Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Low carbon fuels and commodity chemicals from waste gases – systematic approach to understand energy metabolism in a model acetogen†
Esteban
Marcellin‡
a,
James B.
Behrendorff‡
b,
Shilpa
Nagaraju
b,
Sashini
DeTissera
b,
Simon
Segovia
b,
Robin W.
Palfreyman
a,
James
Daniell
bc,
Cuauhtemoc
Licona-Cassani
a,
Lake-ee
Quek
a,
Robert
Speight
ad,
Mark P.
Hodson
a,
Sean D.
Simpson
b,
Wayne P.
Mitchell
b,
Michael
Köpke
 *b and
Lars K.
Nielsen
a
*b and
Lars K.
Nielsen
a
aAustralian Institute for Bioengineering and Nanotechnology (AIBN), The University of Queensland, Brisbane, QLD 4072, Australia
bLanzaTech Inc., Skokie, IL 60077, USA. E-mail: michael.koepke@lanzatech.com
cSchool of Biological Sciences, University of Auckland, Auckland, New Zealand
dQueensland University of Technology (QUT), Brisbane, QLD 4001, Australia
First published on 6th January 2016
Abstract
Gas fermentation using acetogenic bacteria offers a promising route for the sustainable production of low carbon fuels and commodity chemicals from abundant, inexpensive C1 feedstocks including industrial waste gases, syngas, reformed methane or methanol. Clostridium autoethanogenum is a model gas fermenting acetogen that produces fuel ethanol and 2,3-butanediol, a precursor for nylon and rubber. Acetogens have already been used in large scale industrial fermentations, they are ubiquitous and known to play a prominent role in the global carbon cycle. Still, they are considered to live on the thermodynamic edge of life and potential energy constraints when growing on C1 gases pose a major challange for the commercial production of fuels and chemicals. We have developed a systematic platform to investigate acetogenic energy metabolism, exemplified here by experiments contrasting heterotrophic and autotrophic metabolism. The platform is built from complete omics technologies, augmented with genetic tools and complemented by a manually curated genome-scale mathematical model. Together the tools enable the design and development of new, energy efficient pathways and strains for the production of chemicals and advanced fuels via C1 gas fermentation. As a proof-of-platform, we investigated heterotrophic growth on fructose versus autotrophic growth on gas that demonstrate the role of the Rnf complex and Nfn complex in maintaining growth using the Wood–Ljungdahl pathway. Pyruvate carboxykinase was found to control the rate-limiting step of gluconeogenesis and a new specialized glyceraldehyde-3-phosphate dehydrogenase was identified that potentially enhances anabolic capacity by reducing the amount of ATP consumed by gluconeogenesis. The results have been confirmed by the construction of mutant strains.
Introduction
Approximately 10% of the world's energy demand and commodity chemicals are currently produced from renewable feedstocks, primarily using farmed sugars. However, greater volumes of non-food resources need to be accessed in order to address mounting environmental concerns and meet climate targets.1–3 Gas fermentation offers a route to use a wide range of readily available, low-cost C1 feedstocks such as industrial waste gases (e.g. from power plants and steel mills), syngas (e.g. from agricultural waste, industrial waste or municipal solid waste), reformed methane (e.g. from biogas) or methanol into chemicals and fuels.4,5 Believed to be one of the first biochemical pathways to emerge on earth,6 the Wood–Ljungdahl (WL) pathway enables acetogenic clostridia to fix C1 gases such as carbon monoxide (CO) and the greenhouse gas (GHG) carbon dioxide (CO2) into acetyl-CoA.7,8Clostridium autoethanogenum, in particular, offers a robust and flexible platform for fermentation of gases and has been deployed at industrial scale.4,9 Gas fermentation of C. autoethanogenum resolves refractiveness of the Fischer-Tropsch processes, as it offers higher selectivity towards a target product, has high tolerance towards gas contaminants, and is economically viable even with small volume gas streams.9–11C. autoethanogenum natively produces acetate, ethanol, 2,3-butanediol (2,3-BDO) and lactate.12–14 Ethanol is an established fuel molecule, while the four-carbon molecule 2,3-BDO can be converted to 1,3-butadiene, a precursor of rubber and nylon.15 Recently, it has been shown that the product specificity can be adjusted through fermentation optimisation and acetic acid formation can be avoided altogether.16 If energetic impediments can be overcome, synthetic biology promises to enhance the product spectrum of C. autoethanogenum and other acetogens.5
Acetogenic bacteria are widespread in nature and play a major role in the global carbon cycle. They are responsible for fixing about 20% of CO2 on earth and accounting for a minimum of 1012 kg of acetate production per year.8,17 Nevertheless, they are considered to live on the thermodynamic edge of life.18 The biochemical mechanisms for building the WL pathway intermediates are well understood;7,8 for example the coupling sites for most of the enzymes involved have been recently identified,18–20 with the exception of the methylene-tetrahydrofolate reductase.18,19,21 Yet, a globally coherent picture of energy generation and conservation and its regulation remains elusive. In general, energetic constraints are considered as major challenge for developing new products from gas fermentation.22 Conversion of CO2 to acetate provides no net ATP gain. Clearly, additional energy must be therefore generated from differences between major redox couples (Fdox/Fdred–2, NADP+/NADPH and NAD+/NADH). Carbon monoxide dehydrogenase (CODH), the recently identified electron-bifurcating enzymes,18,19,23 and the membrane-bound Rnf complex in combination with the F1Fo ATP synthase (ATPase)19,24–27 are known to play a central role. However, the exact mechanisms and their interconnection is poorly understood.
In this study, we provide the first systemic description of the metabolism of C. autoethanogenum at the transcriptional, translational and metabolome level, contextualized by a mathematical genome-scale metabolic model. This study confirmed the role of the Rnf complex that showed dramatic regulatory changes between autotrophic and heterotrophic conditions and also led to the identification of a novel glyceraldehyde-3-phosphate dehydrogenase (GAPDH) that potentially enhances anabolic capacity by reducing the amount of ATP consumed by gluconeogenesis. Furthermore, we demonstrate the value of this model-platform to direct genome modifications aimed at understanding specific aspects of C. autoethanogenum metabolism.
Results
Bacterial strains and culturing conditions
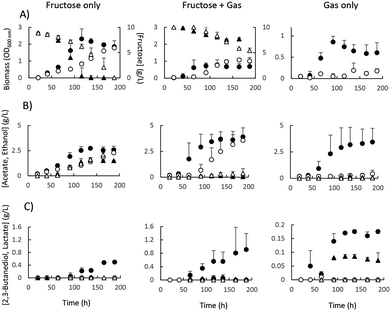
Firstly we compared the C. autoethanogenum metabolic response to energy derived from sugar (heterotrophy) and CO, CO2 and H2 gas (autotrophy). Anaerobic batch fermentations in bottles were conducted in a medium containing either fructose or a defined gas mixture composed of 45% CO, 20% CO2 and 2% H2 that reflects a basic oxygen furnace (BOF) gas stream typically found in the steel-making process. During heterotrophic growth, cells rely mainly on the Embden–Meyerhof–Parnas (EMP) glycolysis pathway, whereas under autotrophic conditions the WL pathway is exclusively used. Growth was 35% faster on fructose (μ = 0.077 ± 0.07 h−1) than on gas (μ = 0.057 ± 0.04 h−1). Acetate and ethanol were the main fermentation products alongside traces of 2,3-BDO (Fig. SI 1†).Comparative genomics
C. autoethanogenum is a close relative to Clostridium ljungdahlii,25,28,29 but several studies highlight divergent phenotypes including more efficient ethanol and 2,3-BDO production in C. autoethanogenum.13,30,31 A comparative genomic alignment revealed the two species to be remarkably similar meaning that these disparities are unexplained by differences in DNA sequence and architecture (Fig. SI 2†). Only 217 homologs (∼5%) were not found in one of the two species (Fig. SI 2†). Moreover, the majority of the proteins unique to C. autoethanogenum are annotated as hypothetical proteins or CRISPR.28 Surprisingly, despite genomic similarities, the two strains behave very differently at the transcriptional level as discussed below.Genome-scale model reconstruction and flux simulations
To help understand metabolic differences a genome-scale metabolic model (GEM), based on the recently closed assembly of the C. autoethanogenum genome,29,32 was generated using KBase and ModelSEED,33 and compared to the published model for C. ljungdahlii.34 The model was curated against existing literature and biochemical data. Gaps in the WL pathway were filled and key reactions were balanced and corrected for cofactor usage and the directionality of several reactions was defined to avoid mathematically unbounded ATP generation and free interconversion of redox cofactors.35 The model contains 1002 reactions and 1075 metabolites, represented by 805 unique genes (dataset Tables SI 7 and SI 8†).Growth and by-product formation on fructose or gas were simulated with the GEM using flux balance analysis36,37 (dataset Table SI 7†). During heterotrophic growth, acetate was the preferred by-product, creating a redox surplus that was used to refix CO2 into additional biomass (43 to 48 mgDW per mmol fructose), in contrast to autotrophic growth where acetate was further reduced to ethanol. The Rnf complex (described below), was used only for proton balancing, and not for ATP production. Overall, for heterotrophic growth, the system relied on substrate-level phosphorylation (SLP) to produce ATP, with reduced ferredoxin mainly used to recover CO2 as well as for NADPH production via the Nfn complex. Nfn catalyzes the reversible synthesis of NADPH from NADH and reduced ferredoxin.19,20,38 The addition of an aldehyde dehydrogenase reaction increased biomass yield to 54 mgDWmmol−1 fructose because it supplants the aldehyde:ferredoxin oxidoreductase (AOR) reaction, diverting reduced ferredoxin into the Rnf complex to produce extra ATP.20,39,40
During autotrophic growth, CO was the preferred substrate over CO2 (plus H2) (5.6 vs. 2.8 mgDW C-mol−1)20 due to more reduced ferredoxin being available for the Rnf complex through CODH and the electron-bifurcating Hyt hydrogenase. As a result, more carbon is shifted into products rather than biomass with ethanol being the preferred product. Ethanol yield on CO and CO2 (plus H2) was 79 and 90% C-mol respectively. While NADPH generation during heterotrophic growth predominantly relies on the Nfn complex, the model simulations predict that the electron-bifurcating Hyt hydrogenase is the preferred enzyme for NADPH production during autotrophic growth when H2 is available. During growth on pure CO or when oxidation of H2 is limited the model predictions suggest that Nfn plays an ancillary role, providing the necessary NADPH. Accordingly, in strains where the gene encoding the Nfn complex (CAETHG_1580) was disrupted (Fig. SI 3†), growth was observed on BOF gas that is rich in CO but limited in H2 but under conditions of high hydrogen the maximum biomass was less than 0.1. Similarly, hardly any growth occurred during growth on CO2/H2. ATP synthase produced over half of the total ATP, the rest was produced by the acetate kinase. Due to ATP limitations, 2,3-BDO can be produced at a higher yield if it is produced concomitantly with other by-products. Importantly, we observed a reduction in the overall biomass yield when we altered the by-products spectrum of the fermentation, thus demonstrating an intricate link between ATP generation, redox balance and intracellular resource apportionment.
Metabolomics
To validate our model predictions, energy and redox carriers were quantified, with several extraction protocols tested to achieve rapid quenching and efficient extraction.41 Bio-triplicated samples were compared by measuring 55 intracellular metabolites from three fermentations under both autotrophic and heterotrophic growth conditions (Fig. SI 4†), revealing minimal differences for the majority of compounds during both conditions despite lower autotrophic energy availability as evidenced by the slower growth rate. Energy and redox metabolite concentration displayed no significant differences. Except for NAD+ (higher when cells grew on gas) no changes were observed for redox carriers, or ATP (Fig. 2). The NADP+/NADPH ratio was also conserved. The obvious exceptions were for glycolytic intermediates which were more abundant when cells grew on fructose (Fig. SI 5†).The metabolomics also revealed presence of overflow products of biotechnological interest like lactate,13 but also succinate (from the incomplete TCA cycle) which has not been reported before (Fig. SI 4 and 5†).
Proteomics
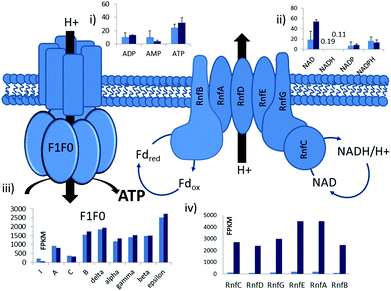
Protein synthesis is the most energy-intensive process in prokaryotic cells, consuming ∼50% of ATP used for cellular proliferation.42,43 iTRAQ was used to quantify proteomic differences, which were significant between autotrophic and heterotrophic conditions. 540 proteins were detected (>2 peptides 95% confidence), and 84 (15.6%) were significantly up or down translated (p < 0.05) (dataset Tables SI 4 and SI 5†).Conspicuous changes were observed for proteins belonging to the Rnf complex, consistent with the prominent role of Rnf in model simulations. Three subunits, RnfG, RnfD and RnfC, were highly up-regulated under autotrophic conditions with a log2 fold change of ∼4.5 (p < 0.009 for all three genes) (Fig. 2). The Rnf complex is a membrane-associated flavin-based electron bifurcation complex for energy conservation which in C. autoethanogenum couples the oxidation of reduced ferredoxin to the reduction of NAD(P)+, pumping a proton out of the cell to establish a transmembrane electrochemical gradient.26,27 Resultant proton motive force is harnessed via the F1F0 ATPase to drive ATP synthesis.
Several central carbon metabolism enzymes were found to have significantly altered protein levels. Up-regulated during autotrophic growth were two aldehyde:ferredoxin oxidoreductases (AOR) CAETHG_0092 and CAETHG_0102 with a log2 fold change of 11.3 and 1.05 respectively (p = 4.95 × 10−11 and p = 1.3 × 10−4) (see discussion below) and an alcohol dehydrogenase CAETHG_1841 (1.05 up, p = 1.3 × 10−4). In contrast during heterotrophic growth, a different alcohol dehydrogenases CAETHG_3954 was upregulated with a log2 fold change of 9.5 (p = 7.6 × 10−6). Amongst the most upregulated during heterotrophic growth were the fructose phosphotransferase system (PTS) CAETHG_0142 (20.0 fold change, p = 1.6 × 10−3), a uroporphyrinogen decarboxylase (14.1 fold change, p = 6.6 × 10−3). Gatekeepers phosphoenolpyruvate carboxykinase (PCK) and the phosphopentomutase were both upregulated during heterotrophic growth with a log2 fold change of 5 and 3.4 respectively (p = 6.1 × 10−6 and 3.3 × 10−5), as was one glyceraldehyde-3-phosphate (G3P) dehydrogenase (GAPDH) CAETHG_1760 (1.05 up p = 2.8 × 10−7). D-3-Phosphoglycerate dehydrogenase on the other hand was upregulated during autotrophic growth (15 fold log2 change, p = 1.5 × 10−5) (Fig. SI 6†).
D-3-Phosphoglycerate dehydrogenase controls important branch points to amino acids (such as lysine and serine) and to a range of cofactors; PCK converts oxaloacetate to PEP using a molecule of ATP and controls the rate-limiting step of gluconeogenesis in E. coli.44 The role of PCK and GAPDH in gluconeogenesis in C. autoethanogenum was investigated by constructing disruption mutants of these genes using the ClosTron methodology.45 In the genome of C. autoethanogenum gene CAETHG_2721 is annotated as an ATP-dependent PCK. A mutant with disrupted CAETHG_2721 (Fig. SI 3†) showed impaired growth when cultured on gas only. The growth of CAETHG_2721 was restored in the presence of both gas and fructose, implying the role of PCK in gluconeogenesis and supply of PEP for other cellular activities. The role of GAPDH in energy metabolism is further discussed below.
Transcriptomics
RNA-sequencing was performed in bio-triplicate for both growth conditions (Fig. SI 7†). A total of 145 million reads were mapped to the genome, achieving a 269-fold coverage. Approximately 50% of the genes showed significant differences at the transcriptional level between the two conditions, displaying a dynamic transcriptional range from 5.4 to −6.4 log2 fold change (dataset Table SI 3 and Fig. SI 6†).Expression of genes involved in energy metabolism showed the greatest difference between the two conditions tested (Fig. 1 and 2). All six genes from the Rnf complex displayed a significant up-regulation under autotrophic growth conditions (log2 ∼ 4.5 fold, p < 0.00005, q < 0.00153) with an average fragment per kilobase of transcript (FPKM) expression of 2285 FPKM compared with 105 FPKM during heterotrophic growth. In the closely related acetogen C. ljungdahlii, the Rnf complex has been shown to be essential for autotrophic growth.24 However, two independent transcriptome studies in C. ljungdahlii did not show significant differences in expression of any of the Rnf genes under similar conditions.34,46 In contrast in this work C. autoethanogenum displayed significant autotrophic-specific Rnf expression both at RNA and protein expression level (Fig. SI 6†).
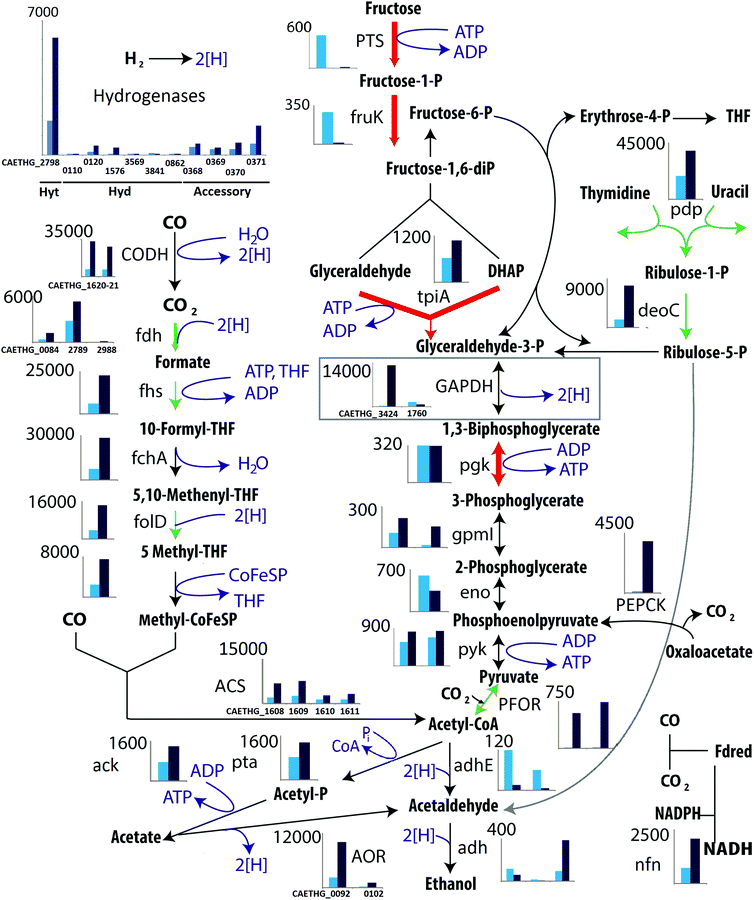 | ||
Fig. 1 Pathway of the central energy metabolism of C. autoethanogenum. The WL (left hand side), EMP (right hand side) and ethanol synthesis (bottom) pathways are shown. Clustered column charts show transcriptomics data of genes associated with each reaction. Data is displayed for autotrophic (dark blue) and heterotrophic (light blue) conditions. Y-axis represents average FPKM data from RNA-sequencing. Charts with more than one pair of bars indicate reactions for which isoenzymes are present. For key isoenzyme reactions gene numbers are given. A full list and complete data can be found in ESI Table 8.† Coloured arrows indicate enzymes with significantly changed expression found in proteomics, red (down during autotrophic growth) and green (up during autotrophic growth). The thickness of the coloured arrows is proportional to the proteomics (iTRAQ) fold change. Proteomics and transcriptomics showed differences in all genes involved in the reductive branch of the WL pathway which were significantly up-regulated under autotrophic growth as opposed to the down regulation of all glycolytic enzymes except for one of the two GAPDH enzymes (grey square). One of the two GAPDH enzymes displayed a major transcriptional increase when cells grew on gas compared to growth on fructose (FPKM ∼14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 vs. <1000 for all other glycolytic enzymes) (see Table SI 8† for details). 000 vs. <1000 for all other glycolytic enzymes) (see Table SI 8† for details). | ||
Ferredoxin oxidoreduction is coupled to the Rnf activity, and changes were also observed for genes involved in ferredoxin utilization and biosynthesis. AOR gene CAETHG_0092 was highly expressed under autotrophic, but not heterotrophic conditions, as was the operon containing three iron–sulfur ferredoxin genes (CAETHG_2796-2799).
Expression of genes involved in the WL pathway and the Rnf complex were downregulated but not repressed by growth on fructose. Cells apparently were able to use reducing equivalents generated during glycolysis to recapture CO2 produced during the pyruvate:ferredoxin oxidoreductase (PFOR) reaction and in other metabolic pathways such as 2,3-BDO production through the action of acetolactate synthase and decarboxylase (Fig. 1 and 2).13,14 The ability to fix free CO2 during heterotrophic growth confers the advantage of increased biomass and product yields.47
Up-regulation, up to 2.3 log2 fold, of all genes involved in the reductive branch of the WL pathway was observed under autotrophic growth conditions (Fig. 1). The bifunctional enzyme CO dehydrogenase/acetyl-CoA synthase (CODH/ACS) showed the largest differential expression highlighting the importance of this enzyme in carbon fixation. Genes belonging to the EMP glycolytic pathway also displayed significant transcriptional changes (Fig. 1). The largest transcriptional fold change in the glycolytic enzymes observed was for one of two GAPDH genes present in C. autoethanogenum.
The roles of each GAPDH-encoding gene was investigated by separately disrupting their coding sequences using the ClosTron method.45 CAETHG_3424 was successfully disrupted, and growth of the mutant was measured on gas, fructose and a combination of both in comparison to the wild-type strain (Fig. 3). In contrast, despite four independent attempts, disruptions of CAETHG_1760 were not obtained, regardless of selection on fructose or BOF gas (or a combination thereof). CAETHG_3424 deficient strains grew on fructose (and gas plus fructose) but were unable to grow on gas only (Fig. 3 and SI 1, SI 8†). In the presence of fructose, a deletion of CAETHG_3424 reduced the growth rate without affecting final biomass yield, while reducing production of 2,3-BDO and lactate to below detection limits (Fig. 3A and B). The CAETHG_3424 deficient strain could not grow autotrophically on gas as the sole carbon and energy source (Fig. 3C).
In silico GEM knock-outs of CAETHG_3424, and particularly the experimentally unobtainable knock-out of CAETHG_1760, were simulated to examine the possibility that CAETHG_1760 being a conventional GAPDH contra CAETHG_3424, a GAPDH variant capable of converting 3-phosphoglycerate to G3P using either NAD+ or NADP+.48 Simulation of growth on fructose showed that maximum biomass yield of CAETHG_1760-deficient strain was reduced due to the loss of ATP production by phosphoglycerate kinase. CAETHG_3424 is not required for growth on fructose, but if CAETHG_3424 is not expressed during high glycolytic flux, then CAETHG_1760 disruption is lethal. This may explain why CAETHG_1760-deficient strains could not be obtained experimentally on fructose medium, but does not explain its essential role in C1 metabolism.
Simulations of the CAETHG_3424 knock-out growing on gas showed only a small reduction of maximum biomass yield to 96%. The 4% difference being due to not saving ATP to drive gluconeogenesis (for comparison 55% of ATP generated is used in the WL pathway). The resulting hypothesis is that reversible interconversion of glycolytic intermediates (G3P, 1,3-bisphosphoglycerate, 2,3-bisphosphoglycerate and 3-phosphoglycerate) may promote futile cycling that significantly depletes ATP, and CAETHG_3424's role is to modulate undesirable side effects of gluconeogenesis by supporting such turnover without an ATP cost.
Discussion
The potential spectrum of products that can be produced by acetogens is ultimately dictated by energetics. Energy metabolism in acetogens is complex and only partially understood despite their important role in the global carbon cycle and their biotechnological potential. The tight coupling between redox and ATP is essential under autotrophic conditions, but the resulting constraints imposed on energy yield are not well understood. The system level study described herein has revealed that the energy yield (ATP, and redox state) is indeed unaffected between heterotrophic and autotrophic growth. Furthermore, succinate was identified as a new overflow product of acetogenic metabolism.The study described here provides measurements of the concentrations of mRNA and the most abundant proteins under two given conditions (Fig. 1 and SI 6†). As described in other bacteria, the intracellular concentrations of mRNAs correlates with the protein abundances, but not strongly. The data shows a squared Pearson correlation coefficient of 0.56, which is in accordance with the ∼0.4 correlation reported in other systems.49 The low correlation has been explained by the lower mRNA production rate compared to the protein production rate; on average, two copies of a given mRNA are produced per hour, whereas dozens of copies of the corresponding protein per mRNA are produced. Likewise, recent studies suggest a perhaps undervalued role for post-transcriptional, translational and degradation regulation in bacterial metabolism, contributing at least as much as transcription itself which needs to be further understood.
Our model predicted, and experimental measurements with genetic mutants confirmed, that fructose grown cells balanced reducing equivalents via the WL pathway, allowing CO2 reassimilation and redirecting the large pool of fructose-6-phosphate into the pentose phosphate pathway. In contrast, the redox requirements of cells grown on gas were balanced through the action of hydrogenases, the electron-bifurcating transhydrogenase Nfn complex20,38 and alternative pathways such as ethanol formation. Consequently, a disruption mutant of the Nfn gene struggled to grow under autotrophic conditions with high hydrogen. Our analysis showed over-expression (both on transcriptional as well as protein level) of an AOR. The AOR39 was previously suspected to play a major role in ethanol formation by reducing acetate to acetaldehyde.20,25 Under autotrophic conditions, this reaction can be coupled with CO oxidation via the CODH that yields reduced ferredoxin required for the AOR reaction. A previously described stoichiometric model suggests that, under energy-limited conditions, AOR provides a significant advantage.40 A similar enzyme has recently also been demonstrated to play a major role in ethanol formation in Pyrococcus furiosus.50
C. autoethanogenum possess two types of GAPDH enzymes possibly providing a thermodynamic advantage through the gain of an additional ATP as in archaea where a glycolytic GAPDH variant controls the interconversion of G3P and 3-phosphoglycerate.48 Flux balance analysis showed that although the net ATP gain is small, cofactor specificity plays a key role in controlling the cells metabolism. The coexistence of a GAPDH specialized in glycolysis, and another one in gluconeogenesis allows acetogens to preserve the high energetic efficiency of EMP glycolysis through an otherwise less energy efficient pathway. Supporting evidence comes from the fact that close homologues of CAETHG_3424 are only found in other acetogens C. ljungdahlii,25,51C. carboxidivorans,52C. drakei52 and C. difficile53,54 as well as C. kluyveri55 which grows on ethanol and therefore also relies heavily on gluconeogenesis. PCK was significantly upregulated during autotrophic growth, controlling the rate-limiting step of gluconeogenesis and could be a major source for PEP, a high energy phosphate group donor in many anabolic pathways.
Conclusion
Gas fermentation offers a promising sustainable route to a range of low carbon fuels and chemicals from a range of C1 feedstocks. To tap into the full potential of gas fermentation and expand the product spectrum, it is important to understand acetogenic organisms on a systems level. Systems biology approaches have been widely adopted for the study and optimization of model organisms such as yeast and E. coli. In contrast, system-wide analysis of anaerobic C1 gas fermentation have not been performed prior to this study. There are only a few transcriptomics datasets describing acetogenic gas fermentation, and even fewer comprehensive frameworks analysing and integrating such data. Herein we have demonstrated that a systems biology approach may be used to understand energy metabolism and expand the product spectrum in acetogens by guiding design of new strains.Energy metabolism in acetogens is complex and only partially understood. Nonetheless, it dictates the potential product spectrum of these organisms and limits our ability to engineer strains for production of commodity chemicals and advanced drop in fuels from C1 substrates. Through the development of the first integrated systems biology toolbox for an acetogen, we revealed an otherwise hidden layer of complexity in the C. autoethanogenum metabolism; not only can cells auto-regulate their metabolism through specialized glycolytic/gluconeogenic enzymes but cells can also achieve consistent ATP levels during autotrophic and heterotrophic growth. Our system level study revealed that the energy yield (ATP, and redox state ratios) is essentially unchanged between heterotrophic and autotrophic growth, despite the stark disparity in supply-and-demand between autotrophic and heterotrophic growth, the former condition starting with a more oxidized substrate (cf. CO versus sugar). This consistency is achieved through a dramatic up-regulation of the Rnf, Nfn and PCK genes which play a key role in the cells’ ability to balance requirements for reducing equivalents and for ATP and PEP energy supplies. In addition a new GAPDH was identified. These hypotheses were confirmed with constructed mutant strains.
The data presented here, contributes to our understanding of acetogenic energy metabolism. With the provided model and protocols, this information can be used to simulate and develop new strategies to expand the product spectrum of what can be produced via gas fermentation.
Experimental
Strain and culture conditions
C. autoethanogenum DSM 10061 was obtained from DSMZ. Media and cultivation were the same as described earlier.13 For heterotrophic growth conditions, the carbon source was 5 g fructose per L. For autotrophic conditions the gas composition was: carbon monoxide (35%), carbon dioxide (10%), hydrogen (2%) and the remainder was nitrogen. Growth studies were carried our in small scale batch fermentations and samples for omics studies were collected during mid-log phase. A detailed description is found in the ESI.†Proteomics
Proteins were extracted from cell pellets sampled in triplicate as described elsewhere.56 Quantitation was performed using iTRAQ as described elsewhere.56 All statistical analysis was done using Protein Pilot 4.5.Transcriptomics
RNA extraction was performed as described elsewhere.57 Reads were aligned to the genome using Bowtie 2,58 with two mismatches allowed per read alignment. Before mapping to the genome the 100 base pair reads were trimmed to 75 base pair to avoid reading errors. Transcript abundance was estimated using FPKM using upper-quantile normalization, Cuffdif59 was used to estimate differentially expressed transcripts.Metabolomics
Intracellular metabolites were extracted from 5 mL of culture after removing media by centrifugation at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700g for 2 minutes at −20 °C as described elsewhere (41).
700g for 2 minutes at −20 °C as described elsewhere (41).
Genome-scale model assembly
The genome-scale metabolic reconstruction was adapted from the method described earlier.60 The core of the genome-scale model was reconstructed using the SEED model annotation pipeline.33 The model was manually gap filled in Excel for ease of annotation and commenting. Constraint-based reconstruction and analysis was performed using the COBRA toolbox.61,62 The XML model can be downloaded from the ESI.†Mutant strain construction and analysis
ClosTron constructs were designed using the web-based design tool (http://clostron.com/clostron2.php) and the intron targeting plasmids were synthesized by DNA 2.0 (USA). Plasmids were transferred into C. autoethanogenum by bacterial conjugation according to previously-published methods.20 A list of primers and intron targeting sequences are available in (Tables SI 1 and SI 2†). The desired genetic disruptions were confirmed by colony PCR and Sanger sequencing across the targeted locus (Macrogen, USA).Author contributions
E. M, M. K., J. B. B., R. S., W. P. M, S. D. S., and L. K. N. designed the experiments. S. S., S. D., and J. B. B. performed all fermentation and extractions. E. M., L. Q. and J. D. constructed the genome-scale model and performed simulations, E. M. and C. L. C. performed proteomics, transcriptomics and metabolomics analyses. R. P. performed alignment and transcript assembly. M. P. H. analysed the metabolomics samples. J. B. B. and S. N. created and characterized the mutant strains. E. M., M. K., J. B. B. and L. K. N. wrote the manuscript.Conflict of interest
The authors declare no competing financial interests. LanzaTech has interest in commercial gas fermentation with C. autoethanogenum.Acknowledgements
We thank Amanda Nouwens for her valuable help with mass spectrometry. We thank Helena Tellier for help with the GAPDH and Kaspar Valgepea for reviewing the manuscript and the following funding sources: the University of Queensland Collaboration and Industry Engagement Fund (UQ CIEF), ARC LP140100213 and The Queensland Government for the Accelerate Fellowship to E. M. We also thank the UQ Dow Centre for Sustainable Engineering Innovation for funding to E. M. We thank the following investors in LanzaTech's technology: Sir Stephen Tindall, Khosla Ventures, Qiming Venture Partners, Softbank China, the Malaysian Life Sciences Capital Fund, Mitsui, Primetals, CICC Growth Capital Fund I, L. P. and the New Zealand Superannuation Fund.References
- M. Kircher, Ind. Biotechnol., 2014, 10, 11–18 CrossRef.
- M. Kircher, Curr. Opin. Chem. Biol., 2015, 29, 26–31 CrossRef CAS PubMed.
- P. Friedlingstein, R. M. Andrew, J. Rogelj, G. P. Peters, J. G. Canadell, R. Knutti, G. Luderer, M. R. Raupach, M. Schaeffer, D. P. van Vuuren and C. Le Quéré, Nat. Geosci., 2014, 7, 709–715 CrossRef CAS.
- M. Köpke, C. Mihalcea, J. C. Bromley and S. D. Simpson, Curr. Opin. Biotechnol., 2011, 22, 320–325 CrossRef PubMed.
- H. Latif, A. a. Zeidan, A. T. Nielsen and K. Zengler, Curr. Opin. Biotechnol., 2014, 27, 79–87 CrossRef CAS PubMed.
- M. J. Russell and W. Martin, Trends Biochem. Sci., 2004, 29, 358–363 CrossRef CAS PubMed.
- H. G. Wood, FASEB J., 1991, 10, 156–163 Search PubMed.
- H. L. Drake, K. Küsel and C. Matthies, in The Prokaryotes, ed. M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer and E. Stackebrandt, Springer, New York, NY, 3rd edn, 2006, pp. 354–420 Search PubMed.
- J. Daniell, M. Köpke and S. Simpson, Energies, 2012, 5, 5372–5417 CrossRef CAS.
- D. W. Griffin and M. A. Schultz, Environ. Prog. Sustainable Energy, 2012, 31, 219–224 CrossRef CAS.
- J. L. Vega, K. T. Klasson, D. E. Kimmel, E. C. Clausen and J. L. Gaddy, Appl. Biochem. Biotechnol., 1990, 24–25, 329–340 CrossRef CAS.
- J. Abrini, H. Naveau and E.-J. Nyns, Arch. Microbiol., 1994, 161, 345–351 CrossRef CAS.
- M. Köpke, C. Mihalcea, F. Liew, J. H. Tizard, M. S. Ali, J. J. Conolly, B. Al-Sinawi and S. D. Simpson, Appl. Environ. Microbiol., 2011, 77, 5467–5475 CrossRef PubMed.
- M. Köpke, M. L. Gerth, D. J. Maddock, A. P. Mueller, F. Liew, S. D. Simpson and W. M. Patrick, Appl. Environ. Microbiol., 2014, 80, 3394–3403 CrossRef PubMed.
- M. Köpke and A. Havill, Catal. Rev., 2014, 27, 7–12 Search PubMed.
- H. N. Abubackar, M. C. Veiga and C. Kennes, Int. J. Environ. Res. Public Health, 2015, 12, 1029–1043 CrossRef CAS PubMed.
- B. P. Tracy, S. W. Jones, A. G. Fast, D. C. Indurthi and E. T. Papoutsakis, Curr. Opin. Biotechnol., 2012, 23, 364–381 CrossRef CAS PubMed.
- K. Schuchmann and V. Müller, Nat. Rev. Microbiol., 2014, 12, 809–821 CrossRef CAS PubMed.
- S. Wang, H. Huang, J. Kahnt, A. P. Mueller, M. Köpke and R. K. Thauer, J. Bacteriol., 2013, 195, 4373–4386 CrossRef CAS PubMed.
- J. Mock, Y. Zheng, A. P. Mueller, S. Ly, L. Tran, S. Segovia, S. Nagaraju, M. Köpke, P. Dürre and R. K. Thauer, J. Bacteriol., 2015, 197, 2965–2980 CrossRef CAS PubMed.
- J. Mock, S. Wang, H. Huang, J. Kahnt and R. K. Thauer, J. Bacteriol., 2014, 196, 3303–3314 CrossRef PubMed.
- J. Bertsch and V. Müller, Biotechnol. Bioeng., 2015, 1–12 Search PubMed.
- W. Buckel and R. K. Thauer, Biochim. Biophys. Acta, 2013, 1827, 94–113 CrossRef CAS PubMed.
- P. Tremblay, T. Zhang, S. A. Dar, C. Leang and D. R. Lovley, mBio, 2012, 4 Search PubMed.
- M. Köpke, C. Held, S. Hujer, H. Liesegang, A. Wiezer, A. Wollherr, A. Ehrenreich, W. Liebl, G. Gottschalk and P. Dürre, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 13087–13092 CrossRef PubMed.
- V. Müller, F. Imkamp, E. Biegel, S. Schmidt and S. Dilling, Ann. N. Y. Acad. Sci., 2008, 1125, 137–146 CrossRef PubMed.
- E. Biegel and V. Müller, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 18138–18142 CrossRef CAS PubMed.
- S. D. Brown, S. Nagaraju, S. Utturkar, S. De Tissera, S. Segovia, W. Mitchell, M. L. Land, A. Dassanayake and M. Köpke, Biotechnol. Biofuels, 2014, 7, 40 CrossRef PubMed.
- S. M. Utturkar, D. M. Klingeman, J. M. Bruno-Barcena, M. S. Chinn, A. M. Grunden, M. Köpke and S. D. Brown, Sci. Data, 2015, 2, 150014 CrossRef CAS PubMed.
- J. L. Cotter, M. S. Chinn and A. M. Grunden, Bioprocess Biosyst. Eng., 2009, 32, 369–380 CrossRef CAS PubMed.
- M. E. Martin, H. Richter, S. Saha and L. T. Angenent, Biotechnol. Bioeng., 2015 DOI:10.1002/bit.25827.
- S. D. Brown, S. M. Utturkar, T. S. Magnuson, A. E. Ray, F. L. Poole, W. A. Lancaster, M. P. Thorgersen, M. W. W. Adams and D. A. Elias, Genome Announc., 2014, 2, e00881–e00814 Search PubMed.
- C. S. Henry, M. DeJongh, A. A. Best, P. M. Frybarger, B. Linsay and R. L. Stevens, Nat. Biotechnol., 2010, 28, 977–982 CrossRef CAS PubMed.
- H. Nagarajan, M. Sahin, J. Nogales, H. Latif, D. R. Lovley, A. Ebrahim and K. Zengler, Microb. Cell Fact., 2013, 12, 118 CrossRef PubMed.
- L.-E. Quek, S. Dietmair, M. Hanscho, V. S. Martínez, N. Borth and L. K. Nielsen, J. Biotechnol., 2014, 184, 172–178 CrossRef CAS PubMed.
- J. D. Orth, I. Thiele and B. Ø. Palsson, Nat. Biotechnol., 2010, 28, 245–248 CrossRef CAS PubMed.
- C. H. Schilling, J. S. Edwards, D. Letscher and B. Ø. Palsson, Biotechnol. Bioeng., 71, 286–306 Search PubMed.
- S. Wang, H. Huang, J. Moll and R. K. Thauer, J. Bacteriol., 2010, 192, 5115–5123 CrossRef CAS PubMed.
- H. White and H. Simon, Arch. Microbiol., 1992, 158, 81–84 CrossRef CAS PubMed.
- A. G. Fast and E. T. Papoutsakis, Curr. Opin. Chem. Eng., 2012, 1–16 Search PubMed.
- E. Marcellin, L. K. Nielsen, P. Abeydeera and J. O. Krömer, Biotechnol. J., 2009, 4, 58–63 CrossRef CAS PubMed.
- R. A. Cox, Microbiology, 2004, 150, 1413–1426 CrossRef CAS PubMed.
- A. H. Stouthamer, Antonie van Leeuwenhoek, 1973, 39, 545–565 CrossRef CAS PubMed.
- Y. P. Chao, R. Patnaik, W. D. Roof, R. F. Young and J. C. Liao, J. Bacteriol., 1993, 175, 6939–6944 CAS.
- J. T. Heap, S. a. Kuehne, M. Ehsaan, S. T. Cartman, C. M. Cooksley, J. C. Scott and N. P. Minton, J. Microbiol. Methods, 2010, 80, 49–55 CrossRef CAS PubMed.
- Y. Tan, J. Liu, X. Chen, H. Zheng and F. Li, Mol. BioSyst., 2013, 9, 2775–2784 RSC.
- A. G. Fast, E. D. Schmidt, S. W. Jones and B. P. Tracy, Curr. Opin. Biotechnol., 2015, 33, 60–72 CrossRef CAS PubMed.
- N. A. Brunner, H. Brinkmann, B. Siebers and R. Hensel, J. Biol. Chem., 1998, 273, 6149–6156 CrossRef CAS PubMed.
- C. Vogel and E. M. Marcotte, Nat. Rev. Genet., 2012, 13, 227–232 CAS.
- M. Basen, G. J. Schut, D. M. Nguyen, G. L. Lipscomb, R. A. Benn, C. J. Prybol, B. J. Vaccaro, F. L. Poole, R. M. Kelly and M. W. W. Adams, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 17618–17623 CrossRef CAS PubMed.
- R. S. Tanner, L. M. Miller and D. Yang, Int. J. Syst. Bacteriol., 1993, 43, 232 CrossRef CAS PubMed.
- J. S.-C. Liou, D. L. Balkwill, G. R. Drake and R. S. Tanner, Int. J. Syst. Evol. Microbiol., 2005, 55, 2085–2091 CrossRef CAS PubMed.
- M. Köpke, M. Straub and P. Dürre, PLoS One, 2013, 8, e62157 Search PubMed.
- M. Sebaihia, B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeño-Tárraga, H. Wang, M. T. G. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell and J. Parkhill, Nat. Genet., 2006, 38, 779–786 CrossRef PubMed.
- H. Seedorf, W. F. Fricke, B. Veith, H. Brüggemann, H. Liesegang, A. Strittmatter, M. Miethke, W. Buckel, J. Hinderberger, F. Li, C. Hagemeier, R. K. Thauer and G. Gottschalk, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 2128–2133 CrossRef CAS PubMed.
- C. A. Orellana, E. Marcellin, B. L. Schulz, A. S. Nouwens, P. P. Gray and L. K. Nielsen, J. Proteome Res., 2015, 14, 609–618 CrossRef CAS PubMed.
- E. Marcellin, T. R. Mercer, C. Licona-Cassani, R. W. Palfreyman, M. E. Dinger, J. A. Steen, J. S. Mattick and L. K. Nielsen, BMC Genomics, 2013, 14, 15 CrossRef CAS PubMed.
- B. Langmead and S. L. Salzberg, Nat. Methods, 2012, 9, 357–359 CrossRef CAS PubMed.
- C. Trapnell, A. Roberts, L. Goff, G. Pertea, D. Kim, D. R. Kelley, H. Pimentel, S. L. Salzberg, J. L. Rinn and L. Pachter, Nat. Protoc., 2012, 7, 562–578 CrossRef CAS PubMed.
- C. G. de Oliveira Dal'Molin, L.-E. Quek, R. W. Palfreyman, S. M. Brumbley and L. K. Nielsen, Plant Physiol., 2010, 152, 579–589 CrossRef PubMed.
- S. A. Becker, A. M. Feist, M. L. Mo, G. Hannum, B. Ø. Palsson and M. J. Herrgard, Nat. Protoc., 2007, 2, 727–738 CrossRef CAS PubMed.
- J. Schellenberger, R. Que, R. M. T. Fleming, I. Thiele, J. D. Orth, A. M. Feist, D. C. Zielinski, A. Bordbar, N. E. Lewis, S. Rahmanian, J. Kang, D. R. Hyduke and B. Ø. Palsson, Nat. Protoc., 2011, 6, 1290–1307 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: In addition proteomics DATASETS can be downloaded from PRIDE. RNAseq data can be downloaded from GEO. The SBML for the genome scale model. See DOI: 10.1039/c5gc02708j |
| ‡ Equal contributors. |
| This journal is © The Royal Society of Chemistry 2016 |