Open Access Article
Open Access ArticleGenetic engineering and production of modified fatty acids by the non-conventional oleaginous yeast Trichosporon oleaginosus ATCC 20509†
Christian
Görner
,
Veronika
Redai
,
Felix
Bracharz
,
Patrick
Schrepfer
,
Daniel
Garbe
and
Thomas
Brück
*
Fachgebiet Industrielle Biokatalyse, Department Chemie, Technische Universität München, Lichtenbergstraße 4, 85748 Garching, Germany. E-mail: brueck@tum.de
First published on 25th November 2015
Abstract
The oleaginous yeast Trichosporon oleaginosus ATCC 20509 can accumulate up to 70% (w/DCW) triglycerides when cultivated on chemically diverse agricultural or food waste streams. In contrast to other lipogenic yeasts T. oleaginosus is able to efficiently convert constituents of hemicellulose and chitin hydrolysates into lipids. This study focused on establishing the genetic accessibility of T. oleaginosus aimed at manipulating lipid biosynthesis in order to generate high value lipids from waste streams. We demonstrate the first transformation protocol for T. oleaginosus based on Agrobacterium tumefaciens. Strong heterologous gene expression of a codon optimized YFP reporter protein was achieved using the constitutive promotor from the endogenous glyceraldehyde-3-phosphate dehydrogenase gene. Subsequently, we evaluated the ability of T. oleaginosus to generate non-natural fatty acid profiles by heterologous expression of several fatty acid modifying enzymes. De novo lipid generation of these recombinant strains was evaluated on diverse carbon sources. Compared to the wild type, recombinant yeast strains showed an increase of α-linolenic acid production from 2.8% to 21% with respect to the total cellular fatty acid content (TFA). Further, we designed yeast strains able to generate the non-native, polyunsaturated very long chain fatty acids eicosatrienoic (16% TFA) and eicosadienoic acid (9% TFA), respectively. Alternatively, T. oleaginosus was engineered to produce the non-native (E-10, Z-12) conjugated linoleic acid, which was generated up to 2.6% TFA. This work demonstrates, that T. oleaginosus ATCC 20509 can be used as versatile biotechnology platform to transform industrial waste streams into designed, high value fatty acids.
Introduction
Driven by demands of the food, chemical and pharmaceutical industry, production of plant and animal based lipids increased by approximately 65% in the last decade.1,2 While about 70% are still used for food purposes, the application of natural oils for biofuels, biogenic oleochemicals and bioactive substances is expanding rapidly.2–4 Despite advances in plant oil engineering, pharmaceutically relevant lipids, such as the very long chain omega-3 (ω-3) polyunsaturated fatty acids (VLC-PUFAs) eicosapentaenoic acid (EPA; 20:5 Δ5,8,11,14,17) and docosahexaenoic acid (DHA; 22:6 Δ4,7,10,13,16,19), are mainly derived from marine resources.1,5–7 Particularly, PUFA sourcing from fish and crustaceans has a significant negative impact on the marine food chain thereby increasing pressure on these delicate ecosystems.5,8Hence, development of sustainable sources for VLC-PUFAs that are economically competitive could be a chance to preserve marine eco systems. Consequently, multiple approaches focused on providing ω-3 VLC-PUFAs from plant, algae and yeast biomass.9–11 These production platforms need to be subjected to further economical optimisation to enable replacement of marine oils. At present, the utilisation of cost efficient food and agricultural waste streams for the de novo, fermentative production of ω-3 VLC-PUFAs has not been exploited.
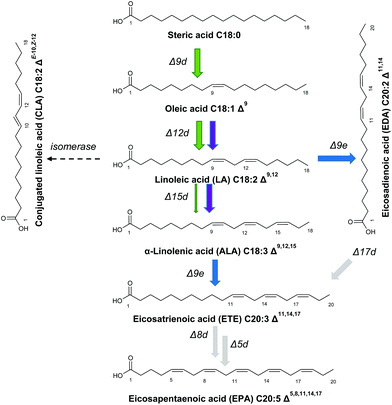
Other fatty acids with beneficial pharmacological effects comprise conjugated linoleic acids (CLA).12 CLA comprise linoleic acid (LA) isomers with conjugated double bonds (Fig. 2), which are currently produced by isomerization of biogenic linoleic acid using an energy intensive and rather unselective chemical process.13 In the absence of CLA rich natural sources, biotechnological CLA production processes represent a sustainable alternative. In this respect, the in vitro enzymatic transformation of free linoleic acid to CLA with isomerases from Clostridium sporogenes and Propionibacterium acnes has been reported.14
Recent advances in process and metabolic engineering of oleaginous microorganisms now offer a sustainable production approach for non-food lipids.15,16 Fast growth rates, high intracellular lipid content and the use of chemically complex waste biomass feedstocks designate oleaginous yeasts as potential fermentation platform organism for generation of tailor made lipids.17 However, strain specific limitations in genome engineering, limited data on biochemical mechanisms involved in lipogenesis and organism-specific substrate preferences currently prohibit industrial deployment of these technologies.18–20 To date, advanced molecular engineering of lipid biosynthesis and high yield production of designer lipids has only been realized in Yarrowia lipolytica.21 Nonetheless, Y. lipolytica is predominantly an ex novo lipid producer, which requires triglyceride or fatty acid containing growth media.17 Generally, data on de novo lipid producing yeasts is scarce. However, the recent establishment of genetic accessibility in Rhodosporidium toruloides is a milestone towards tailored, de novo lipid production in yeasts. As this yeast strain lacks the ability to convert chemically complex biogenic waste streams into lipids, its economic applicability at industrial scale remains to be demonstrated.22 Moreover, the dependence of engineered, de novo lipid production on the applied nutrient limitation and the nature of the supplied carbon source has not been elucidated.
Trichosporon oleaginosus ATCC 20509 has recently been assigned to the basidomycetous genus Trichosporon. The organism was previously referred to as Cryptococcus curvatus, Candida curvata or Apiotrichum curvatum.23 It has been extensively studied for its ability to accumulate up to 70% triglycerides as dry cell weight (DCW) resembling a cocoa butter-like fatty acid composition.17T. oleaginosus can be grown in high cell density cultures using various waste material feedstocks, such as whey permeate,24 crude glycerol,25 sweet sorghum bagasse,26 pectin-derived sugar acids,27 wheat straw hydrolysate28 and N-acetylglucosamine29 from chitin hydrolysate. Therefore, T. oleaginosus is distinct from other lipogenic yeasts in that it can utilise highly diverse carbon sources for de novo lipid production. Further, fermentation inhibitors, such as furfural, which are constituents of waste biomass hydrolysates, i.e. wheat straw hydrolysate, do not affect biomass and lipid formation in T. oleaginosus cultivation.28 Therefore, T. oleaginosus is ideal for fermentative high value lipid production on cost efficient waste biomass hydrolysates or alternative biotechnological waste streams such as crude glycerol from biodiesel production.
In the presence of excess carbon, nitrogen or phosphorus limitation in the growth medium T. oleaginosus conventionally attenuates growth and triggers intracellular lipid accumulation.18,19,30 In contrast to other oleaginous yeasts, T. oleaginosus tolerates higher nitrogen concentrations in the medium without adverse effects on de novo lipid biosynthesis. Its innate metabolic versatility designate T. oleaginosus as an ideal fermentative platform organism and a valuable candidate for genetic engineering.
Generation of tailored fatty acid profiles in T. oleaginosus required the initial development of an effective transformation and genetic engineering protocol. Previously, only the generation of methionin as well as unsaturated fatty acid auxotrophic T. oleaginosus mutants have been generated by random UV-based mutagenesis.31,32 These mutant strains were subsequently combined by intraspecific spheroplast fusion to create strains with an increased saturated fatty acid profile.33 To the best of our knowledge, no targeted transformation and heterologous expression has been published for T. oleaginosus ATCC 20509.
We have recently reported detailed genome and transcriptome data for a Trichosporon oleaginosus strain, which facilitated the development of a targeted genetic engineering approach for this organism.30 In this study we present a proof of concept that establishes T. oleaginosus as a flexible production platform for the recombinant generation of tailor made lipids derived from various monomeric sugars that are major constituents of waste biomass streams. Specifically, we addressed recombinant production of the very long chain fatty acids eicosadienoic acid (EDA, C20:3 Δ11,14) and eicosatrienoic acid (ETE, C20:3 Δ11,14,17) which are important intermediates towards the synthesis of EPA and DHA. Further, we present the first in vivo process for the specific production of E-10, Z-12 conjugated linoleic acid (CLA) utilizing the free cytosolic LA pool present in T. oleaginosus. We report on the CLA production performance of recombinant strains containing a bacterial isomerase on different carbon sources. This in vivo CLA production process has the potential for continuous de novo production of this pharmaceutically important fatty acid. For the first time we examine the dependence of engineered lipid formation with respect to the applied nutrient limitation and the nature of the supplied carbon source.
Experimental
Strains and media
Trichosporon oleaginosus ATCC 20509 (DSM-11815) was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DMSZ) (Braunschweig, Germany), Agrobacterium tumefaciens strain AGL1 (BAA-101) was obtained from American Type Culture Collection (ATCC) (Manassas, VA, USA) and Escherichia coli strain XL-1 Blue was obtained from Novagen/Merck Millipore (Schwalbach, Germany). Media for cultivation and lipid production were LB medium (tryptone, 10 g L−1; yeast extract, 5 g L−1; NaCl, 10 g L−1), YPD medium (glucose, 20 g L−1; tryptone, 20 g L−1; yeast extract, 10 g L−1), nitrogen limitation medium (glucose or xylose, 30 g L−1; yeast extract, 0.5 g L−1; (NH4)2SO4, 0.3 g L−1; MgSO4·7H2O, 1.5 g L−1; KH2PO4, 2.4 g L−1; Na2HPO4 0.91 g L−1; CaCl2·H2O, 0.22 g L−1; ZnSO4·7H2O, 0.55 μg L−1; MnCl2·4H2O, 22.4 μg L−1; CuSO4·5H2O, 25 μg L−1; FeSO4·7H2O, 25 μg L−1, pH 6.1), and phosphorus limitation medium (N-acetylglucosamine, 30 g L−1; yeast extract, 0.5 g L−1; NH4Cl, 0.5 g L−1; MgSO4·7H2O, 1.5 g L−1; KH2PO4, 0.11 g L−1; Na2HPO4, 38.7 mg L−1; CaCl2·2H2O, 0.22 g L−1; ZnSO4·7H2O, 0.55 μg L−1; MnCl2·4H2O, 22.4 μg L−1; CuSO4·5H2O, 25 μg L−1; FeSO4·7H2O, 25 μg L−1). Media for the Agrobacterium tumefaciens mediated transformation were liquid induction medium with acetosyringone (L-IMAS) (K2HPO4, 2.05 g L−1; KH2PO4, 1.45 g L−1; NaCl, 0.15 g L−1; MgSO4·7H2O, 0.5 g L−1; CaCl2·2H2O, 67.0 mg L−1; 2- 4-morpholineethanesulfonic acid monohydrate (MES), 7.8 g L−1; glucose, 1.8 g L−1; acetosyringone, 39.24 mg L−1; FeSO4·7H2O, 2.5 mg L−1; (NH4)2SO4, 0.5 g L−1; glycerol 5% (v/v); trace elements solution (100 mg Na2MoO4, MnSO4·H2O, ZnSO4·7H2O, CuSO4·5H2O, H3BO3 in 1 L ddH20, 5% (v/v), pH 5.6) and solid induction medium with acetosyringone (S-IMAS) (equivalent to L-IMAS, without glucose and supplemented with 18 g L−1 agar).Cloning
The plasmid pRF-HU2 containing the T-DNA for the A. tumefaciens mediated transformation was obtained from the Fungal Genetics Stock Center (FGSC) (Manhattan, KS, USA).34 Genes were synthesized by Life Technologies (Carlsbad, CA, USA) (Fig. S21–24†). For transformation, the tryptophan promotor (Aspergillus nidulans) from the hygromycin B resistance cassette in the vector pRF-HU2 was exchanged with a 390 bp long fragment from the glyceraldehyde-3-phosphate dehydrogenase (GPD) promotor of T. oleaginosus (Fig. S3†). As cloning method, the USER Cloning from New England Biolabs (Ipswich, MA, USA) was applied. Therefore the pRF-HU2 vector was linearized by PCR using the PfuTurbo Cx Hotstart DNA Polymerase from Agilent Technologies (Santa Claram, CA, USA) and the primers 5′-ATG AAA AAG CCU GAA CTC ACC GCG-3′ and 5′-ATT AAT GCC UAT CGA TGG GCC CGC TGA G-3′. The GPD promotor gene was amplified from genomic DNA of T. oleaginosus, which was isolated using the Yeast DNA Extraction Reagent Kit from Thermo Scientific (Waltham, MA, USA) according to the manufacturer procedure. Primers used were: 5′-AGG CTT TTT CAU TGT TGA TCA AGT TGA TTT TTG GG-3′ and 5′-AGG CAT TAA UCC TCC TCC GGC ACC-3′. Ligation was carried out following USER Cloning standard protocols and yielded the vector pRF-HU2-(GPD) (Fig. S3†). For protein expression, pRF-HU2-(GPD) was extended harbouring a second gene cassette. Genes for YFP reporter protein, Δ9 elongase IgASE2 from Isochrysis galbana, Δ12/ω3 desaturase Fm1 from Fusarium moniliforme and linoleic acid isomerase PAI from Propionibacterium acnes were codon optimized based on the preferred codon usage table for the glyceraldehyde-3-phosphate dehydrogenase (GPD) (Genebank AF126158.1). All genes were synthesised as cassettes containing 800 bp of the GPD promotor and 600 bp of the GPD terminator derived from the NCBI genome JZUH00000000.1 (Fig. S21–24†). The expression cassettes were integrated into the pRF-HU2-(GPD) in the same orientation than the hygromycin B resistance cassette on its 3′ end (Fig. S4–7†). Cloning occurred in analogy to the creation of the pRF-HU2-(GPD) vector. Therefore pRF-HU2-(GPD) was linearized using the primers 5′-ATT AAA CCC UAT GCC TCA GCA CTA GTC-3′ and 5′-ATT AAG ACC UGA CCT CAG CAA GCT TCG TGA C-3′. The diverse gene expression cassettes were amplified using the primers 5′-AGG TCT TAA UAT CCG CTG ACA TTG GAC CTT-3′ and 5′-AGG GTT TAA UGG GGA TTG GCG TCA TCA AGT GC-3′. USER ligation of the PCR products from the pRF-HU2-(GPD) and cassettes yielded the vectors pRF-HU2-(GPD)-YFP, pRF-HU2-(GPD)-IgASE2, pRF-HU2-(GPD)-Fm1 and pRF-HU2-(GPD)-PAI (Fig. S4–7†). For the production of ETE, the vector pRF-HU2-(GPD)-IgASE2 was extended by the Fm1 expression cassette to create the vector pRF-HU2-(GPD)-IgASE2-Fm1 (Fig. S8†). The Fm1 cassette was integrated in the same orientation subsequent to the 3′ end of the IgASE2 cassette. Cloning was completed by ligation of the Fm1 cassette into pRF-HU2-(GPD)-IgASE2 using the HindIII restriction site. Therefore the Fm1 cassette was flanked with a HindIII restriction site using the primers 5′-ATA TAA GCT TGG GGA TTG GCG TCA TCA AGT-3′ and 5′-ATT AAA GCT TAT CCG CTG ACA TTG GAC CTT TTG G-3′.Transformation of Trichosporon oleaginosus
The vectors pRF-HU2-(GPD)-YFP, pRF-HU2-(GPD)-IgASE2, pRF-HU2-(GPD)-Fm1, pRF-HU2-(GPD)-PAI and pRF-HU2-(GPD)-IgASE2-Fm1 were introduced into A. tumefaciens by a standard transformation procedure. An overnight culture (LB medium supplemented with 30 μg mL−1 kanamycin at 28 °C) was used to inoculate (OD600) a 10 mL shake flask culture with L-IMAS medium and cultivated at 28 °C for 6 hours. Next, the A. tumefaciens was mixed at an equal ratio with T. oleaginosus cells resuspended in L-IMAS medium at OD600 0.5. Therefore, an overnight culture of T. oleaginosus in YPD was centrifuged (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) and re-suspended in an appropriate volume of L-IMAS medium. 100 μl of the A. tumefaciens and T. oleaginosus cell mixture were plated on top of an Amersham Hybond-N+ blotting membrane from GE Healthcare (Little Chalfont, Buckinghamshire, UK) that was placed on S-IMAS agar plates. The plates were incubated at 24 °C for 48 hours and subsequently the membranes were transferred to YPD agar plates supplemented with 200 μg mL−1 hygromycin B and 300 μg mL−1 cefotaxime and incubated at 28 °C for 5 days.
000g) and re-suspended in an appropriate volume of L-IMAS medium. 100 μl of the A. tumefaciens and T. oleaginosus cell mixture were plated on top of an Amersham Hybond-N+ blotting membrane from GE Healthcare (Little Chalfont, Buckinghamshire, UK) that was placed on S-IMAS agar plates. The plates were incubated at 24 °C for 48 hours and subsequently the membranes were transferred to YPD agar plates supplemented with 200 μg mL−1 hygromycin B and 300 μg mL−1 cefotaxime and incubated at 28 °C for 5 days.
Selection of clones and cultivation conditions
After transformation single colonies were picked and cultivated in 5 mL YPD medium supplemented with 300 μg mL−1 cefotaxime. To find the best YFP producer 30 colonies were cultivated at 28 °C for 2 days. Subsequently, the cells were pelleted, washed with double distilled water (ddH2O) and resuspended in ddH2O. The cell solution was transferred to MaxiSorp F96 plates from Thermo Scientific (Waltham, MA, USA) and analysed on a plate reader EnSpire 2 from Perkin Elmer (Waltham, MA, USA). Fluorescence intensity was measured at 527 nm (excitation 490 nm). For the production of modified fatty acids 30 colonies and for the production of ETE (pRF-HU2-(GPD)-IgASE2-Fm1) 100 colonies were cultivated and screened. Cultivation was executed for 3 days at 28 °C. The cells were pelleted and flash frozen for further analysis.To analyse the lipid content and fatty acid distribution yeast strain cultivation was carried out in 500 mL baffled shake flasks at 28 °C for 7 days in triplicate. Cultivation media (100 mL) used were YPD, nitrogen limitation medium with glucose/xylose and phosphorus limitation medium with N-acetylglucosamine. Cultivation was started by inoculation from an overnight culture (YPD medium supplemented with 300 μg mL−1 cefotaxime) at OD600 0.5 in YPD medium. 15 mL samples were taken after 24, 72 and 168 hours to determine the dried cell mass, lipid content and fatty acid composition.
Cell mass yield and lipid yield
Cellular dry weight was determined by centrifugation of the cells at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min. Subsequently, the cell pellet was washed with ddH2O and dried at 60 °C to constant weight. The cellular total lipid was determined by extraction with chloroform and methanol according to the protocol of Folch et al.35
000g for 10 min. Subsequently, the cell pellet was washed with ddH2O and dried at 60 °C to constant weight. The cellular total lipid was determined by extraction with chloroform and methanol according to the protocol of Folch et al.35
Fatty acid composition analysis
For the fatty acid analysis, cells from 2 ml cultivation medium were pelleted and washed with ddH2O. The cell biomass was directly converted into fatty acid methyl esters (FAME) by methanol transesterification according to the protocol of Griffiths et al.36 FAMEs were analysed on a GC-2010 Plus gas chromatograph from Shimadzu (Nakagyo-ku, Kyōto, Japan) with flame ionisation detector. 1 μl sample was applied by AOC-20i auto injector (Shimadzu) onto a ZB-WAX column (30 m, 0.32 mm ID; 0.25 μm df; phenomenex (Torrance, CA, USA)). The initial column temperature was 150 °C (maintained for 1 min). A temperature gradient was applied from 150 °C–240 °C (5 °C min−1), followed by 6 min maintenance at 240 °C. Fatty acids were identified according to retention times of authentic standards.Fluorescence microscopy
Microscopic photographs were acquired on an Axio Lab. A1, fluorescence microscope equipped with an Axio Cam ICm1 (Zeiss, Oberkochen, Germany). For microscopy, cells were washed and resuspended in ddH2O. Nile Red staining was conducted by addition of 25 μL DMSO and 25 μL (0.1 mg mL−1 in DMSO) Nile Red to a cell solution. After 10 minutes incubation, the cells were washed with ddH2O twice.Results and discussion
Establishing genetic accessibility of T. oleaginosus
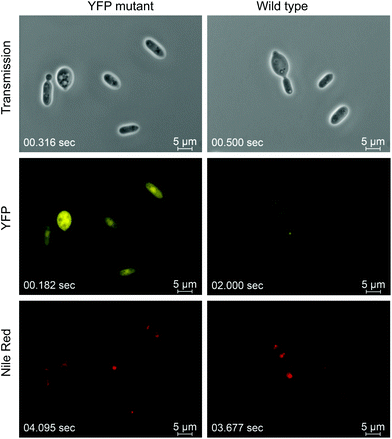
Initially, we established a robust A. tumefaciens mediated transformation protocol (ATMT) using hygromycin B as dominant selection marker. Similar protocols have been reported for the genomic integration of heterologous genes in other non-conventional fungi and yeasts.22,37,38 To enable strong expression of heterologous genes, the constitutive promotor of the glyceraldehyde-3-phosphate dehydrogenase (GPD, EC1.2.1.12) gene was chosen.39 While for expression of the hygromycin B resistance encoding gene (hph) a truncated version (390 bp) of the GPD promotor was sufficient, an 800 bp elongated GPD promotor was required for efficient expression of a codon optimized YFP reporter protein (Fig. 1).Interestingly, screening numerous T. oleaginosus transformants harbouring the yellow fluorescence protein (YFP) expression cassette, indicated a wide distribution of the YFP fluorescence strength. Subsequently, strains displaying the highest YFP fluorescence levels were quantified by a microtiter plate reader. Since ATMT causes random integration in the genome, differences in expression level are most likely caused by variable genomic copy numbers and loci. Consequently, we applied a screening method to all our transgenic strains. Genomic integration was stable during mitosis and the YFP fluorescence levels remained constant after multiple sub-cultivations in absence of the selection marker. Comparative growth curves indicate that both wt and recombinant T. oleaginosus strains have identical growth kinetics (Fig. S1†). The fluorescence microscopy images of T. oleaginosus expressing YFP are shown in Fig. 1. The strain was cultivated for 48 hours in rich media (YPD) and additionally stained with Nile Red to visualize intracellular lipid bodies. Lipid quantification indicated that even in complex, N-rich media, such as YPD, T. oleaginosus accumulates small quantities of lipids (about 5–10% dry cell weight (DCW)).
Engineering tailor made fatty acid biosynthesis in T. oleaginosus
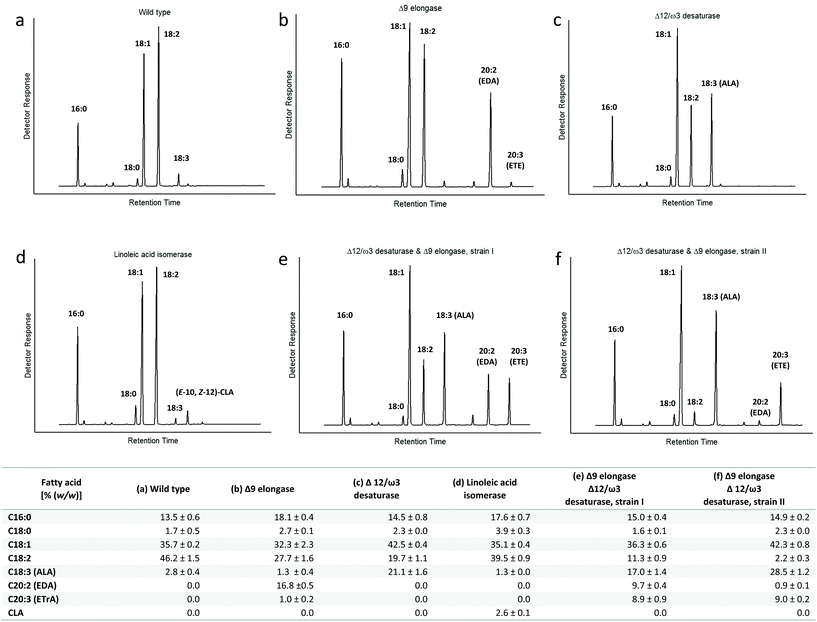
The native lipid profile of T. oleaginosus is dominated by oleic acid, which constitutes approximately 50% of the total fatty acid content. Based on this large metabolic oleic acid pool, we devised methodologies to redirect fatty acid biosynthesis towards alternative product profiles according to Fig. 2.In order to increase the intracellular ALA pool, we created an integration vector harbouring two expression cassettes containing Fm1 and IgASE2, respectively. To identify the best ETE producer, we screened numerous transformants. Interestingly, we could identify two transformants with varying fatty acid profiles. The first transformant accumulated 17% (TFA) ALA and 9% (TFA) for EDA and ETE, respectively (Fig. 3e). By contrast, the fatty acid distribution of the second transformant encompassed 28% (TFA) ALA, 0.8% (TFA) EDA and 9% (TFA) ETE (Fig. 3f). Despite the difference in the intracellular ALA pool, the amount of ETE was equivalent in both strains, indicating that ETE is likely subject to rapid intracellular turnover.
Comparable data for Y. lipolytica, which accumulates high concentrations of free fatty acids intracellularly, indicates the CLA yields (5.9% TFA) are generally quite low.14
Therefore, cumulative data suggests, that yeasts such as T. oleaginosus, can accumulate sufficient free fatty acids that can be converted to CLA by PAI. However, the resulting CLA product is either rapidly degraded or used as substrate for further fatty acid modifying enzymes.
Influence of carbon source on recombinant lipid productivity
In subsequent experiments we evaluated the lipid production capacity of transgenic T. oleaginosus strains in complex (YPD) and defined media containing different carbon sources. In the latter, lipid accumulation was triggered by either nitrogen or phosphorus limitation using the C-6 sugars glucose, N-acetylglucosamine and the C-5 sugar xylose as carbon source. All transgenic strains were cultivated for 7 days in 500 ml shake flask containing 100 ml of the respective medium. Time dependent lipid production and the associated fatty acid distribution were monitored after 24, 72 and 168 hours and referenced to the wild type T. oleaginosus control.We found no significant difference in the growth kinetics and biomass yield. Cultivations in all media resulted in biomass (DCW) production between 10–12 g L−1 after 24 hours and increased to 12–14 g L−1 after 72 to 168 hours. By contrast, the lipid yield varied with growth conditions and the nature of the applied carbon source. The amount of lipids per DCW reached about 10% (72 h) in rich media and increased significantly when N- or P-limitation was applied in defined media.9 Lipid biosynthesis was triggered by N-limitation in cultivations based on xylose and glucose as the carbon source. The C/N ratios 102 of both cultivation approaches were equivalent.43 The highest amount of stored lipids were recovered after 72 hours in media supplemented with xylose 57% (DCW) and glucose 48% (DCW).
By contrast, when T. oleaginosus was grown on N-acetylglucosamine only P-limitation could effectively be applied to induce lipid biogenesis as its metabolism results in release of free ammonium, which can be taken up by cell and reused as a nitrogen source.29,40 Lipid induction was triggered by a C/P ratio of 388 as reported previously.40 As a result, the highest lipid accumulation was 35% (DCW) after 72 hours. The lipid content in all evaluated media, remained constant and did not increase after 168 hours. At 24 hours about 50% of the final lipid content was already present in all media.
It was not surprising that relatively low lipid yields were obtained when T. oleaginosus was grown on N-acetylglucosamine containing medium because P-limitation is reported to be a weaker inducer of lipid biosynthesis compared to N-limiting conditions. Additional work to uncover the regulatory metabolic network governing lipid biogenesis is required to decouple T. oleaginosus based lipid formation from specific nutrient depletion.
Interestingly, we detected significant changes in the fatty acid profiles of wt and transgenic T. oleaginosus cells cultivated in either complex (YPD) or nutrient depleted media. Consequently, we summarized the relative and absolute yields for each of the recombinantly produced fatty acids in Tables S1 and S2,† respectively.
Generally, an increase (ca. 10–20% TFA) of saturated, palmitic (C16:0) and steric (C18:0) acids was observed, when a specific nutrient limitation (N or P) was applied (Table S1†). At the same time the linoleic acid (LA) content decreased from 46% (TFA) in complex (YPD) medium to 6–14% (TFA) in nutrient limited lipid production media (Table S1†). Interestingly, the maximal LA content was reached after 24 h in nutrient limited media. By contrast, the intracellular LA concentration increased continuously from 37.7% TFA (24 h) to 46% TFA (168 h) in cultivation in complex YPD medium.
As the metabolic linoleic acid pool also governs EDA, CLA, ALA and ETE biosynthesis, the production titres of these engineered fatty acids decreased proportionally (Table S1†). The highest titres for all engineered fatty acids were detected when xylose was used as carbon source.
The elevated content of modified fatty acids is consistent with the observation that the wild type displayed a minor increase in linoleic acid when cultivated on xylose. For instance when the wild type strain was cultivated for 168 hours to reach the maximal lipid content, the linoleic acid yield was 8% (TFA) in xylose compared to 7% (TFA) in glucose or 6% (TFA) in N-acetylglucosamine (Table S1†).
With respect to the total lipid content the same trend for LA formation is observed translating into values of 4.6% (DCW), 3. 3% (DCW) and 2.1% (DCW) for xylose, glucose and N-acetylglucosamine based cultivations, respectively (Table S2†). Interestingly, when cultivation is done in YDP rich media the total lipid content for LA was about 4.7% after 168 hours. This indicates, that the intracellular LA pool is not replenished or increased when lipid production was conducted in nutrient limited, defined media.
We primarily addressed the intracellular LA pool of T. oleaginosus by creating a mutant expressing the elongase IgASE2, which provides for conversion of LA to EDA. Under nutrient limiting conditions, the highest relative EDA yield was 4.3% (TFA) after 168 hours, when xylose was applied as a carbon source (Table S1†). By contrast, in complete YPD medium the relative EDA yield was enhanced to 17% (TFA) at the same time point. However, when data was put into context of the total lipid yield, 1.7% (DCW) and 2.4% (DCW) were obtained in YPD respectively xylose containing, nutrient deficient media (Table S2†).
Analogous to EDA formation, we introduced the PAI isomerase into T. oleaginosus, which enables LA conversion to CLA. The PAI expressing transformant produced a relative CLA titre of 0.6% (TFA) after 24 hours in nutrient deficient media containing either xylose or N-acetylglucosamine as the carbon source. With both carbon sources, a time dependent decrease in the CLA pool was observed with progressive cultivation. However, the decrease with N-acetylglusoamine was attenuated after 72 h (0.1% TFA), while a progressive CLA decrease up to the end of the cultivation (168 h) was observed when xylose (0.2% TFA) was applied as the carbon source (Table S1†). When data for YPD, xylose and N-acetylglucosamine based cultivations were related to the total lipid yield, no performance difference (0.1% DCW) was observed (Table S2†).
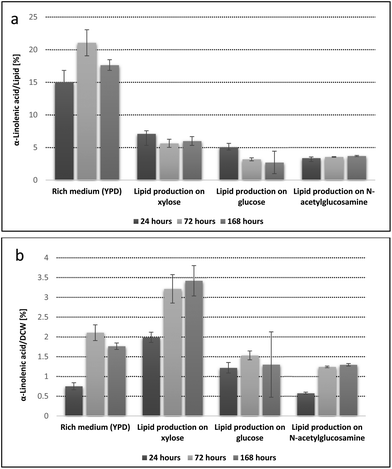
Interestingly, the modified fatty acid contents barely changed during the cultivation when nutrient limited media (N or P) was applied. Exemplary, Fig. 4 graphically summarizes the α-linolenic acid (ALA) formation in the T. oleaginosus mutant expressing the desaturase Fm1 on various carbon sources. Yields of target fatty acid are expressed either with reference to the total amounts of lipids (relative yields, Fig. 4a) or with respect to the total dried biomass (absolute yields, Fig. 4b). In YPD medium relative ALA yields are superior to those obtained in its nutrient limited counterparts (Fig. 4a and Table S1†). When data was put into context with total lipid yields, the total ALA concentration approximately doubled in xylose based cultivations (3.4% DCW, 168 h) compared to YPD (1.8% DCW, 168 h) equivalents (Fig. 4b).
When xylose was used as a carbon source a total ALA yield of 3.4% (DCW) was obtained after 168 h (Fig. 4b). The average biomass yield was 13 g L−1 in shake flask experiments. This biomass contained 57% (DCW) or 7.41 g L−1 total lipids. Consequently, the volumetric ALA yield was calculated as 442 mg L−1. These figures compare well with recombinant lipid production in other model organisms such as E. coli and Y. lipolytica.21,44 Current literature reports with other model organisms mostly focused on improving total yields of naturally occurring lipids. By contrast, our proof of concept study not only aimed at improving substrate dependent lipid yields but also focused on shifting the natural T. oleaginosus fatty acid profile towards generation of non-native fatty acids. Moreover, under fermentative conditions T. oleaginosus biomass formation can be significantly increased (ca. 100 g L−1), which potentially enables a 10 fold improvement in production of our recombinant target fatty acids.25
Unexpectedly, the ALA pool could be further increased by addition of the I. galbana elongase IgASE2 to our engineered T. oleaginosus Fm1 system. For all tested lipid production media, the highest amount of ALA (9% TFA) was obtained with the T. oleaginosus double mutant strain II in xylose supplemented medium after 24 h cultivation (Table S1†). However, at the end of the cultivation (168 hours) the ALA content was reduced to 6% (TFA). As at this point only about 1% ETE is formed, the remaining 2% may be catabolically degraded. The double mutant strain I produced lower amounts of ALA (6.4% TFA, 24 h) in xylose containing media compared to strain II. The ALA decrease after 168 h (4.6% TFA) can be rationalized by complete conversion of ALA to ETE (ca. 2% TFA).
However, when strain II was cultivated on xylose, total ALA formation (24 h) was 1.5 fold higher under nutrient limiting conditions (2.5% DCW) compared to the equivalent experiment under non-limiting conditions with YDP (ca. 1.5% DCW, Table S2†).
Our data suggests that despite the inherent Δ12 desaturase activity of Fm1, which can potentially replenish the intracellular linoleic acid pool, the reactivity of this enzyme is insufficient to compensate the large decrease in linoleic acid when N- or P- limitation is applied to induce lipogenesis. Consequently, for efficient production of very long chain polyunsaturated fatty acids further work on increasing the linoleic acid concentration in nutrient limiting media has to be conducted.
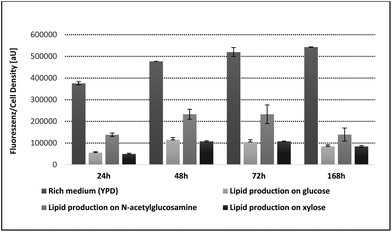
As the recombinant production of tailored fatty acids is highly dependent on the promotor strength driving heterologous gene expression, we evaluated the GPD promotor under different cultivation conditions. In order to quantify GDP promoter strength, the YFP reporter protein expression levels were determined in complex YPD and defined media under the previously applied cultivation conditions. The results are illustrated on Fig. 5. The highest YFP expression signal was detected in complex YPD medium that is consistent with previously observed production titres of tailor made fatty acids. When cultivation was carried out in P-limited medium containing N-acetylglucosamine as a carbon source, the YFP fluorescence signal was reduced by at about two third compared to the YDP medium cultivation reference. Surprisingly, compared to the YPD control, the YFP signal further decreased to one fifth when cultivations were carried out under N-limiting conditions using either xylose or glucose as the carbon source. These findings indicate that the GPD promotor strength is upregulated in media with high nitrogen content. Our cumulative data indicate that the composition of the cultivation media regulate the strength of the constitutive GPD promoter, which reflects in lipid production titres in complex and defined, nutrient limited media. However, the increased GPD promotor strength in N-acetylglucosamine media compared to glucose and xylose had no effect on production titres and therefore other metabolic regulators have to be considered.
In summary, the constitutive GPD promotor appears to be an adequate choice for heterologous expression of recombinant genes in nitrogen and carbon complex media. However, due to its reduced strength in N- or P-limited lipid production media it is reasonable to seek alternative promoters, which are independent or up-regulated under these cultivation conditions.
Conclusions
We have established the genetic accessibility of T. oleaginosus and demonstrated the feasibility to engineer its fatty acid metabolism in order to generate tailor made lipids. In consensus with previous reports we could demonstrate, that T. oleaginosus accumulated superior amounts of lipids under nutrient limiting conditions compared to cultivation in complete media, such as YPD. Our data indicates that lipid accumulation is highly dependent on the nutrient limitation (N or P) applied and on the nature of the carbon source present during cultivation. The highest total lipid yield (57% DCW) was obtained using N-limiting conditions in the presence of a xylose carbon source excess. Therefore, xylose rich biomass waste streams, such as wheat germ or straw hydrolysate, may be considered to realize cost efficient, high yield production of tailor made fatty acids with engineered T. oleaginosus production systems.Our data indicates that in YDP media the relative yields of engineered fatty acids was higher than in defined, nutrient deficient media. However, when this data was analysed with respect to the total intracellular lipid content, nutrient deficient media containing xylose as the carbon source did provide superior total fatty acid yields compared to complete media, such as YPD. This effect was most prominent after an initial cultivation period of 24 h. The effect was commonly reversed at 168 h, when cultivation sized. The time dependent decrease of target fatty acid concentrations can be attributed to both metabolic fatty acid remodelling and catabolic degradation. The exact peak of target fatty acid formation is dependent on growth conditions and has to be considered when cellular harvesting is optimized during fermentative production processes. Further, studies are required to uncover and subsequently modify specific metabolic cues that lead to intracellular degradation of target lipids (fatty acids) during the cultivation procedure. Unfortunately, our previous targeted transcriptomics data did not yield any information on metabolic signals that could modulate the intracellular stability of desired fatty acid pools.30
However, our genome data allowed utilization of the constitutive glyceraldehyde-3-phosphate dehydrogenase promotor gene (GPD) to efficiently drive heterologous gene expression in carbon and nitrogen rich media, but also showed reduced expression rates in media optimized for lipid production. To enable a further increase in production titres of tailor made fatty acids it is reasonable to evaluate alternative promotors that are either not affected or upregulated when nutrient deficiency is applied to induce lipogenesis. Additionally, detailed systems biology studies to uncover the complex regulatory networks that govern lipid biosynthesis in T. oleaginosus would ultimately provide tools to uncouple lipid formation from nutrient limiting cultivation conditions. This would enable high content, intracellular lipid accumulation without compromising cellular growth. Only these conditions would provide an economic framework for high yield and high value fatty acid production targeted at food, chemical and pharmaceutical applications.
Specifically, the intracellular linoleic acid pool needs to be increased to provide high yield production of long chain fatty acids. This could be achieved by overexpression of an endogenous Δ12 desaturase from T. oleaginosus. By reference, lipid production has been increased in Y. lipolytica by knocking out the peroxisomal β-oxidation pathway to prevent degradation and overexpression of acyl-CoA:diacylglycerol acyltransferases (DGA1p, DGA2p) to facilitate lipid production.21 An equivalent strategy could be applied to increase T. oleaginosus lipid production levels when cultivated on complex waste biomass streams. Nonetheless, it is noteworthy, that in our hands target selective gene knockouts by homologous recombination have been challenging when T. oleaginosus has been the target organisms. Consequently, we are currently developing a CRISPR/Cas9 based technique to generate T. oleaginosus mutants. The enhancement of genetic engineering tools and a detailed knowledge of metabolic interactions governing T. oleaginosus lipogenesis will enable more sophisticated synthetic biology approaches for sustainable production of designer lipids. The advancement of genetic knowledge is a prerequisite for this endeavour as exemplified by various reports in E. coli and Y. lipolytica.21,44,45 In this respect the increasing malonyl-CoA biosynthesis can be seen as a milestone concerning engineered lipogenesis in these model organisms.
Acknowledgements
CG, VR, DG and TB would like to acknowledge the financial support by the European commission through FP7 grant 289284. Information concerning the granted “ChiBio” project can be found under: http://www.chibiofp7.fraunhofer.de/.FB and TB would like to thank the German Federal Ministry of Education and Research for supporting the “Advanced Biomass – Value” research project (Grant Number: 03SF0446A). Information concerning the project is stated under: http://www.ibbnetzwerk-gmbh.com/de/service/pressebereich/pm-06052013-advanced-biomass-value/.
Finally, the authors would like to express their gratitude to Dipl. Ing. (FH) Martina Haack for her kind help addressing analytical questions.
Notes and references
- C. Lu, J. A. Napier, T. E. Clemente and E. B. Cahoon, Curr. Opin. Biotechnol., 2011, 22, 252–259 CrossRef CAS PubMed.
- B. F. Pfleger, M. Gossing and J. Nielsen, Metab. Eng., 2015, 29, 1–11 CrossRef CAS PubMed.
- U. R. Fritsche, R. E. Sims and A. Monti, Biofuels, Bioprod. Biorefin., 2010, 4, 692–704 CrossRef CAS.
- E. F. Lambin and P. Meyfroidt, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 3465–3472 CrossRef CAS PubMed.
- T. C. Adarme-Vega, S. R. Thomas-Hall and P. M. Schenk, Curr. Opin. Biotechnol., 2014, 26, 14–18 CrossRef CAS PubMed.
- E. J. Brunner, P. J. Jones, S. Friel and M. Bartley, Int. J. Epidemiol., 2009, 38, 93–100 CrossRef PubMed.
- R. J. Deckelbaum and C. Torrejon, J. Nutr., 2012, 142, 587S–591S CrossRef CAS PubMed.
- T. E. Lewis, P. D. Nichols and T. A. McMeekin, Mar. Biotechnol., 1999, 1, 580–587 CrossRef CAS PubMed.
- W. Barclay, K. Meager and J. Abril, J. Appl. Phycol., 1994, 6, 123–129 CrossRef CAS.
- B. Qi, T. Fraser, S. Mugford, G. Dobson, O. Sayanova, J. Butler, J. A. Napier, A. K. Stobart and C. M. Lazarus, Nat. Biotechnol., 2004, 22, 739–745 CrossRef CAS PubMed.
- Z. Xue, P. L. Sharpe, S.-P. Hong, N. S. Yadav, D. Xie, D. R. Short, H. G. Damude, R. A. Rupert, J. E. Seip and J. Wang, Nat. Biotechnol., 2013, 31, 734–740 CrossRef CAS PubMed.
- I. Churruca, A. Fernández-Quintela and M. P. Portillo, Biofactors, 2009, 35, 105–111 CrossRef CAS PubMed.
- T. Mounts, H. Dutton and D. Glover, Lipids, 1970, 5, 997–1005 CrossRef CAS.
- B. Zhang, C. Rong, H. Chen, Y. Song, H. Zhang and W. Chen, Microb. Cell Fact., 2012, 11, 51 CrossRef CAS PubMed.
- J. Xu, W. Du, X. Zhao, G. Zhang and D. Liu, Biofuels, Bioprod. Biorefin., 2013, 7, 65–77 CrossRef CAS.
- R. Subramaniam, S. Dufreche, M. Zappi and R. Bajpai, J. Ind. Microbiol. Biotechnol., 2010, 37, 1271–1287 CrossRef CAS PubMed.
- J. M. Ageitos, J. A. Vallejo, P. Veiga-Crespo and T. G. Villa, Appl. Microbiol. Biotechnol., 2011, 90, 1219–1227 CrossRef CAS PubMed.
- S. Papanikolaou and G. Aggelis, Eur. J. Lipid Sci. Technol., 2011, 113, 1031–1051 CrossRef CAS.
- S. Papanikolaou and G. Aggelis, Eur. J. Lipid Sci. Technol., 2011, 113, 1052–1073 CrossRef CAS.
- C. Huang, X.-f. Chen, L. Xiong, L.-L. Ma and Y. Chen, Biotechnol. Adv., 2013, 31, 129–139 CrossRef CAS PubMed.
- J. Blazeck, A. Hill, L. Liu, R. Knight, J. Miller, A. Pan, P. Otoupal and H. S. Alper, Nat. Commun., 2014, 5, 3131 Search PubMed.
- Y. Liu, C. M. J. Koh, L. Sun, M. M. Hlaing, M. Du, N. Peng and L. Ji, Appl. Microbiol. Biotechnol., 2013, 97, 719–729 CrossRef CAS PubMed.
- P. Gujjari, S.-O. Suh, K. Coumes and J. J. Zhou, Mycologia, 2011, 103, 1110–1118 CrossRef PubMed.
- P. Akhtar, J. Gray and A. Asghar, J. Food Lipids, 1998, 5, 283–297 CrossRef CAS.
- P. Meesters, G. Huijberts and G. Eggink, Appl. Microbiol. Biotechnol., 1996, 45, 575–579 CrossRef CAS.
- Y. Liang, T. Tang, T. Siddaramu, R. Choudhary and A. L. Umagiliyage, Renewable Energy, 2012, 40, 130–136 CrossRef CAS.
- Y. Wang, Z. Gong, X. Yang, H. Shen, Q. Wang, J. Wang and Z. K. Zhao, Process Biochem., 2015, 50, 1097–1102 CrossRef CAS.
- X. Yu, Y. Zheng, K. M. Dorgan and S. Chen, Bioresour. Technol., 2011, 102, 6134–6140 CrossRef CAS PubMed.
- S. Wu, C. Hu, X. Zhao and Z. K. Zhao, Eur. J. Lipid Sci. Technol., 2010, 112, 727–733 CrossRef CAS.
- R. Kourist, F. Bracharz, J. Lorenzen, O. N. Kracht, M. Chovatia, C. Daum, S. Deshpande, A. Lipzen, M. Nolan and R. A. Ohm, mBio, 2015, 6, e00918–e00915 CrossRef PubMed.
- A. Ykema, E. C. Verbree, I. I. Verwoert, K. H. van der Linden, H. J. J. Nijkamp and H. Smit, Appl. Microbiol. Biotechnol., 1990, 33, 176–182 CrossRef CAS.
- M. Hassan, P. J. Blanc, L.-M. Granger, A. Pareilleux and G. Goma, Process Biochem., 1996, 31, 355–361 CrossRef CAS.
- I. I. Verwoert, A. Ykema, J. A. Valkenburg, E. C. Verbree, H. John, J. Nijkamp and H. Smit, Appl. Microbiol. Biotechnol., 1989, 32, 327–333 CrossRef CAS.
- R. J. Frandsen, J. A. Andersson, M. B. Kristensen and H. Giese, BMC Mol. Biol., 2008, 9, 70 CrossRef PubMed.
- J. Folch, M. Lees and G. Sloane-Stanley, J. Biol. Chem., 1957, 226, 497–509 CAS.
- M. Griffiths, R. Van Hille and S. Harrison, Lipids, 2010, 45, 1053–1060 CrossRef CAS PubMed.
- L. Ji, Z.-D. Jiang, Y. Liu, C. M. J. Koh and L.-H. Zhang, Fungal Genet. Biol., 2010, 47, 279–287 CrossRef CAS PubMed.
- M. D. van de Rhee, P. M. Graca, H. J. Huizing and H. Mooibroek, Mol. Gen Genetics, 1996, 250, 252–258 CAS.
- C. Tristan, N. Shahani, T. W. Sedlak and A. Sawa, Cell. Signalling, 2011, 23, 317–323 CrossRef CAS PubMed.
- G. Zhang, W. T. French, R. Hernandez, J. Hall, D. Sparks and W. E. Holmes, J. Chem. Technol. Biotechnol., 2011, 86, 642–650 CrossRef CAS.
- M. Li, X. Ou, X. Yang, D. Guo, X. Qian, L. Xing and M. Li, Biotechnol. Lett., 2011, 33, 1823–1830 CrossRef CAS PubMed.
- H. G. Damude, H. Zhang, L. Farrall, K. G. Ripp, J.-F. Tomb, D. Hollerbach and N. S. Yadav, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 9446–9451 CrossRef CAS PubMed.
- X.-F. Chen, C. Huang, X.-Y. Yang, L. Xiong, X.-D. Chen and L.-L. Ma, Bioresour. Technol., 2013, 143, 18–24 CrossRef CAS PubMed.
- P. Xu, Q. Gu, W. Wang, L. Wong, A. G. Bower, C. H. Collins and M. A. Koffas, Nat. Commun., 2013, 4, 1409 CrossRef PubMed.
- P. Xu, L. Li, F. Zhang, G. Stephanopoulos and M. Koffas, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 11299–11304 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5gc01767j |
| This journal is © The Royal Society of Chemistry 2016 |