip, 2006, 6, 724–734 RSC.
- P. M. van Midwoud, E. Verpoorte and G. M. Groothuis, Integr. Biol., 2011, 3, 509–521 RSC.
- Y. Kim and P. Rajagopalan, PLoS One, 2010, 5, e15456 Search PubMed.
- A. Van der Meer, A. Poot, M. Duits, J. Feijen and I. Vermes, J. Biomed. Biotechnol., 2009, 2009, 823148 CrossRef CAS PubMed.
- A. Van der Meer, A. Poot, J. Feijen and I. Vermes, Biomicrofluidics, 2010, 4, 011103 CrossRef CAS PubMed.
- J. B. Lee and J. H. Sung, Biotechnol. J., 2013, 8, 1258–1266 CrossRef CAS PubMed.
- A. Sin, K. C. Chin, M. F. Jamil, Y. Kostov, G. Rao and M. L. Shuler, Biotechnol. Prog., 2004, 20, 338–345 CrossRef CAS PubMed.
- M. A. Guzzardi, C. Domenici and A. Ahluwalia, Tissue Eng., Part A, 2011, 17, 1635–1642 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2016 |