Open Access Article
Open Access ArticleQuantification of purple non-sulphur phototrophic bacteria and their photosynthetic structures by means of total reflection X-ray fluorescence spectrometry (TXRF)
Joanna
Fiedor
*,
Beata
Ostachowicz
,
Monika
Baster†
,
Marek
Lankosz
and
Květoslava
Burda
Department of Medical Physics and Biophysics, Faculty of Physics and Applied Computer Science, AGH-University of Science and Technology, al. A. Mickiewicza 30, Kraków 30-059, Poland. E-mail: fiedor@agh.edu.pl; Fax: +48 12 634 0010; Tel: +48 12 617 2984
First published on 2nd September 2016
Abstract
Phototrophic bacteria are metabolically diverse microorganisms that attract substantial attention due to their growing potential in scientific and industrial applications. In the present study, total reflection X-ray fluorescence (TXRF) spectrometry was applied to perform a comprehensive analysis of the trace elements content of purple non-sulphur phototrophic bacteria, their chromatophores and selected photosynthetic structures in response to alterations in oxygen growth conditions. There is a lack of consistent information on the content of microelements, their distribution and in particular correlations between them. This analysis, carried out on the Rhodobacter sphaeroides species aims to rectify this. Qualitative examination revealed the presence of microelements generally not considered as basic in the bacterial ionome. Quantitative inspection pointed to Fe as the major trace element in this phototrophic species irrespective of growth conditions (sample type). The K/Rb and Ca/Sr ratios were determined for the first time for bacteria and their photosynthetic membranes. Finally, the ionomic approach to elemental accumulation followed by statistical analysis revealed intriguing relationships between the elements within cells and phototrophic membranes. The vast potential and usefulness of the TXRF technique in a wide range of biological and environmental applications is underlined.
Introduction
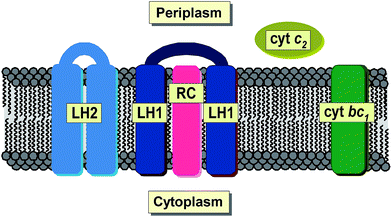
Total reflection X-ray fluorescence spectrometry (TXRF) is a well-established analytical technique used for simultaneous qualitative and quantitative determination of a wide range of chemical elements. Its major advantages include the versatility of its application and the relative simplicity of sample preparation. Furthermore, the extension of the detection limit to one part per billion1 and the need for only 10−6 to 10−9 g of material makes TXRF an excellent technique for the analysis of microsamples irrespective of their original physical state. So far, TXRF has been successfully applied in physical, environmental, chemical, biological, medical, pharmaceutical, nutritional, geological and art-historical studies, not to mention nanotechnology or industry.2–9 Hitherto, TXRF has been used to investigate a variety of microorganisms.10 However, not much has been done on bacterial systems. One of the very few examples is the work of Wittershagen et al. in which TXRF was shown to be useful in determining the metal cofactors of two enzymes in respiratory chain complexes from the soil bacterium Paracoccus denitrificans.11 More recently, TXRF was used for the evaluation of the kinetic behavior of chromium bioaccumulation in Acinetobacter beijerinckii,12 Gram-negative, strictly aerobic bacteria found in human and animal tissues as well as of other environmental origins.13 By applying other techniques that are based on X-ray radiation, i.e. EDXRF (energy dispersive X-ray fluorescence), the accumulation of some metals in the cells of the aerobic acidophilic proteobacterium Acidiphilium rubrum was investigated.14 More recently, SRXFM (synchrotron X-ray fluorescence microscopy) was used to determine changes in the elemental composition of the chemolithoautotrophic ammonia oxidizing bacterium, Nitrosomonas europea as a result of exposure to copper stress and as a function of the physiological batch growth phase.15Purple non-sulphur photosynthetic bacteria constitute one of the major groups of phototrophic microorganisms capable of growing in habitats of widely varying oxygen pressure and light intensity.16 They can be found in all kinds of water reservoirs and in terrestrial environments characterized by a low level of sulfide.17 Some species are known to exhibit rare metabolic features that allow them to exist in extreme environments such as hot, cold, salty, alkaline or acidic habitats.18 Under anaerobic conditions, in the presence of light, purple non-sulphur bacteria are capable of changing their metabolism from aerobic respiration into phototrophic, developing de novo photosynthetic apparatus. During this process, bacterial cells build up an extensive system of intracytoplasmic membranes, known as chromatophores, and synthesize photosynthetic pigments, bacteriochlorophylls (magnesium containing porphyrin derivatives, BChls) and carotenoids (Crts). BChls are primarily responsible for light-harvesting, while Crts carry out various functions among which (photo)protection is considered the most important.19–22 All pigments are non-covalently bound to membrane proteins, that is the two types of light-harvesting pigment–protein complexes (LH1 and LH2) and the reaction center (RC), which together with the cytochrome bc1 complex and ATP synthase are regarded as the basic bacterial photosynthetic unit23 (Fig. 1). Bacterial photosynthesis, in contrast to plant photosynthesis, occurs exclusively under anaerobic conditions. It results in extremely effective conversion of light energy into chemical energy, which is further used for the biosynthesis of primary biomolecules.
Among purple non-sulphur phototrophic bacteria one of the most often studied species is Rhodobacter (Rb.) sphaeroides. This is a Gram-negative, metabolically diverse microorganism that can grow heterotrophically or photosynthetically, fixing carbon dioxide or nitrogen.16,24Rhodobacter sphaeroides serves as a universal model organism for investigations of different aspects of photosynthesis as well as other metabolic processes. Under appropriate conditions it has been shown to produce hydrogen from a variety of organic compounds at high yields.25 Moreover, its efficacy in the production of extracellular 5-aminolevulinic acid, commonly used in agriculture as a herbicide, insecticide and a plant growth-promoting factor has been reported.26 Its versatile biotechnological potential has also been demonstrated in the case of the production of an efficient antioxidant, coenzyme Q10,27 or polyhydroxybutyrate, an example of a fully biodegradable polyester.28 Finally, cultures of Rb. sphaeroides have proved useful in bioremediation with a large potential to remove heavy metals (mercury, chromium or cadmium) as well as radioactive isotopes of uranium, cobalt, cesium or strontium from both synthetic sewage wastewater and natural water reservoirs.29
Taking into account the broad range of the potential scientific and industrial applications of phototrophic bacteria, a fundamental knowledge of their elemental profiling confirmed by TXRF spectrometry is of particular relevance. The lack of consistent and systematic data prompts us to present a detailed qualitative and quantitative analysis of the trace elements present in intact cells of Rb. sphaeroides 2.4.1 in response to alterations in environmental (oxygen) growth conditions. To complement the work, there is a detailed analysis of the data, followed by the evaluation of the distribution of trace elements across chromatophores and selected photosynthetic structures. An ionomic approach to elemental accumulation made it possible to uncover relationships between elements within cells and photosynthetic membranes. The findings aim to deliver, for the first time, a unique view of the distribution of chemical microelements in bacterial photosynthetic structures and correlations between them. This should extend our knowledge in this field of science, and, also corroborate the usefulness of TXRF spectrometry in this field of study.
Experimental
Culturing of bacteria
A strain of purple non-sulphur photosynthetic bacteria Rb. sphaeroides 2.4.1 was grown first non-photosynthetically (in darkness) and then photosynthetically at 27 °C in a modified Hutner medium.30 Bacteria, inoculated on agar plates, were grown under aerobic conditions in the dark for 7 days. Subsequently, they were transferred to liquid cultures and kept in the dark for an additional 12 hours. After that time, cultures were illuminated continuously with white light (55 μmol × m−2 × s−1). The cells were harvested by centrifugation (Centurion Scientific K241R, 2600 × g, 20 min, 10 °C) after 7 days (late log phase) of illumination, and washed twice with a 0.01 M TRIS–HCl buffer (pH 7.8). After harvesting, the cells were kept at −25 °C.Isolation of photosynthetic structures
Chromatophores were prepared from Rb. sphaeroides 2.4.1 cells grown anaerobically. Briefly, bacterial cells suspended in a 0.02 M TRIS–HCl buffer (pH 7.8) were disrupted by twofold passage through a French pressure cell (SLM-Aminco FA-031, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 psi) in the presence of a small amount of DNase I. To remove cell debris, the homogenate was centrifuged (Sigma 3K30, 18
000 psi) in the presence of a small amount of DNase I. To remove cell debris, the homogenate was centrifuged (Sigma 3K30, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 30 min, 10 °C). The chromatophores were separated from cytochromes and other low-mass contaminants by high-speed centrifugation (Beckmann L7-65, 70
000 × g, 30 min, 10 °C). The chromatophores were separated from cytochromes and other low-mass contaminants by high-speed centrifugation (Beckmann L7-65, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 90 min, 4 °C), and then purified by triple washing with a 0.02 M TRIS–HCl buffer (pH 7.8) followed by ultracentrifugation. The bacterial photosynthetic membranes were stored at −25 °C until further use.
000 × g, 90 min, 4 °C), and then purified by triple washing with a 0.02 M TRIS–HCl buffer (pH 7.8) followed by ultracentrifugation. The bacterial photosynthetic membranes were stored at −25 °C until further use.
Reaction centers were prepared from the chromatophores of Rb. sphaeroides 2.4.1 as described by Orzechowska et al. with some modifications.31 In brief, the purified chromatophores were incubated in a 0.02 M TRIS–HCl buffer (pH 7.8) containing 0.25% LDAO, for 30 min in the dark at room temperature. Afterwards, the suspension was diluted 2-fold and subjected to ultracentrifugation (Sorvall WX Ultra 100, 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 60 min, 10 °C). The final purification of the RCs was achieved by ion-exchange chromatography on a DEAE-cellulose column (DE52, Whatman). The reaction centers were eluted as a brown band with 0.125–0.15 M NaCl in a 0.02 M TRIS–HCl/0.08% LDAO buffer (pH 7.8) and intensively dialyzed (24 hours, 4 °C).
000 × g, 60 min, 10 °C). The final purification of the RCs was achieved by ion-exchange chromatography on a DEAE-cellulose column (DE52, Whatman). The reaction centers were eluted as a brown band with 0.125–0.15 M NaCl in a 0.02 M TRIS–HCl/0.08% LDAO buffer (pH 7.8) and intensively dialyzed (24 hours, 4 °C).
The purification of a water-soluble fraction of cytochromes was performed according to Samyn et al. with some modifications.32 Briefly, the crude cytochromes were subjected to a DEAE-cellulose column and eluted as a purple band with 0.065–0.075 M NaCl in a 0.02 M TRIS–HCl buffer (pH 7.8), followed by dialysis (24 hours, 4 °C). The cytochromes and RCs were concentrated by the use of Amicon Ultra-15 centrifugal filters. In each case the purification steps were repeated twice. The quality of photosynthetic structures was checked electrophoretically by SDS-PAGE according to the Laemmli procedure.33 The purified and concentrated samples were kept at −25 °C until further use. To minimize degradation, all laboratory operations were carried out under dim white light.
Total reflection X-ray fluorescence spectrometry
The analysis of elemental composition was done with a benchtop TXRF spectrometer (Nanohunter, Rigaku) equipped with a Mo X-ray tube as an excitation source, a multilayer monochromator and a silicon drift detector with energy resolution of 180 eV for energy 6.4 keV. All operations were carried out at 50 kV and 0.8 mA. A fixed acquisition time of 2 × 103 seconds was applied. As an internal standard gallium (Merck) was used. Its final concentration was set at ∼100 ppm. The standard sample preparation protocol involved pipetting 5 μl of the investigated solution onto the middle of a glass reflector. Samples were left to dry on a leveled hot-plate under a fume hood. In each case, the number of analyzed samples of a particular type (aerobically and anaerobically grown bacteria, chromatophores, RCs and cytochrome c2 (cyt c2)) was set to eight.UV-VIS spectroscopy
Steady state absorption spectra were recorded with a Cary50 Bio UV-VIS spectrophotometer (Varian). In order to determine BChl concentrations, 10 μl of each pigmented sample was diluted with 1 ml of acetone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol solution (30
methanol solution (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 v/v) and their absorption spectra were measured. For the calculations an approximate BChl molar absorption coefficient of ε770 = 7 × 104 M−1 cm−1 was used. This was based on the ε values of the BChl in pure solvents.34 To estimate RC and cytochrome concentrations ε802 = 0.288 M−1 cm−1 and ε550 = 0.029 M−1 cm−1 were applied, respectively.35,36
70 v/v) and their absorption spectra were measured. For the calculations an approximate BChl molar absorption coefficient of ε770 = 7 × 104 M−1 cm−1 was used. This was based on the ε values of the BChl in pure solvents.34 To estimate RC and cytochrome concentrations ε802 = 0.288 M−1 cm−1 and ε550 = 0.029 M−1 cm−1 were applied, respectively.35,36
Atomic force microscopy
Freshly cleaved mica (grade V5, Electron Microscopy Sciences) attached to microscope cover slips was used as a support. Briefly, a drop of adsorption buffer was placed in the middle of a mica scrap to which an aliquot of the sample was added.37 The preparation was left in the dark for about one hour.Topographical analysis was performed with Agilent 5500 atomic force microscopy apparatus (Agilent Technologies) equipped with a multi-purpose top-down scanner (8 μm × 8 μm). Standard silicon nitride cantilevers (nominal spring constant of 0.01 N m−1; Bruker AFM Probes) were used. The images were obtained using contact mode. All operations were carried out at ambient temperature. AFM images were processed using PicoView 4.0 and Gwyddion 2.38 software.
Statistical analysis
A non-parametric Mann–Whitney U-test was applied to reveal statistically significant differences between the elemental composition of (i) photosynthetically versus aerobically grown bacteria; (ii) photosynthetic cells versus photosynthetic membranes; and (iii) RCs versus cyt c2. The elemental relationship was evaluated by the application of the r Pearson's correlation coefficient and the coefficient of determination R2. The analysis was performed using Statistica 10.0 software.Results and discussion
Characterization of biological material
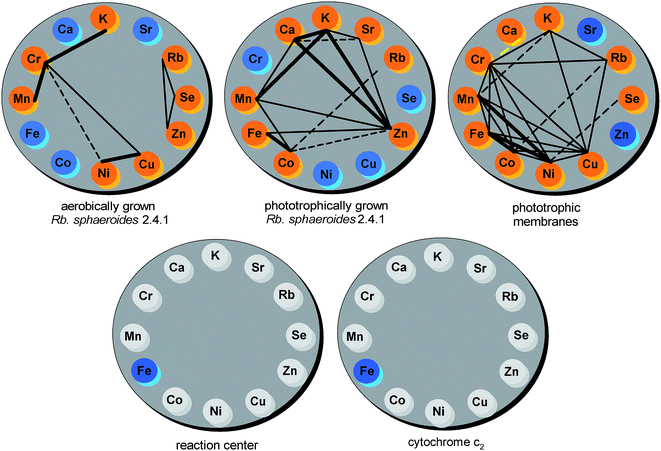
Rhodobacter sphaeroides 2.4.1 was cultured both anaerobically and aerobically as described in the Experimental section. Photosynthetic growth resulted in the formation of brownish coccobacillus (elliptical-like) cells with dimensions typical of this bacterial strain.38 An AFM topographical image of an example cell 1.85 μm in length, 1.05 μm in width and 0.19 μm in height is presented in Fig. 2A.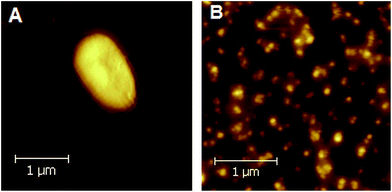 | ||
| Fig. 2 AFM images of Rb. sphaeroides 2.4.1 cell (A), and a large-view of several clusters of photosynthetic membranes (chromatophores) (B). | ||
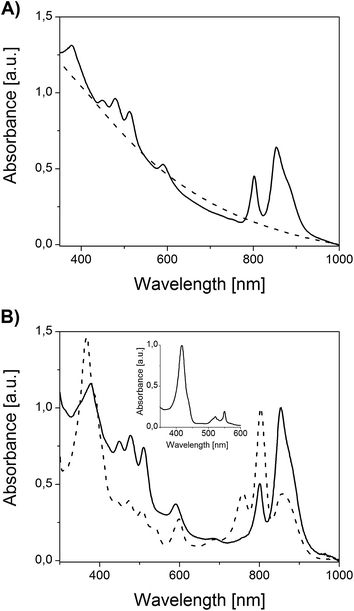
The absorption spectrum of an anoxygenically grown bacterial suspension is shown in Fig. 3A. The spectrum has features typical of purple non-sulphur bacteria such as two absorption bands in the long-wavelength region as well as three bands in the range of 430–550 nm. All of them correspond to photosynthetic pigment–protein complexes containing BChls and Crts, respectively. Under aerobic conditions, in the dark, the formation of photosynthetic membranes is suppressed in the presence of oxygen, so that heterotrophic culturing yields pinkish, otherwise unpigmented bacterial colonies. Correspondingly, there are no signals of photosynthetic origin in the absorption spectrum of the non-phototrophic bacterial suspension (Fig. 3A).
Chromatophores were prepared from phototrophically grown Rb. sphaeroides 2.4.1 cells by two passages through a French pressure cell. A large-view image of the clusters of intracytoplasmic membranes is presented in Fig. 2B. It is generally accepted that chromatophores of Rb. sphaeroides are present in the form of discrete autonomous vesicles hosting the entire photosynthetic machinery39 (Fig. 1). This organization is expected to reflect their functioning, maintenance and biogenesis,40 yet there are still a number of open questions concerning their intrinsic architecture. The UV-VIS absorption spectrum of Rb. sphaeroides chromatophores in a 0.02 M TRIS–HCl buffer (pH 7.8) is presented in Fig. 3B. In the spectrum, characteristic bands arising from the presence of LH2 complexes dominate, exhibiting maxima at 853 nm, 800 nm (BChls), and 509 nm, 476 nm, 448 nm (Crts). The contribution of LH1 antennae is visible as a shoulder with a maximum at ∼880 nm, while that of RCs as a weak band with a maximum at ∼760 nm. The shorter-wavelength bands with maxima at 590 nm and 376 nm also originate from BChl as a consequence of the pigments' electronic structure.
Two components from the bacterial photosynthetic apparatus, the RCs and cyt c2, were subjected to closer inspection. The LH antennae were excluded from further analysis since they, except for polypeptide chains, are exclusively composed of Mg-containing BChls and metal-free Crts. Magnesium belongs to the group of alkaline earth metals, with a characteristic X-ray radiation of 1.254 keV (Kα1) and 1.297 keV (Kβ1), respectively, too weak to be excited and detected by our experimental TXRF setup.
A reaction center is a pigment–protein complex composed of the cofactors of an electron transport chain embedded within 3 polypeptide chains.41 It utilizes solar energy to drive reactions eventually leading to the translocations of protons across the photosynthetic membrane, thereby generating the proton-motive force for ATP synthesis.42 The absorption spectrum of an isolated RC is presented in Fig. 3B. In the spectrum, bands originating from pigment-based cofactors are clearly seen. Hence, BChl and bacteriopheophytin (BPhe), its demetallated derivative, give rise to three sets of absorption bands in the Soret region, the QX region (maxima at 598 nm and 536 nm, respectively), and the QY region, with maxima at 758 nm (BPhe), 802 nm (BChl monomer), and 858 nm (BChl dimer). The contribution of Crts is seen in the range of 430–520 nm. This spectrum is in good agreement with previously reported spectra.31,42 The purity of the RC was additionally verified by the application of two independent methods: (i) spectroscopically, by checking the ratio of protein absorbance at 280 nm to BChl absorbance at 802 nm, and (ii) electrophoretically. In the first case the satisfactory ratio of 1.5 was achieved, while the SDS-PAGE separation revealed only the presence of proteins characteristic of RCs, further corroborating the quality of preparation (not shown).
Another structure investigated in the course of the study was the water-soluble, mobile electron carrier, cyt c2. This is a haemoprotein responsible for efficient electron transfer between two membrane bound proteins, i.e. cyt bc1 and the RC43 (Fig. 1). The absorption spectrum of the reduced form of cyt c2 is presented in Fig. 3B. The spectrum exhibits cytochrome bands typical of c-types, with maxima located at 550 nm, 521 nm, and 417 nm (α, β and γ band, respectively).36,44 The purity of cyt c2 was verified spectroscopically by checking the α/β absorbance ratio (α/β = 1.5) and by SDS-PAGE electrophoresis, which delivered satisfactory results consistent with previous reports (not shown).
TXRF analysis
The analysis of the composition of chemical elements present in the aerobically and phototrophically grown bacteria and their photosynthetic structures was performed by the application of the TXRF technique. In Fig. 4 a typical TXRF spectrum of anaerobically grown Rb. sphaeroides cells superimposed onto a chromatophore spectrum is shown. | ||
| Fig. 4 TXRF profiles of elemental composition of phototrophically grown Rb. sphaeroides cells (dotted line) and chromatophores (solid line). | ||
In the course of the examination the presence of the following elements was revealed: K, Ca, Cr, Mn, Fe, Co, Ni, Cu, Zn, Se, Rb and Sr. The same set of elements was detected in the aerobically grown bacteria, while in the case of RCs and cyt c2 the presence of Fe was established. Apart from the above listed elements, in all types of samples tiny amounts of Br were also found. However, this chemical element was not involved in further analysis as it was regarded as an unstable contamination accompanying the chloride ions which were applied to the material at the stage of culturing and sample preparation.
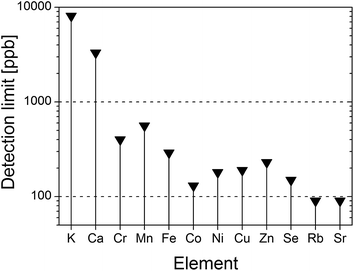
On the basis of the recorded TXRF spectra and according to the formula suggested by Currie45 the detection limits (LDs) of all determined elements were estimated (Fig. 5). Hence, LD ranges from 90 ppb in the case of Sr or Rb up to 3.3 ppm and 8 ppm for Ca and K, respectively. The calculated relative errors due to the measurement-counting statistics for each element reached as much as: K – 0.8%, Ca – 0.7%, Cr – 7.7%, Mn – 6.8%, Fe – 2.3%, Co – 5.1%, Ni – 4.8%, Cu – 5.6%, Zn – 2.5%, Se – 9.8%, Rb – 4.4%, Sr – 1.7%. The precision of deposition of the sample was calculated on the basis of measurements of 8 residues from the same sample type. The relative precisions for each element are as follows: K – 2.4%, Ca – 1%, Cr – 3.8%, Mn – 2.1%, Fe – 3.6%, Co – 2.3%, Ni – 2.6%, Cu – 1.4%, Zn – 0.9%, Se – 3.3%, Rb – 3.3%, Sr – 1.1%.
The main aim of the study was not only to identify but to quantify the elements present in purple, non-sulphur phototrophic bacteria and their photosynthetic structures. For this purpose the concentrations of typical microelements (Cr, Mn, Fe, Co, Ni, Cu, Zn, Se), and macroelements (K and Ca) and their chemical analogues (Rb and Sr, respectively) were established. In order to exclude potential effects of TRIS–HCl on elemental quantity, additional measurement of the plane buffer was performed. This revealed the presence of negligible amounts of Ca, consequently not included in further analysis. The results of TXRF analysis together with evaluated measurement uncertainties are summarized in Table 1. In each case the element concentration is expressed as a median with the standard error of the mean taken as the estimated uncertainty.
| Sample | Element | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fe | u(Fe) | Zn | u(Zn) | Co | u(Co) | Ni | u(Ni) | Mn | u(Mn) | Cr | u(Cr) | Cu | u(Cu) | Se | u(Se) | Ca | u(Ca) | K | u(K) | Sr | u(Sr) | Rb | u(Rb) | |
| [ppm] | [ppm] | [ppm] | [ppm] | [ppm] | [ppm] | [ppm] | [ppm] | [ppm] | [ppm] | [ppm] | [ppm] | |||||||||||||
| a nd* – not determined. | ||||||||||||||||||||||||
| Aerobic cells | 4.17 | 0.17 | 1.66 | 0.02 | 0.61 | 0.02 | 0.56 | 0.02 | 0.62 | 0.01 | 0.55 | 0.02 | 0.40 | 0.01 | 0.080 | 0.003 | 200 | 2 | 166 | 4 | 2.19 | 0.03 | 0.30 | 0.01 |
| Phototrophic cells | 7.78 | 0.27 | 2.16 | 0.18 | 1.01 | 0.03 | 0.59 | 0.01 | 0.59 | 0.03 | 0.55 | 0.02 | 0.64 | 0.06 | 0.080 | 0.002 | 53 | 7 | 51 | 7 | 0.27 | 0.02 | 0.14 | 0.01 |
| Phototrophic membranes | 4.95 | 0.37 | 1.22 | 0.03 | 0.81 | 0.04 | 0.70 | 0.04 | 0.54 | 0.04 | 0.59 | 0.08 | 0.49 | 0.03 | 0.050 | 0.003 | 34 | 1 | 7.55 | 0.44 | 0.16 | 0.01 | 0.080 | 0.004 |
| Reaction center | 2.52 | 0.13 | nd* | — | nd | — | nd | — | nd | — | nd | — | nd | — | nd | — | nd | — | nd | — | nd | — | nd | — |
| Cytochrome c2 | 2.33 | 0.79 | nd | — | nd | — | nd | — | nd | — | nd | — | nd | — | nd | — | nd | — | nd | — | nd | — | nd | — |
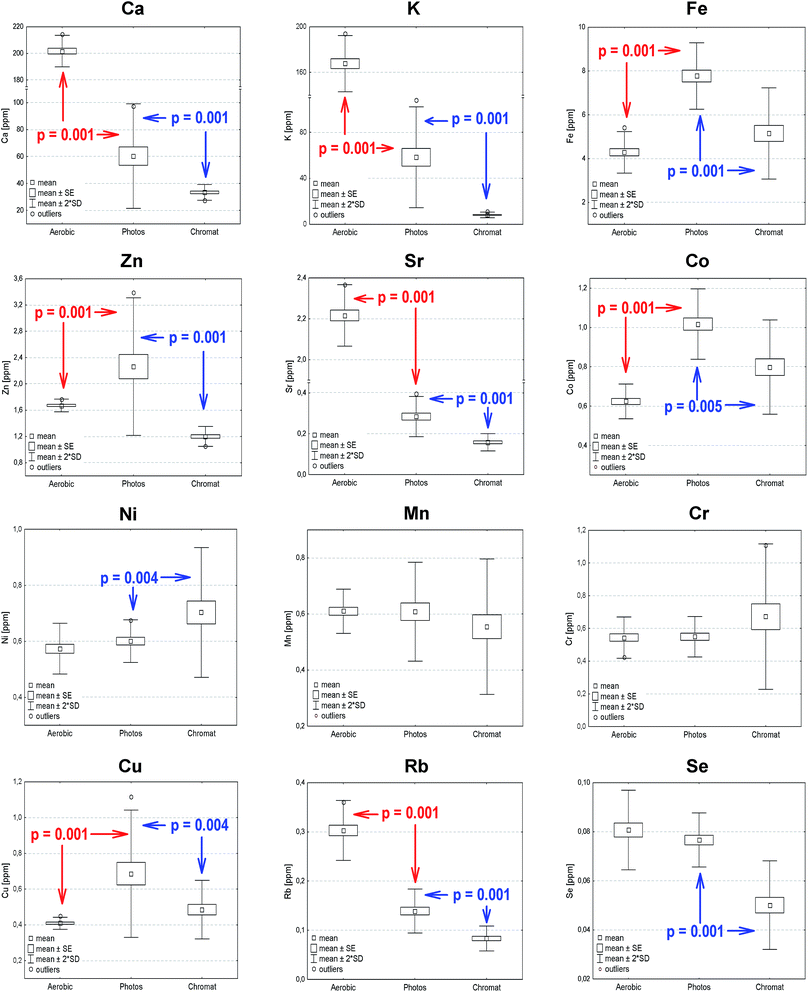
Closer inspection of the results summarized in Table 1 indicates that the concentrations of elements detected in the aerobic cells decreased roughly in the following order: Ca > K ≫ Fe > Sr > Zn > Co ≈ Mn ≈ Ni ≈ Cr > Cu ≥ Rb > Se whereas in the case of anaerobic cells a Ca ≈ K ≫ Fe > Zn > Co > Mn ≈ Ni ≈ Cr ≈ Cu > Sr > Rb > Se sequence could be established. Interestingly, irrespective of the growing conditions the amount of Ca or K exceeds the quantity of iron by an order of magnitude (in fact, in aerobic cells by even 40-(potassium) or 50-(calcium) times, respectively). In bacteria cultured under anaerobic conditions the concentrations of Ca and K are similar. If compared to the cells from the aerobic culture their amount decreased by a factor of about 3.8 and 3.3, respectively. At the same time, the concentration of strontium in phototrophically grown cells decreased by approximately 10 times if compared to aerobically kept bacteria. On the other hand, the amount of Sr is only about twice as high as that of Rb, whose content also decreased by a factor of two in phototrophic cells. The concentrations of elements determined in chromatophores “obeyed” the same order as that established for photosynthetic cells with one major exception, the concentrations of potassium and iron being similar. In this case K content decreased by almost 7 times as compared to its amount in photosynthetic cells, while at the same time the quantity of Fe was only slightly changed. It is also worth noting that in isolated membranes the concentrations of Ca, Sr and Rb are reduced by a similar factor of ∼1.7. In all cases, selenium exhibited the lowest concentration of all those elements.
For the clarity of further analysis and discussion, the elements were divided into two groups: (i) microelements (Fe, Zn, Co, Ni, Mn, Cr, Cu, Se) and (ii) macroelements along with their analogues (Ca–Sr and K–Rb). Iron is the major microelement. Its concentration is the highest of all determined trace elements, irrespective of the type of sample. The amount of Fe in chromatophores and phototrophic cells exceeds that in aerobic cells, in the latter case almost 2-fold. One possible reason for such differences between anaerobic and aerobic cells is the de novo formation of the photosynthetic apparatus. In the presence of light, and the simultaneous absence of oxygen, bacteria adjust their metabolism to new environmental conditions synthesizing a complicated light-harvesting apparatus (Fig. 3A). During this process genes encoding metalloproteins active in photosynthetic reactions are expressed leading eventually to a number of element-containing proteins.46 These findings also reflect the observation derived from plants that, depending on the tissue, while the amount of iron varies strongly, it is still the most abundant among the transition metals.47 In biological systems iron is predominantly present as a constituent of non-haem iron containing complexes (e.g. bacterial RC) or haem-proteins (e.g. cytochromes). Indeed, it is the only microelement detected in the bacterial substructures investigated here, RCs and cyt c2. In RCs, a ferrous non-haem iron located between two ubiquinone-10 molecules forms an iron–quinone complex, but it does not participate directly in the electron transport between the quinone acceptors. The biological role of the iron atom in RCs still remains ambiguous, although some structural function is considered the most probable.42 Recently, by the application of Mössbauer spectroscopy and the nuclear inelastic scattering of synchrotron radiation it was established that non-haem iron from Rb. sphaeroides can be stabilized both in a high and low spin ferrous state, which suggests that it may be responsible for the dynamic properties of the acceptor side of RCs.31,48 Cytochrome c2 belongs to the family of c-type cytochromes. In contrast to RCs, it is characterized by the presence of iron in the form of haem covalently attached to a polypeptide chain via a thioether bond.43 In the present study the concentration of iron present in cyt c2 was comparable to its amount in RCs, yielding a Fe-cyt c2/Fe-RC ratio of 0.92. This finding corresponds very well with data from co-crystallization studies, which report the stoichiometry of cyt c2/RC to be ∼0.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.43
1.43
Apart from Co, whose amount was found to be somewhat higher in photosynthetic cells and membranes than in aerobic cells, the quantities of other trace elements, depending on their nature, were comparable among particular sample types.
Microelements, also called “micronutrients”, are a very important group of chemical elements known to be present in tiny amounts in all living organisms where they realize a variety of functions. For instance, they have been found to serve as catalytically active cofactors in metalloenzymes and hence to influence diverse biochemical reactions eventually regulating e.g. metabolic and redox processes or osmotic pressure. They are also known to perform a structural role by stabilizing macromolecules and biological complexes through electrostatic interactions.46,49,50 Any discussion on the localization of microelements accumulated within bacterial structures, their chemical fate as well as their functions goes beyond the scope of this publication and will be presented elsewhere. Depending on the bacterial organism the composition of trace elements may differ substantially. From a physiological point of view the elements present in bacteria are divided into three groups: (i) essential metals characterized by low toxicity (Fe, Mo, Mn); (ii) metals with moderate importance as trace elements of potential toxicity (Zn, Ni, Cu, V, Co, W and Cr); (iii) metals without any biological functions of high toxicity (Ag, Hg, Cd, Sb and Pb). Non-metals of high importance include Se, whereas non-essential metalloids comprise As and Te.46 Nowadays, in the case of most bacteria Mn, Co, Zn, Cu and Mo are commonly regarded as a basic microelemental set present in all species. In the present study, the occurrence of all except for molybdenum was verified, both in bacterial cells and chromatophores. Moreover, apart from the above mentioned, the presence of additional key elements (Fe, Ni, Cr and Se) was revealed. One should also expect to confirm the presence of Mo but, due to the use of a Mo X-ray tube as an excitation source, the detection of Mo or Cd was not possible. However, it has been shown that Cd has a high affinity binding site in the vicinity of the non-haem iron in RCs,51,52 hence its presence in the culturing medium would result in its accumulation in the system investigated, in particular in anaerobically grown cells and chromatophores. The elements determined in Rb. sphaeroides match the results of early studies aimed to evaluate manganese content in comparison to other metals (Cu, Zn, Fe) in some photosynthetic bacterial strains.53 In the case of other (non-phototrophic) bacterial types a list of 16 elements comprising B, Al, Ti, Cr, Mn, Fe, Ni, Cu, Zn, Sr, Ag, Sn, Ba, Pb, V and Mo has been reported. They were found in the cells of Escherichia (E.) coli and Sphaerotilus (S.) natans (both are examples of Gram-negative aerobic bacteria, E. coli can be found in human intestines, S. natans occurs in slime and sewage), Micrococcus roseus (Gram-positive, strictly aerobic bacteria, found in dust) or Bacillus cereus (Gram-positive aerobic bacteria responsible for clinical infections and food poisoning).54 More recent analysis of the elemental composition of 10 methanogenic bacteria (strict anaerobes typically using H2 and CO2 as energy and carbon sources, respectively) corroborated the presence of only 7 microelements (Fe, Ni, Co, Mo, Zn, Cu and Mn).55 However, studies on Acidiphilium rubrum, a chemoheterotrophic acidophilic aerobe, the only example in some way related to the Rb. sphaeroides species, revealed merely 4 microelements (Fe, Cr, Ni and Zn).14
In all living organisms the basic set of macroelements usually comprises Na, Mg, K, Ca, P, S and Cl. Here, we would like to focus on those elements determined by our experimental system. Thus, the second group of elements analyzed consisted of K and Ca and their chemical analogues, Rb and Sr, respectively. Rubidium, along with K, belongs to the group of alkali metals. In spite of having similar properties to K, Rb is considered a biologically non-essential trace element. In the same manner, Ca and Sr are members of the alkaline earth metals, but from a biological point of view, Ca is regarded a much more important element. The uptake of Rb or Sr by plants is known to depend on the physicochemical properties of soil such as the K (Ca) availability, acidity, adsorption properties or organic matter content.56 The K/Rb and Ca/Sr ratios have been estimated for a number of plant species and for their particular parts (roots, foliage etc.).57,58 So far, no comparable studies on any type of bacteria, including phototrophic species and in particular photosynthetic membranes, have been undertaken. Ratios of the respective elements based on the findings of the present study are summarized in Table 2.
| Ratio | Sample | ||
|---|---|---|---|
| Aerobic cells | Phototrophic cells | Phototrophic membranes | |
| K/Rb | 553 ± 4 | 365 ± 7 | 94 ± 0.4 |
| Ca/Sr | 91 ± 2 | 196 ± 7 | 212 ± 1 |
The highest K/Rb ratio was found for aerobically grown Rb. sphaeroides. It is 1.5-fold higher than in phototrophic cells and almost 6-fold higher than in phototrophic membranes, respectively. This effect is related mainly to significant changes in the concentration of K in the systems investigated. In the case of Ca/Sr the result is contrary: the ratio is comparable for photosynthetic cells and membranes and about 2 times higher than for aerobic cells. This phenomenon results from a much greater decrease in Sr concentration than in Ca in phototrophic cells in comparison to aerobic cells as already discussed.
Statistical analysis
In order to verify statistically significant differences between the elemental composition of Rb. sphaeroides cells, chromatophores and selected photosynthetic structures, a Mann–Whitney U test at p < 0.05 was carried out. This is a non-parametric, rank sum test regarded as an alternative to the t-test for two samples, which allows a comparison of the median scores of two unpaired groups whose values do not have to be normally distributed. In the present study the following groups were subjected to the U-test: (1) photosynthetically versus aerobically grown bacteria; (2) photosynthetically grown bacteria versus photosynthetic membranes; (3) RCs versus cyt c2. In each case the null hypothesis assumed that the distributions of both groups were identical. Evaluations were conducted on the basis of the data summarized in Table 1, and their results are overlaid onto the box-and-whisker plots shown in Fig. 6 (statistically significant differences between aerobically and anaerobically grown bacterial cells as well as photosynthetically grown bacteria and chromatophores).According to the results of the U-test analysis in the case of aerobically and anaerobically grown bacteria no statistically significant changes were observed for Ni, Mn, Cr and Se. On the other hand, the highest statistically important differences were noted for Ca, Co, Cu, Fe, K, Rb and Sr. For the photosynthetically grown bacteria and photosynthetic membranes there were no differences in the case of Mn and Cr, while the statistically most important changes were seen for Ca, K, Rb, Se, Sr and Zn. Finally, in the case of RCs and cyt c2, no significant differences were observed.
The r Pearson correlation coefficient was applied as a statistical measure of the strength and direction of the linear relationship between particular pairs of macro- and microelements across the samples. In line with the required data assumptions, all outliers (indicated in Fig. 6) were removed before running the analysis. Simultaneously, in each case a test of significance was conducted, with the null hypothesis that there is no linear correlation between a pair of elements. The results of the analysis were further corroborated by the R2 coefficient of determination. On the basis of the acquired data a set of correlation wheels showing significant correlations (p < 0.05, R2 > 0.39) for each pairwise combination of elements within a particular sample was constructed (Fig. 7).
For aerobically grown Rb. sphaeroides cells 66 comparisons were made, out of which a very strong positive correlation was found only in the case of Cr and Mn (R2 = 0.9652), K and Cr (R2 = 0.9266), and Ni and Cu (R2 = 0.8082), respectively. For photosynthetically grown bacteria (66 comparisons) such strong positive correlations were established for the following pairs of elements: K and Ca (R2 = 0.93), K and Zn (R2 = 0.9229), Ca and Zn (R2 = 0.8845), Fe and Co (R2 = 0.8227), and K and Mn (R2 = 0.8086), respectively. In the case of photosynthetic membranes (66 comparisons) very strong positive correlations were found for combinations of the following transition metals: Fe and Co (R2 = 0.9497), Mn and Ni (R2 = 0.8231), and Fe and Ni (R2 = 0.8155), respectively.
The results of the analysis show considerable differences in the elemental correlations between anaerobically and aerobically grown bacteria as well as in photosynthetic membranes. Pairs of elements most significantly correlated in aerobic bacteria (Mn–Cr, K–Cr, Cu–Ni) were only moderately correlated in photosynthetic membranes and no correlation was observed in the case of anaerobically grown cells. The pair Fe–Co was shown to be the only strongly correlated both in phototrophic bacteria and chromatophores. Finally, Mn–K, strongly correlated in phototrophic cells, was only weakly correlated in photosynthetic membranes. Other pairs of elements that were strongly correlated in phototrophic bacteria were weakly correlated in chromatophores and vice versa. Of all these elements Cr, Zn, Cu, Co, Mn and Ni turned out to be highly correlated. In aerobic cells Cr is correlated with Mn, K, Cu and weakly with Ni. In chromatophores it is correlated with Mn, K, Cu, Ni and additionally with Fe, Co, Rb and these correlations are roughly of the same degree. In phototrophic cells Zn has a strong correlation with Ca, a medium one with Mn, Fe, K and Sr, and a weak one with Co. Remarkably, only positive correlations were found for all but one pair that is Cr–Ca in chromatophores.
A comprehensive approach leading to a detailed understanding of the relationships in these systems would involve not only analyzing mutual correlations but also overall networks of correlations like those depicted in Fig. 7. For example these are the triangular motifs formed by correlations between three elements such as Ni–Cu–Cr and Zn–Se–Rb present in aerobically grown cells. In the case of photosynthetically grown bacteria and chromatophores even more of these triangular motifs can be found, but there are still elements which are not correlated to any others, such as Cu, Ni, Se and Cr (phototrophic cells) or Zn and Sr (phototrophic membranes). On the other hand, in the case of anaerobic cells, there are positive correlations for any pair taken from the group of Mn, Zn, Ca and K. In addition, each element from the pairs Zn–K, Zn–Ca and Ca–K is correlated to Sr whereas only from Zn–Mn to Co. The strongly positive correlated pair Fe–Co forms a triangular motif with Zn out of which only Co is additionally significantly correlated to Rb. In chromatophores triangular motifs are formed between any bi-correlated elements among Fe, Co, Mn, Ni, Cu and Cr. Moreover, each element from the pairs Mn–Cr and Cu–Cr exhibits bi-correlations with K and, in the latter case also with Rb. Closer examination of the Cr–Rb pair points to other triangular motifs including additionally Co and K. In phototrophic membranes Se is exclusively weakly correlated with Ni. Finally, bearing in mind that some elements do not show measurable correlations in the networks depicted in Fig. 7 it is interesting to compare bi-correlations of Co with other elements in the two phototrophic systems (cells and membranes). In both cases Co correlates with Fe, Mn and Rb, in photosynthetic cells additionally with Zn, whereas in membranes with Cu, Ni and Cr. Lastly, one has to stress that there are no triangular motifs common to phototrophic cells and chromatophores.
Conclusion
The application of TXRF spectrometry made it possible for the first time to provide a clear elemental profile of purple non-sulphur phototrophic bacteria and their photosynthetic structures. The method proved its value in studies where the samples are highly limited in amount and concentration. The qualitative and quantitative analysis of trace elements provided a fuller picture of the bacterial ionome and gave hints of its possible regulation at different levels, which needs to be further investigated. Among the trace elements found in bacterial cells, four seem to be of special significance to the species in question. Quantitative analysis pointed to Fe as a major trace element both in phototrophic structures and bacterial cells irrespective of growth conditions. The relatively high concentration of this microelement is primarily due to its diverse functions in photosynthesis, serving as a cofactor of electron transport chains and as a component of other biologically important structures. For the first time the bacterial elemental ratio of the chemical analogues Ca/Sr and K/Rb was established. Such data are of special concern since they may be treated as useful markers of elemental requirement along with their potential environmental availability. The statistical analysis revealed interesting relationships between specific elements within cells and photosynthetic membranes. The correlation between particular pairs of elements was shown to be highly environmentally determined, which suggests a certain degree of flexibility in the bacterial ionome (leading to their physiological adaptation) controlled not only on the genetic but also definitely on the environmental level. Furthermore, the correlations revealed may serve as convenient indicators of assimilation dependencies between particular elements, both macro- and microelements. In addition to mutual correlations, the analysis of an entire network of correlations delivered a comprehensive picture of the relationships between all the elements. In the present study, triangular motifs were identified and analyzed, and an analysis of e.g. quadrangular motifs could also be possible. Finally, the new dataset adds to available resources and contributes to our knowledge and understanding of the natural variation of the bacterial ionome and hence, indirectly, of photosynthesis and its related processes. So far no such data were accessible for any phototrophic bacteria.Acknowledgements
The authors would like to thank Mrs Dominika Najberg for her help at the initial stage of this project. The work was supported by the National Science Center (NCN, Poland) and its grant for Scientific Research, no. 11.11.220.01. The studies were performed within the BIONAN project.References
- M. West, A. T. Ellis, P. J. Potts, C. Streli, C. Vanhoof, D. Węgrzynek and P. Wobrauschek, J. Anal. At. Spectrom., 2011, 26, 1919–1963 RSC.
- J. Spanke, A. Bohlen von, R. Klockenkaemper, A. Quentmeier and D. Klockow, J. Anal. At. Spectrom., 2000, 15, 673–679 RSC.
- B. Ostachowicz, M. Lankosz, B. Tomik, D. Adamek, P. Wobrauschek, C. Streli and P. Kregsamer, Spectrochim. Acta, Part B, 2006, 61, 1210–1213 CrossRef.
- P. Janik, G. Tylko, B. Ostachowicz and K. Turnau, Microsc. Res. Tech., 2010, 73, 1134–1142 CrossRef CAS PubMed.
- E. Margui, G. H. Floor, M. Hidalgo, P. Kregsamer, G. Roman-Ross, C. Streli and I. Queralt, Spectrochim. Acta, Part B, 2010, 65, 1002–1007 CrossRef.
- K. Kocot, B. Zawisza, E. Margui, I. Queralt, M. Hidalgo and R. Sitko, J. Anal. At. Spectrom., 2013, 28, 736–742 RSC.
- M. B. B. Guerra, C. E. G. R. Schaefer, G. G. A. Carvalho de, P. F. Souza de, D. S. Junior, L. C. Nunes and F. J. Krug, J. Anal. At. Spectrom., 2013, 28, 1096–1101 RSC.
- R. Klockenkaemper and A. Bohlen von, Spectrochim. Acta, Part B, 2014, 99, 133–137 CrossRef CAS.
- M. W. Lankosz, M. Grzelak, B. Ostachowicz, A. Wandzilak, M. Szczerbowska-Boruchowska, P. Wróbel, E. Radwańska and D. Adamek, Spectrochim. Acta, Part B, 2014, 101, 98–105 CrossRef CAS.
- N. Szoboszlai, Z. Polgari, V. G. Mihucz and G. Zaray, Anal. Chim. Acta, 2009, 633, 1–18 CrossRef CAS PubMed.
- A. Wittershagen, P. Rostam-Khani, V. Zickermann, I. Zickermann, S. Gemeinhardt, B. Ludwig and B. O. Kolbesen, Fresenius. J. Anal. Chem., 1998, 361, 326–328 CrossRef CAS.
- R. Fernandez-Ruiz, M. Malki, A. I. Morato and I. Marin, J. Anal. At. Spectrom., 2011, 26, 511–516 RSC.
- A. Nemec, M. Musilek, M. Maixnerova, T. Baere de, T. J. Reijden van der, M. Vaneechoutte and L. Dijkshoorn, Int. J. Syst. Evol. Microbiol., 2009, 59, 118–124 CrossRef CAS PubMed.
- S. Itoh, M. Iwaki, N. Wakao, K. Yoshizu, A. Aoki and K. Tazaki, Plant Cell Physiol., 1998, 39, 740–744 CrossRef CAS.
- R. Yu, B. Lai, S. Vogt and K. Chandran, PLoS One, 2011, 6, e21255 CAS.
- M. T. Madigan and D. O. Jung, in The purple phototrophic bacteria, ed. C. N. Hunter, F. Daldal, M. C. Thurnauer and J. T. Beatty, Springer, Dordrecht, 2009, pp. 1–15 Search PubMed.
- N. Pfenning, in The photosynthetic bacteria, ed. R. K. Clayton and W. R. Sistrom, Plenum Press, New York, 1978, pp. 3–18 Search PubMed.
- M. T. Madigan, Photosynth. Res., 2003, 76, 157–171 CrossRef CAS PubMed.
- H. Scheer, in Light-harvesting antennas in photosynthesis, ed. B. R. Green and W. W. Parson, Kluwer Academic Publishers, 2003, pp. 29–81 Search PubMed.
- J. Fiedor, L. Fiedor, R. Haessner and H. Scheer, Biochim. Biophys. Acta, 2005, 1709, 1–4 CrossRef CAS PubMed.
- J. T. Landrum, Carotenoids: physical, chemical, and biological functions and properties, CRC Press, Boca Raton, FL, USA, 2010 Search PubMed.
- J. Fiedor, A. Sulikowska, A. Orzechowska, L. Fiedor and K. Burda, Acta Biochim. Pol., 2012, 59, 61–64 CAS.
- R. J. Cogdell and A. W. Roszak, Nature, 2014, 508, 196–197 CrossRef CAS PubMed.
- A. Vermeglio and P. Joliot, Trends Microbiol., 1999, 7, 435–440 CrossRef CAS PubMed.
- N. Basak and D. Das, World J. Microbiol. Biotechnol., 2007, 23, 31–42 CrossRef CAS.
- K. Sasaki, M. Watanabe, T. Tanaka and T. Tanaka, Appl. Microbiol. Biotechnol., 2002, 58, 23–29 CrossRef CAS PubMed.
- H. W. Yen and T. Y. Shih, Bioprocess Biosyst. Eng., 2009, 32, 711–716 CrossRef CAS PubMed.
- K. Sangkharak and P. Prasertsan, J. Biotechnol., 2007, 132, 331–340 CrossRef CAS PubMed.
- K. Sasaki, H. Morikawa, T. Kisibe, K. Takeno, A. Mikami, T. Harada and M. Ohta, Adv. Biosci. Biotechnol., 2013, 4, 6–13 CrossRef CAS.
- G. Cohen-Bazire, W. R. Sistrom and R. Y. Stanier, J. Cell. Comp. Physiol., 1957, 49, 25–58 CrossRef CAS.
- A. Orzechowska, M. Lipińska, J. Fiedor, A. Chumakov, M. Zając, T. Ślęzak, K. Matlak, K. Strzałka, J. Korecki, L. Fiedor and K. Burda, Biochim. Biophys. Acta, 2010, 1797, 1696–1704 CrossRef CAS PubMed.
- B. Samyn, J. Fitch, T. E. Meyer, M. A. Cusanovich and J. J. Beeumen van, Biochim. Biophys. Acta, 1998, 1384, 345–355 CrossRef CAS.
- U. K. Laemmli, Nature, 1970, 227, 680–685 CrossRef CAS PubMed.
- B. H. Davies and H.-P. Köst, in CRC Handbook of Chromatography, Plant Pigments, Fat-soluble Pigments, ed. H.-P. Köst, CRC Press, Inc., Boca Raton, FL, USA, 1988, vol. 1, pp. 3–185 Search PubMed.
- S. C. Straley, W. W. Parson, D. C. Mauzerall and R. K. Clayton, Biochim. Biophys. Acta, 1973, 305, 597–609 CrossRef CAS.
- Cytochrome c. Biological aspects, ed. G. W. Pettigrew and G. R. Moore, Springer-Verlag, Berlin, 1987 Search PubMed.
- S. Scheuring, in Biophysical Techniques in Photosynthesis, ed. T. J. Aartsma and J. Matysik, Springer, Dordrecht, 2008, vol. II, pp. 1–11 Search PubMed.
- P. M. Slovak, G. H. Wadhams and J. P. Armitage, J. Bacteriol., 2005, 187, 54–64 CrossRef CAS PubMed.
- D. E. Chandler, J. Hsin, C. B. Harrison, J. Gumbart and K. Schulten, Biophys. J., 2008, 95, 2822–2836 CrossRef CAS PubMed.
- S. Scheuring, R. Nevo, L.-N. Liu, S. Mangenot, D. Charuvi, T. Boudier, V. Prima, P. Hubert, J. N. Sturgis and Z. Reich, Biochim. Biophys. Acta, 2014, 1837, 1263–1270 CrossRef CAS PubMed.
- M. Y. Okamura, R. Y. Stanier and G. Feher, Biochemistry, 1974, 13, 1394–1403 CrossRef CAS PubMed.
- M. R. Jones, Biochem. Soc. Trans., 2009, 37, 400–407 CrossRef CAS PubMed.
- H. L. Axelrod and M. Y. Okamura, Photosynth. Res., 2005, 85, 101–114 CrossRef CAS PubMed.
- W. P. Laratta, M. J. Nanaszko and J. P. Shapleigh, Microbiology, 2006, 152, 1479–1488 CrossRef CAS PubMed.
- L. A. Currie, Anal. Chem., 1968, 40, 586–593 CrossRef CAS.
- F. Borsetti, P. L. Martelli, R. Casadio and D. Zannoni, in The purple phototrophic bacteria, ed. C. N. Hunter, F. Daldal, M. C. Thurnauer and J. T. Beatty, Springer, Dordrecht, 2009, pp. 655–689 Search PubMed.
- S. S. Merchant, Plant Physiol., 2010, 154, 512–515 CrossRef CAS PubMed.
- A. Hałas, A. Orzechowska, V. Derrien, A. I. Chumakov, P. Sebban, J. Fiedor, M. Lipińska, M. Zając, T. Ślęzak, K. Strzałka, K. Matlak, J. Korecki, L. Fiedor and K. Burda, Biochim. Biophys. Acta, 2012, 1817, 2095–2102 CrossRef PubMed.
- I. Baxter, Curr. Opin. Plant Biol., 2009, 12, 381–386 CrossRef CAS PubMed.
- R. Haensch and R. R. Mendel, Curr. Opin. Plant Biol., 2009, 12, 259–266 CrossRef CAS PubMed.
- H. Ishikita and E.-W. Knapp, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 16215–16220 CrossRef CAS PubMed.
- M. Lipińska, A. Orzechowska, J. Fiedor, A. Chumakov, T. Ślęzak, M. Zając, K. Matlak, J. Korecki, A. Hałas, K. Strzałka, L. Fiedor and K. Burda, J. Phys.: Conf. Ser., 2010, 217, 012021 CrossRef.
- R. J. Kassner and M. D. Kamen, Biochim. Biophys. Acta, 1968, 153, 270–278 CrossRef CAS.
- M. A. Rouf, J. Bacteriol., 1964, 88, 1545–1549 CAS.
- P. Scherer, H. Lippert and G. Wolff, Biol. Trace Elem. Res., 1983, 5, 149–163 CrossRef CAS PubMed.
- P. Peltola, C. Brun, M. Astrom and O. Tomilina, Chem. Geol., 2008, 257, 92–100 CrossRef CAS.
- C. Reimann, F. Koller, B. Frengstad, G. Kashulina, H. Niskavaara and P. Englmaier, Sci. Total Environ., 2001, 278, 87–112 CrossRef CAS PubMed.
- S. A. Watmough and P. J. Dillon, Nature, 2003, 423, 823–824 CrossRef CAS PubMed.
Footnote |
| † Work performed in partial fulfillment of the M.Sc. degree. |
| This journal is © The Royal Society of Chemistry 2016 |